Abstract
The nuclear receptor PXR (pregnane X receptor) protects the body from hepatotoxicity of secondary bile acids such as lithocholic acid (LCA) by inducing expression of the hydroxylating cytochrome P450 enzyme CYP3A and promoting detoxification. We found that activation of PXR also increases the activity and gene expression of the phase II conjugating enzyme dehydroepiandrosterone sulfotransferase (STD) known to sulfate LCA to facilitate its elimination. This activation is direct and appears to extend to other xenobiotic sulfotransferases as well as to 3′-phosphoadenosine 5′-phosphosulfate synthetase 2 (PAPSS2), an enzyme that generates the donor cofactor for the reaction. Because sulfation plays an important role in the metabolism of many xenobiotics, prescription drugs, and toxins, we propose that PXR serves as a master regulator of the phase I and II responses to facilitate rapid and efficient detoxification and elimination of foreign chemicals.
Cholestatic liver diseases are manifested by progressive intrahepatic retention of bile acids resulting in liver injury, cirrhosis, and frequently death. Amongst various forms of bile acids, lithocholic acid (LCA) is the most toxic. LCA is a hydrophobic secondary bile acid that is primarily formed by intestinal bacteria. Administration of LCA and its conjugates in rodents causes histologic liver damage and other pathological changes (1, 2). Elevated levels of LCA have also been correlated with increased incidence of colorectal cancer (3, 4).
One of the mechanisms that protect our body from LCA toxicity involves the orphan nuclear receptor PXR [pregnane X receptor, also known as the steroid and xenobiotic receptor (SXR) or the pregnane-activated receptor (PAR) in humans] (5–8). PXR is activated by a diverse array of lipophilic chemicals, including endogenous LCA and its direct metabolite 3-keto-LCA (9, 10) as well as many xenobiotic compounds such as prescription drugs, over-the-counter medications, herbal medicines, ingested food derivatives, and industrial pollutants (for reviews, see refs. 11 and 12). Activated PXR in turn induces expression of genes encoding oxidative cytochrome P450 enzymes (CYPs) and drug transporters by binding to the PXR response elements found within the promoter regions of these target genes. Both loss- and gain-of-function mouse genetic experiments have demonstrated that activation of PXR is essential in the catabolism and detoxification of LCA. First, PXR-null mice are more susceptible to LCA-induced liver toxicity than wild-type mice when excess LCA is introduced into the digestive tract, presumably because of a defect of PXR-null mice in mounting a LCA-induced and PXR-mediated protective xenobiotic response (10). Second, activation of PXR by pretreating mice with an prototypic PXR activator and CYP3A inducer pregnenolone-16α-carbonitrile (PCN) or by hepatic expression of an activated form of human PXR transgene (Alb-VP-hPXR, originally described as Alb-VPSXR) confers marked resistance to multiple xenotoxicants, including LCA (9, 10, 13–15).
Since CYP3A enzymes are the originally characterized target genes for PXR, the mechanism of LCA detoxification was attributed to increased hydroxylation by CYP3A and conversion to nontoxic bile acids such as hyodeoxycholic acid or murideoxycholic acid (10). However, CYP3A induction may not fully explain the protection. Inspection of the hepatic level of CYP3A mRNA revealed that LCA elicits similar levels of CYP3A expression in wild-type and PXR-null mice (9, 10). This observation suggests that additional PXR target genes and/or alternative nuclear receptors might be involved in the catabolism of LCA.
In humans, 40–70% of total LCA present in gall bladder bile and feces is in sulfated forms (16, 17). In fact, a number of experiments indicate sulfation as an important detoxification step of LCA, particularly under cholestatic conditions where LCA level is elevated (1, 18). In addition, LCA produces marked cholestasis when introduced in experimental animals, whereas sulfated LCA does not (19). Sulfated LCA also shows less cytotoxicity than LCA when isolated cells are exposed to it (20, 21). Rhesus monkeys, defective in LCA sulfation, accumulate LCA in the enterohepatic circulation upon administration of chenodeoxycholic acid, resulting in marked hepatotoxicity (22, 23). Moreover, sulfation increases water solubility of LCA, thus increasing its biliary and urinary excretion as well as reducing reabsorption through intestinal lumen (24, 25). Together, sulfation appears to play a critical role in detoxification and elimination of LCA (Fig. 1A).
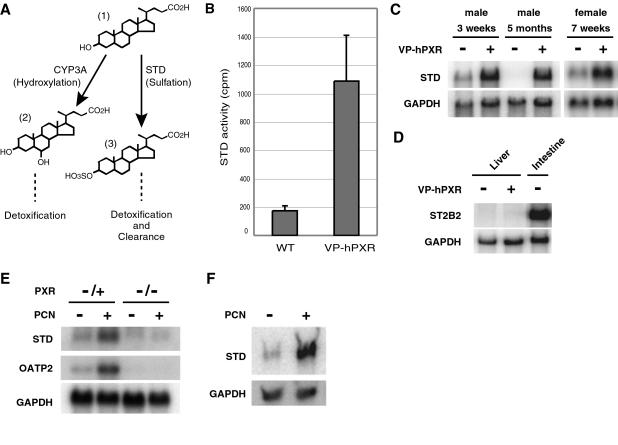
Figure 1.
Activation of PXR induces dehydroepiandrosterone (DHEA) sulfotransferase (STD) activity in rodent livers. (A) Two pathways known to metabolize LCA in the liver. Hydroxylation by CYP3A converts LCA (1) to nontoxic bile acids such as hyodeoxycholic acid or murideoxycholic acid (2), whereas sulfation by STD converts LCA to a less toxic and more water-soluble form (3). (B) Cytosolic liver extracts from transgenic Alb-VP-hPXR male mice or nontransgenic littermates were subjected to enzymatic assays for STD activity on the prototypic substrate DHEA using the cofactor 3′-phosphoadenosine 5′-phospho[35S]sulfate ([35S]PAPS). Shown is the mean ± SD obtained from three pairs of mice. (C) Total liver RNA obtained from transgenic Alb-VP-hPXR mice or nontransgenic littermates of indicated age and sex was subjected to Northern blot analysis using STD and GAPDH (loading control) cDNAs as probes. (D) Northern blot analysis of adult male liver and intestine RNA with ST2B2 cDNA as probe. Note that ST2B2 mRNA is detectable in the intestine, but not in the liver. (E) Total liver RNA from male mice treated with PCN or solvent control was extracted and subjected to Northern blot analysis using STD, OATP2, and GAPDH cDNA as probes. The results show that PCN treatment induces expression of STD mRNA. The film was exposed for 5 times longer than the image shown in C to visualize the relatively lower levels of induction of STD and OATP2 by PCN. OATP2, which encodes Na+-independent organic anion transporter, was previously shown to be a direct target of PXR and thus serves as a positive control for the drug treatment (9). Note that neither STD nor OATP2 is induced by PCN in PXR-null mice. (F) Primary hepatocytes were isolated from rat livers and treated with PCN or solvent control. Total mRNA was isolated and subjected to Northern blot analysis using mouse STD cDNA as a probe.
Sulfation of LCA is mediated primarily by dehydroepiandrosterone sulfotransferase (DHEA SULT or STD), which belongs to a family of conjugating enzymes called cytosolic sulfotransferases (SULTs). SULTs catalyze transfer of a sulfonyl group from the donor molecule 3′-phosphoadenosine 5′-phosphosulfate (PAPS) to hydroxyl or amino groups of lipophilic molecules, forming sulfate or sulfamate conjugates, respectively. At least 10 human isoforms and 11 mouse isoforms of SULTs have been identified to date (for reviews, see refs. 26 and 27). Among SULTs, STD is mainly responsible for the sulfation of hydroxysteroids, including bile acids, of which LCA is one of the most favorable substrates (28–30). The expression and activity of STD has been shown to be induced by PCN in rats (31). PCN is a prototypic activator for rodent PXR (7), and pretreatment with PCN is known to protect the liver from subsequent LCA toxicity (13). These observations suggest that PCN-induced protection is mediated, in part, by increasing STD expression.
In this report, we present the evidence that activation of PXR in mice results in increased STD expression and enzymatic activity. We identified a binding site for PXR within the STD promoter, indicating that STD is a direct transcriptional target of PXR. Moreover, activation of PXR also induces PAPS synthetase 2 (PAPSS2), an enzyme responsible for generating the sulfonate donor PAPS. We propose that induction of SULTs and PAPSS2 by PXR synergistically increases sulfation capacity of liver, resulting in detoxification of bile acids and other xenobiotic chemicals.
Materials and Methods
Animal Studies.
PXR-null mice and Alb-VP-hPXR transgenic mice have been described (14). In Fig. 1E, mice were subjected to daily i.p. injection of 40 mg/kg PCN for 4 days. Three hours after the last treatment, liver total RNA was isolated and subjected to Northern blot analysis. Results shown are representative of three experiments.
Sulfotransferase Assay.
Sulfotransferase was assayed by using [35S]PAPS (DuPont/NEN) essentially as described (32). Specifically, 2.5 μg/ml total liver cytosolic extract was used with 5 μM DHEA as the substrate (Fig. 1B) and 5 μg/ml total extract was used with 2 μM p-nitrophenol as the substrate (Fig. 4B). After the reaction, free [35S]PAPS was removed either by sequential addition of Ba(OH)2 and ZnSO4, which precipitates [35S]PAPS (for Fig. 1B), or by extracting with ethyl acetate (Fig. 4B). Soluble fractions or aqueous phase were then analyzed for radioactivity by using a scintillation counter. Sulfotransferase activity specific to each substrate was calculated by subtracting cpm obtained from control reactions that did not contain substrate. Each reaction was assayed in triplicate, and the mean ± SD of results obtained from three pairs of mice is shown.
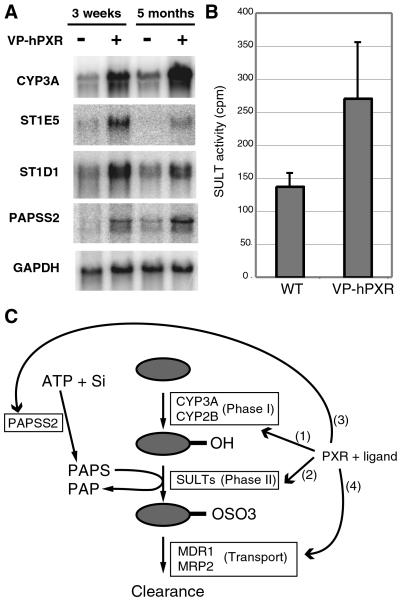
Figure 4.
Coordinate regulation of the phase II sulfonation system by PXR. (A) Northern blot analysis of total liver RNA isolated from transgenic Alb-VP-hPXR male mice or nontransgenic littermates of the indicated age. The same blot was reprobed with the indicated labeled cDNAs. Mice encode two isoforms of PAPSS, of which only PAPPS2, not PAPSS1, is induced by the presence of VP-hPXR (not shown). (B) Cytosolic liver extracts from transgenic VP-hPXR male mice or nontransgenic littermates were subjected to enzymatic assay for phenolic SULT activity by using the prototypic substrate p-nitrophenol and the cofactor [35S]PAPS. Shown is the mean ± SD of results obtained from three pairs of mice. (C) Model for PXR as a regulator of sulfonation pathway and drug clearance. Activation of PXR by LCA or xenobiotic compounds (gray oval) results in coordinate up-regulation of drug-metabolizing pathways through transcriptional induction of genes encoding phase I CYP enzymes (1), phase II SULT enzymes (2), a bifunctional enzyme, PAPSS2, that catalyzes conversion of ATP and inorganic sulfate (Si) into the SULT cofactor PAPS (3), and drug transporters such as MDR1 or MRP2 (4).
Transient Transfection.
The reporter plasmid tk-3A4-luc and the expression vectors for hPXR, mPXR, and hRXRα have been described (6). To construct reporter plasmid rSTD-luc, a fragment encoding 5′ flanking region (−1023 to +38) of rat STD was generated by PCR and inserted into pGL3 (Promega). The mutant derivative was generated by site-directed mutagenesis. tk-IR0-luc was generated by inserting an oligonucleotide encoding three copies of each IR0 element into tk-luc plasmid. CV-1 cells and HepG2 cells were transfected in 48-well format by using N-[1-(2,3-dioleoyloxy)propyl]-N,N,N-trimethylammonium methylsulfate (DOTAP) transfection reagent (Roche) and Lipofectamine 2000 (Invitrogen), respectively. When necessary, cells were treated with LCA (Sigma), rifampicin (10 μM, Sigma), or PCN (10 μM, Sigma) in medium containing 10% steroid-stripped serum.
Primary Rat Hepatocytes.
Rat hepatocytes were freshly isolated essentially as described in ref. 33. Specifically, hepatocytes isolated by collagenase perfusion were plated at a density of 3.3 million per 60-mm dish in our standard Waymouth medium supplemented with 5% FBS. After 4 h of incubation at 37°C, the medium was replaced with serum-free Waymouth medium containing 0.33 mg/ml Matrigel. The cultures were maintained for 62 h in serum-free Waymouth medium before treatment for 24 h with 10 μM PCN. The plates were incubated 30 min at 4°C in 2 ml of 40 mM Tris⋅HCl buffer, pH 7.4, containing 5 mM EDTA and 150 mM NaCl to solubilize the Matrigel before RNA isolation.
Northern Blot Analysis.
RNA was prepared from frozen liver tissue by using the Trizol reagent (Invitrogen) or from rat primary hepatocytes by using a Qiagen kit. Northern hybridization was carried out by the standard method. Fragments used to generate 32P-labeled probes were excised from the following EST clones: STD (IMAGE Consortium no. 1450928), ST1E5 (IMAGE 2649156), ST1D1 (IMAGE 4235817), ST2B2 (IMAGE 403950), and PAPSS2 (IMAGE 336536). The probe for OATP2 was described in ref. 9.
DNA-Binding Analysis.
Electrophoretic mobility-shift assays (EMSAs) were performed as described in ref. 6, using in vitro transcribed and translated proteins (TNT, Promega). For competition binding, 32P-labeled IR0 probe was premixed with unlabeled probe at 0.5, 5, or 50-fold molar excess and then mixed with proteins premixed with binding buffers. The following oligonucleotides were used: rat IR0, TTTGGGGGTCATGAACTTGGGC; IR0 mutant, TTTGGGGGTACCGAACTTGGGC; CYP3A1 DR3, and TAGACAGTTCATGAAGTTCATCTAC.
Results
PXR Induces STD.
We have previously shown that transgenic mice harboring an activated form of human PXR under the albumin promoter (Alb-VP-hPXR) exhibit sustained induction of hepatic CYP3A11 expression (14) and enhanced resistance against LCA toxicity (10). To test whether resistance of Alb-VP-hPXR mice to LCA involves induction of STD (Fig. 1A), we measured STD activity in the liver from transgenic and nontransgenic littermates by using the substrate DHEA. As shown in Fig. 1B, total liver extracts from the Alb-VP-hPXR transgenic mice exhibit enhanced STD activity compared with nontransgenic wild-type mice, suggesting that STD is also under positive control of PXR. This result is in agreement with a previous report that administration of PCN induces STD activity in liver of both male and female rats (31).
In addition to STD, the mouse genome encodes the intestinal ST2B2 gene that is predicted to bear sulfotransferase activity toward hydroxysteroids such as bile acids and DHEA (34). To test whether increased sulfotransferase activity toward DHEA in Alb-VP-hPXR mice is due to transcriptional activation of either gene, we examined the mRNA levels of STD and ST2B2 in Alb-VPhPXR liver by Northern blot analysis. As shown Fig. 1C, a significantly higher level of STD mRNA is present in the Alb-VP-hPXR liver compared with wild-type control in both male and female and both younger and older mice, although the basal STD expression is regulated in age- and sex-dependent manner (35). In contrast, no expression of ST2B2 mRNA was detected in the liver in either genotype (Fig. 1D). These results provide compelling evidence that activation of PXR induces hepatic STD activity through transcriptional induction.
We also found that administration of the PXR agonist PCN induces hepatic expression of STD in the wild-type mice. The liver induction of STD is mediated by PXR because PCN treatment has no effect on STD expression in PXR null mice (Fig. 1E). The level of STD induction by PCN is lower than that in the VP-hPXR transgenic mice, but it is comparable to the induction level of OATP2, a previously identified direct PXR target gene (Fig. 1E) (9, 36). PCN is known to induce expression of mRNA encoding STD in rat livers (31). Indeed, PCN treatment of primary hepatocytes isolated from rats results in strong induction of mRNA that cross-hybridizes to the mouse STD probe (Fig. 1F), suggesting that mechanisms of STD regulation by PXR are conserved between these two rodent species.
STD Is Directly Regulated by PXR.
To ask whether PXR regulates STD mRNA level by controlling its transcription, 5′ flanking regions of PCN-inducible isoforms of rat STD (also known as ST2A1 or SULT2–40/41) were cloned and fused to a firefly luciferase reporter gene. A similar fragment has been shown to support basal transcription in rat hepatocytes as well as in human hepatoma-derived HepG2 cells (37, 38). We also cloned the corresponding region from the 5′ flanking region of the mouse STD gene (known as ST2A4/9), whose nucleotide sequence is highly conserved with its rat counterpart (76% identity, not shown).
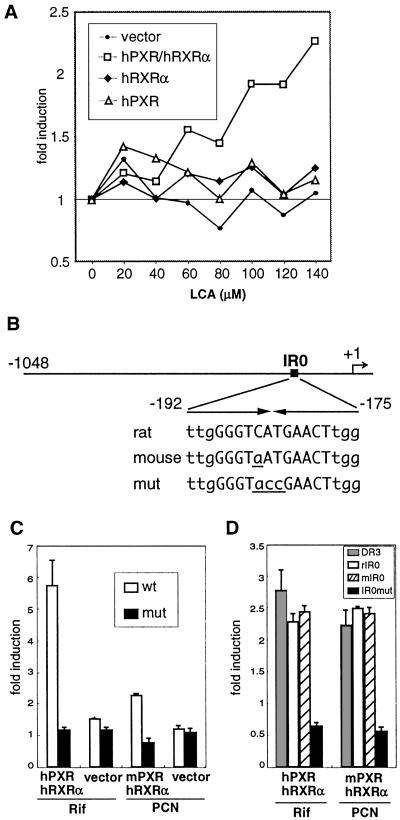
Using an approximately 1-kb fragment of the rat STD promoter, we found that LCA activates transcription in HepG2 cells, but only when both hPXR and its heterodimeric partner retinoid X receptor α (RXRα) were cotransfected (Fig. 2A). Inspection of the rat STD promoter revealed a candidate IR0 (inverted repeats without a spacing nucleotide) response element (37) (Fig. 2B). A similar IR0 element was also found in the conserved mouse STD gene (Fig. 2B). The rat STD IR0 element has previously been described as an atypical response element for farnesoide X receptor (FXR) (30) and required for transcriptional induction by PCN in rat primary hepatocytes (38). We examined whether this element mediates the PXR-dependent transcriptional regulation by generating a reporter construct that disrupts the IR0 sequence. As shown in Fig. 2C, PXR-mediated induction of the STD gene is abolished by the IR0 mutation. This element is not only necessary, but in the context of a synthetic promoter is sufficient to confer responsiveness (Fig. 2D). The IR0 element is conserved in the 5′ flanking region of the mouse STD gene except for a single base change (Fig. 2B). We found that the mouse IR0 element also supports PXR-dependent activation when used in a similar cotransfection assay (Fig. 2D). Moreover, the levels of induction through the IR0 element from either species are comparable to that of the DR3-type PXR-binding site previously identified in the rat CYP3A23. Taking all these observations together, we conclude that the IR0 element mediates PXR-dependent transcriptional activation of STD.
Figure 2.
Regulation of the STD promoter by PXR. (A) HepG2 cells were cotransfected with expression vector for human PXR (hPXR; ▵), human RXRα (hRXRα; ♦), both hPXR and hRXRα (□), or with a control vector (●) together with rSTD-luc, CMX-β-gal. Transfected cells were incubated with media containing increasing concentrations of LCA or solvent control for 24 h before luciferase and β-galactosidase assays. Transcriptional activation was assessed by the relative luciferase activity normalized by β-galactosidase activity. The results are shown as a fold induction over solvent control and represent the average of triplicate experiments. (B) Schematic of the conserved 5′ flanking region from rat and mouse STD genes. Position of the IR0 element is indicated by a filled rectangle. The transcription start site is indicated by the arrow. Also, the sequences of IR0 elements from rat and mouse STDs as well as the mutant IR0 (used in C and D and Fig. 3) are shown with arrows indicating the inverted repeat. The core IR0 sequence is shown in uppercase and flanking nucleotides are shown in lowercase. Nonconserved nucleotides in the mouse IR0 and mutant IR0 are shown in underlined lowercase. (C) HepG2 cells were cotransfected as in A with the indicated receptor expression vectors along with the rSTD-luc vector containing either the wild-type or the mutant IR0 element. Transfected cells were incubated with medium containing rifampicin (Rif; for hPXR), PCN (for mouse PXR, mPXR), or solvent control for 24 h before being assayed. The results are shown as a fold induction over solvent control and represent the mean ± SD from triplicate experiments. (D) A similar experiment in which monkey kidney CV-1 cells were cotransfected with synthetic tk-luc reporters harboring three copies of indicated response elements. A reporter gene harboring the DR3 elements from CYP3A23 was used as a positive control. Similar results were obtained by using LCA or 3-keto-LCA as a ligand (not shown).
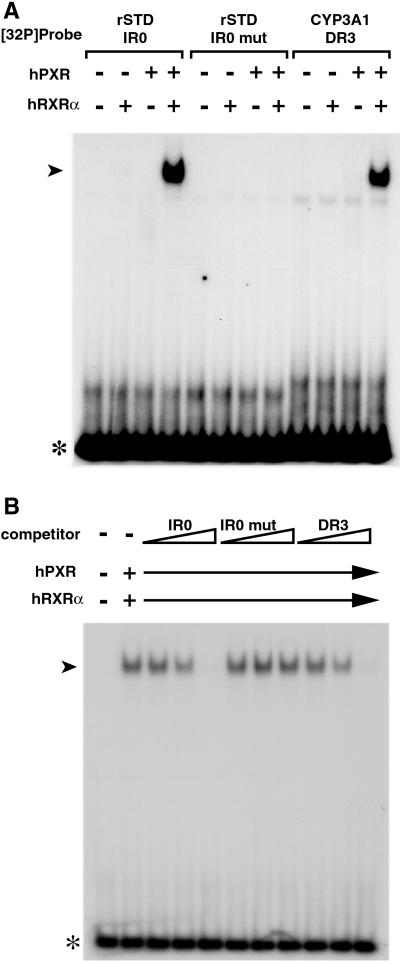
To see whether the IR0 serves as a binding site for PXR, we conducted an electrophoretic mobility-shift assay (EMSA) using in vitro synthesized receptor proteins and a 32P-labeled oligonucleotide probe encoding a single copy of the element. As shown in Fig. 3A, a mobility-shifted complex forms with the STD IR0 and CYP3A1 DR3 probes when both PXR and RXRα are present (lanes 4 and 12). The mutant IR0 abolishes binding of the PXR/RXRα complex in vitro (lane 8), indicating that receptor binding to the IR0 element is essential for induction of STD by LCA or other PXR ligands. To test the specificity and affinity of the binding in a semiquantitative manner, we performed competition experiments. Nonlabeled competitor oligonucleotides encoding the rat STD IR0 element or the rat CYP3A1 DR3 element efficiently compete with binding of the PXR/RXRα heterodimer to the IR0 element, whereas the mutant IR0 competitor does not affect the binding (Fig. 3B). We conclude that the PXR/RXRα heterodimer binds to the STD IR0 element with an affinity similar to that of the previously identified DR3 element.
Figure 3.
PXR/RXR heterodimers bind to IR0 elements. Shown are electrophoretic mobility-shift assays (EMSAs). Each reaction mixture contained reticulocyte lysate programmed to synthesize hPXR and/or hRXRα along with a 32P-labeled oligonucleotide encoding a single copy of the indicated PXR response element. Arrowheads indicate labeled DNA-bound PXR/RXR. Free probes are indicated with asterisks. In B, all reaction mixtures contained 32P-labeled rat STD IR0 probe along with increasing amounts (0.5×, 5×, 50×) of unlabeled oligonucleotide harboring the competitive response element. Note that both IR0 and DR3 competitor probes prevent formation of the slower mobility complex to similar extents, whereas the IR0 mutant competitor does not.
Coordinate Regulation of the Sulfation System by PXR.
Since PXR is thought to function as a sensor for numerous other drugs, including those metabolized by phenolic SULTs, we wondered whether activation of PXR also results in induction of additional SULTs. To test this idea, we compared liver expression of nine other SULT genes between VP-hPXR transgenic mice and nontransgenic control mice. We have found that VP-hPXR in the liver induces ST1D1 (also known as SULT-N) and ST1E5, both encoding SULT enzymes that preferentially sulfonate phenolic compounds (Fig. 4A) (39–41). Consistent with increased mRNA expression of these two phenolic SULTs, total liver extract from Alb-VP-hPXR transgenic mice contains enhanced sulfotransferase activity toward the substrate p-nitrophenol (Fig. 4B). These results suggest that multiple hepatic sulfotransferases with distinct substrate specificities are under transcriptional control of PXR.
In the presence of excess SULT substrates, sulfonation reactions become saturated in part because of the depletion of sulfonyl donor cofactor PAPS (42). PAPS is synthesized from ATP and inorganic sulfate by the activity of a bifunctional enzyme, PAPSS (43, 44). Of the two known PAPSS isozymes in mouse, PAPSS2 is primarily responsible for PAPS biosynthesis in the liver, as mutant mice deficient for the PAPSS2 gene exhibit severely reduced PAPSS activity in the liver, whereas no reduction was observed in brain and skin (45, 46). In search for novel PXR target genes by using a microarray technology (J.M.R., W.X., and R.M.E., unpublished work), we have found that mRNA encoding PAPSS2 is also elevated in the Alb-VP-hPXR liver (Fig. 4A). Taking these findings together, PXR activation in the liver appears to induce two classes of rate-limiting factors for chemical sulfation, SULT enzymes and the cofactor PAPS (Fig. 4C).
Discussion
In this report we have identified the bile acid sulfotransferase STD as a regulatory target of PXR. Activation of PXR in mice or rats results in marked induction of endogenous STD. Previously, the STD promoter has been shown to be transactivated by the bile acid receptor farnesoide X receptor (FXR), although induction of STD by FXR-specific agonists in animals remains to be demonstrated (30). Since LCA is a strong antagonist for FXR, the FXR pathway may play more significant role when the concentration of LCA relative to other bile acids is low (47). In addition, vitamin D receptor (VDR) has recently been shown to be activated by LCA (48), resulting in induction of CYP3A, a previously known PXR target gene (49, 50). Thus, these three distinct nuclear receptors may collaborate to detoxify LCA and other toxic bile acids through induction of overlapping sets of target genes.
A number of studies have implicated sulfation as an important metabolic and detoxification pathway for LCA and other secondary bile acids, especially in cholestatic conditions when high concentration of bile acids accumulate in the liver. Our results suggesting that sulfation of LCA can be up-regulated by PXR agonists may thus have implications in the treatment of cholestatic diseases. In fact, rifampicin, a potent ligand for hPXR, has been reported to be effective in treating chronic cholestasis (51, 52). Our results suggest that rifampicin may exert its effect at least in part by inducing STD. While more sulfated LCA is often observed in patients with cholestasis (18), there is a group of patients that show impaired sulfation and low STD expression level (53–55). Thus, in this group, activation of PXR and induction of STD may be beneficial in reducing intrahepatic levels of toxic bile acids and treating cholestasis.
In addition to STD, our study has identified three other genes involved in sulfonyl conjugation as potential regulatory targets of PXR. Of these, two encode phenolic SULTs, ST1D1 and ST1E5, and one encodes PAPSS2, the enzyme responsible for generation of the sulfotransferase cofactor PAPS in the liver. The induction of PAPSS2 is of particular significance, because its catalytic product PAPS is the rate-limiting factor for sulfonation at high concentrations of substrate (42). Whether these genes are all direct transcriptional targets of PXR has yet to be established, nevertheless our results indicate that activation of PXR results in an increased general capacity of the sulfonation system (Fig. 4C).
The regulation of SULTs by PXR may also be implicated in hormonal homeostasis. The sex hormones and their precursors, including estrogen, androgen, and DHEA, are known substrates for SULTs. With rare exceptions, sulfonated sex hormones are completely inactive (for reviews, see refs. 26 and 56). Thus, changes in sulfotransferase expression and enzymatic activity can markedly alter the bioavailability of steroid hormones and sensitivity of target cells. An influence of estrogen on the occurrence of breast cancer is well established. Interestingly, STD expression is altered in a number of human breast cancer cell lines (57). Because STD is a direct target of PXR, which is also expressed in the breast (58), ligands that allow manipulation of PXR activity may help to reduce cancer risk through enhanced sex hormone sulfation.
Sulfonation by SULTs also plays an important role in metabolism of many foreign chemicals, including the commonly used anti-inflammatory drug acetaminophen, whose excess causes liver injury. In fact, sulfation of acetaminophen has been shown to be increased by PXR agonists such as dexamethasone and PCN in mouse hepatocytes (59). In general, most drugs are metabolized in the liver and intestine by a two-step mechanism. The first is the functionalization step (phase I) that involves oxidation and hydroxylation by CYP or by other oxidative enzymes. This is followed by conjugation to polar moieties such as sulfonyl group, glucuronides, or glutathione (phase II) that facilitates detoxification as well as urinary and fecal excretion. Previously, PXR has been shown to directly induce expression of phase I enzymes CYP3A and CYP2B. The results shown here thus indicate that PXR can coordinately regulate both the phase I and phase II steps of the drug metabolic pathway. Consistent with this idea, recent microarray analysis using the known PXR ligand rifampicin has identified several phase I enzymes, including CYP2A, CYP2C, monoamine oxidase, and flavin-containing monooxidases as well as phase II enzymes such as UDP-glucuronosyltransferases and glutathione S-transferases as putative PXR target genes (60). A similar set of enzymes including SULTs described here were also identified by microarray analysis of liver mRNA from Alb-VP-hPXR mice (J.M.R., W.X., and R.M.E., unpublished work). Together, these results support a proposal that PXR serves as a master regulator of a hepatic drug clearance system that determines sensitivity of animals to a wide variety of endogenously and exogenously derived chemicals.
Acknowledgments
We thank Henry Juguilon for technical assistance and Elaine Stevens and Lita Ong for administrative assistance. J.S. is a fellow of the Damon Runyon Cancer Research Foundation, DRG 1711-02. W.X. is supported by the Competitive Medical Research Fund of The University of Pittsburgh Medical Center Health System. R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute for Biological Studies and March of Dimes Chair in Molecular and Developmental Biology. This work was supported by the Howard Hughes Medical Institute (R.M.E.) and National Institutes of Health Grant ESO5744 (P.S.G.).
Abbreviations
- CYP
cytochrome P450 enzyme
- DHEA
dehydroepiandrosterone
- h-
human
- LCA
lithocholic acid
- PAPS
3′-phosphoadenosine 5′-phosphosulfate
- PAPSS
PAPS synthetase
- PCN
pregnane-16α-carbonitrile
- PXR
pregnane X receptor
- RXR
retinoid X receptor
- STD
dehydroepiandrosterone sulfotransferase
- SULT
cytosolic sulfotransferase
References
- 1.Fisher M M, Magnusson R, Miyai K. Lab Invest. 1971;25:88–91. [PubMed] [Google Scholar]
- 2.Javitt N B. Nature. 1966;210:1262–1263. doi: 10.1038/2101262a0. [DOI] [PubMed] [Google Scholar]
- 3.Owen R W, Dodo M, Thompson M H, Hill M J. Nutr Cancer. 1987;9:73–80. doi: 10.1080/01635588709513914. [DOI] [PubMed] [Google Scholar]
- 4.Kishida T, Taguchi F, Feng L, Tatsuguchi A, Sato J, Fujimori S, Tachikawa H, Tamagawa Y, Yoshida Y, Kobayashi M. J Gastroenterol. 1997;32:306–311. doi: 10.1007/BF02934485. [DOI] [PubMed] [Google Scholar]
- 5.Lehmann J M, McKee D D, Watson M A, Willson T M, Moore J T, Kliewer S A. J Clin Invest. 1998;102:1016–1023. doi: 10.1172/JCI3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blumberg B, Sabbagh W, Jr, Juguilon H, Bolado J, Jr, van Meter C M, Ong E S, Evans R M. Genes Dev. 1998;12:3195–3205. doi: 10.1101/gad.12.20.3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kliewer S A, Moore J T, Wade L, Staudinger J L, Watson M A, Jones S A, McKee D D, Oliver B B, Willson T M, Zetterstrom R H, et al. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- 8.Bertilsson G, Heidrich J, Svensson K, Asman M, Jendeberg L, Sydow-Backman M, Ohlsson R, Postlind H, Blomquist P, Berkenstam A. Proc Natl Acad Sci USA. 1998;95:12208–12213. doi: 10.1073/pnas.95.21.12208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Staudinger J L, Goodwin B, Jones S A, Hawkins-Brown D, MacKenzie K I, LaTour A, Liu Y, Klaassen C D, Brown K K, Reinhard J, et al. Proc Natl Acad Sci USA. 2001;98:3369–3374. doi: 10.1073/pnas.051551698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xie W, Radominska-Pandya A, Shi Y, Simon C M, Nelson M C, Ong E S, Waxman D J, Evans R M. Proc Natl Acad Sci USA. 2001;98:3375–3380. doi: 10.1073/pnas.051014398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie W, Evans R M. J Biol Chem. 2001;276:37739–37742. doi: 10.1074/jbc.R100033200. [DOI] [PubMed] [Google Scholar]
- 12.Goodwin B, Redinbo M R, Kliewer S A. Annu Rev Pharmacol Toxicol. 2002;42:1–23. doi: 10.1146/annurev.pharmtox.42.111901.111051. [DOI] [PubMed] [Google Scholar]
- 13.Selye H. Proc Soc Exp Biol Med. 1972;141:555–558. doi: 10.3181/00379727-141-36821. [DOI] [PubMed] [Google Scholar]
- 14.Xie W, Barwick J L, Downes M, Blumberg B, Simon C M, Nelson M C, Neuschwander-Tetri B A, Brunt E M, Guzelian P S, Evans R M. Nature. 2000;406:435–439. doi: 10.1038/35019116. [DOI] [PubMed] [Google Scholar]
- 15.Staudinger J, Liu Y, Madan A, Habeebu S, Klaassen C D. Drug Metab Dispos. 2001;29:1467–1472. [PubMed] [Google Scholar]
- 16.Palmer R H, Bolt M G. J Lipid Res. 1971;12:671–679. [PubMed] [Google Scholar]
- 17.Podesta M R, Murphy G M, Dowling R H. J Chromatogr. 1980;182:293–300. doi: 10.1016/s0378-4347(00)81477-2. [DOI] [PubMed] [Google Scholar]
- 18.Stiehl A. Digestion. 1974;11:406–413. doi: 10.1159/000197609. [DOI] [PubMed] [Google Scholar]
- 19.Leuschner U, Czygan P, Liersch M, Frohling W, Stiehl A. Z Gastroenterol. 1977;15:246–253. [PubMed] [Google Scholar]
- 20.Takikawa H, Tomita J, Takemura T, Yamanaka M. Biochim Biophys Acta. 1991;1091:173–178. doi: 10.1016/0167-4889(91)90058-6. [DOI] [PubMed] [Google Scholar]
- 21.Garner C M, Mills C O, Elias E, Neuberger J M. Biochim Biophys Acta. 1991;1091:41–45. doi: 10.1016/0167-4889(91)90219-n. [DOI] [PubMed] [Google Scholar]
- 22.Gadacz T R, Allan R N, Mack E, Hofmann A F. Gastroenterology. 1976;70:1125–1129. [PubMed] [Google Scholar]
- 23.Dyrszka H, Chen T, Salen G, Mosbach E H. Gastroenterology. 1975;69:333–337. [PubMed] [Google Scholar]
- 24.Donovan J M, Yousef I M, Carey M C. Biochim Biophys Acta. 1993;1182:37–45. doi: 10.1016/0925-4439(93)90150-y. [DOI] [PubMed] [Google Scholar]
- 25.Cowen A E, Korman M G, Hofmann A F, Cass O W, Coffin S B. Gastroenterology. 1975;69:67–76. [PubMed] [Google Scholar]
- 26.Strott C A. Endocr Rev. 1996;17:670–697. doi: 10.1210/edrv-17-6-670. [DOI] [PubMed] [Google Scholar]
- 27.Nagata K, Yamazoe Y. Annu Rev Pharmacol Toxicol. 2000;40:159–176. doi: 10.1146/annurev.pharmtox.40.1.159. [DOI] [PubMed] [Google Scholar]
- 28.Chen L J, Segel I H. Arch Biochem Biophys. 1985;241:371–379. doi: 10.1016/0003-9861(85)90559-4. [DOI] [PubMed] [Google Scholar]
- 29.Radominska A, Comer K A, Zimniak P, Falany J, Iscan M, Falany C N. Biochem J. 1990;272:597–604. doi: 10.1042/bj2720597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Song C S, Echchgadda I, Baek B S, Ahn S C, Oh T, Roy A K, Chatterjee B. J Biol Chem. 2001;276:42549–42556. doi: 10.1074/jbc.M107557200. [DOI] [PubMed] [Google Scholar]
- 31.Liu L, Klaassen C D. Drug Metab Dispos. 1996;24:854–858. [PubMed] [Google Scholar]
- 32.Rubin G L, Harrold A J, Mills J A, Falany C N, Coughtrie M W. Mol Hum Reprod. 1999;5:995–1002. doi: 10.1093/molehr/5.11.995. [DOI] [PubMed] [Google Scholar]
- 33.Li D, Schuetz E G, Guzelian P S. Methods Enzymol. 1991;206:335–344. doi: 10.1016/0076-6879(91)06103-a. [DOI] [PubMed] [Google Scholar]
- 34.Shimada M, Kamiyama Y, Sato A, Honma W, Nagata K, Yamazoe Y. J Biochem (Tokyo) 2002;131:167–169. doi: 10.1093/oxfordjournals.jbchem.a003083. [DOI] [PubMed] [Google Scholar]
- 35.Klaassen C D, Liu L, Dunn R T., 2nd Chem Biol Interact. 1998;109:299–313. doi: 10.1016/s0009-2797(97)00141-5. [DOI] [PubMed] [Google Scholar]
- 36.Guo G L, Staudinger J, Ogura K, Klaassen C D. Mol Pharmacol. 2002;61:832–839. doi: 10.1124/mol.61.4.832. [DOI] [PubMed] [Google Scholar]
- 37.Song C S, Jung M H, Kim S C, Hassan T, Roy A K, Chatterjee B. J Biol Chem. 1998;273:21856–21866. doi: 10.1074/jbc.273.34.21856. [DOI] [PubMed] [Google Scholar]
- 38.Runge-Morris M, Wu W, Kocarek T A. Mol Pharmacol. 1999;56:1198–1206. doi: 10.1124/mol.56.6.1198. [DOI] [PubMed] [Google Scholar]
- 39.Garcia-Allan C, Lord P G, Loughlin J M, Orton T C, Sidaway J E. J Biochem Mol Toxicol. 2000;14:65–72. doi: 10.1002/(sici)1099-0461(2000)14:2<65::aid-jbt1>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 40.Sakakibara Y, Yanagisawa K, Takami Y, Nakayama T, Suiko M, Liu M C. Biochem Biophys Res Commun. 1998;247:681–686. doi: 10.1006/bbrc.1998.8872. [DOI] [PubMed] [Google Scholar]
- 41.Song W C, Moore R, McLachlan J A, Negishi M. Endocrinology. 1995;136:2477–2484. doi: 10.1210/endo.136.6.7750469. [DOI] [PubMed] [Google Scholar]
- 42.Liu L, Klaassen C D. Toxicol Appl Pharmacol. 1996;139:128–134. doi: 10.1006/taap.1996.0151. [DOI] [PubMed] [Google Scholar]
- 43.Li H, Deyrup A, Mensch J R, Jr, Domowicz M, Konstantinidis A K, Schwartz N B. J Biol Chem. 1995;270:29453–29459. doi: 10.1074/jbc.270.49.29453. [DOI] [PubMed] [Google Scholar]
- 44.Lyle S, Stanczak J, Ng K, Schwartz N B. Biochemistry. 1994;33:5920–5925. doi: 10.1021/bi00185a032. [DOI] [PubMed] [Google Scholar]
- 45.Kurima K, Warman M L, Krishnan S, Domowicz M, Krueger R C, Jr, Deyrup A, Schwartz N B. Proc Natl Acad Sci USA. 1998;95:8681–8685. doi: 10.1073/pnas.95.15.8681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugahara K, Schwartz N B. Arch Biochem Biophys. 1982;214:602–609. doi: 10.1016/0003-9861(82)90065-0. [DOI] [PubMed] [Google Scholar]
- 47.Yu J, Lo J L, Huang L, Zhao A, Metzger E, Adams A, Meinke P T, Wright S D, Cui J. J Biol Chem. 2002;277:31441–31447. doi: 10.1074/jbc.M200474200. [DOI] [PubMed] [Google Scholar]
- 48.Makishima M, Lu T T, Xie W, Whitfield G K, Domoto H, Evans R M, Haussler M R, Mangelsdorf D J. Science. 2002;296:1313–1316. doi: 10.1126/science.1070477. [DOI] [PubMed] [Google Scholar]
- 49.Schmiedlin-Ren P, Thummel K E, Fisher J M, Paine M F, Watkins P B. Drug Metab Dispos. 2001;29:1446–1453. [PubMed] [Google Scholar]
- 50.Thummel K E, Brimer C, Yasuda K, Thottassery J, Senn T, Lin Y, Ishizuka H, Kharasch E, Schuetz J, Schuetz E. Mol Pharmacol. 2001;60:1399–1406. doi: 10.1124/mol.60.6.1399. [DOI] [PubMed] [Google Scholar]
- 51.Bachs L, Pares A, Elena M, Piera C, Rodes J. Lancet. 1989;i:574–576. doi: 10.1016/s0140-6736(89)91608-5. [DOI] [PubMed] [Google Scholar]
- 52.Cancado E L, Leitao R M, Carrilho F J, Laudanna A A. Am J Gastroenterol. 1998;93:1510–1517. doi: 10.1111/j.1572-0241.1998.00472.x. [DOI] [PubMed] [Google Scholar]
- 53.Fischer S, Beuers U, Spengler U, Zwiebel F M, Koebe H G. Clin Chim Acta. 1996;251:173–186. doi: 10.1016/0009-8981(96)06305-x. [DOI] [PubMed] [Google Scholar]
- 54.Iqbal S, Vickers C, Elias E. J Hepatol. 1990;11:37–42. doi: 10.1016/0168-8278(90)90269-w. [DOI] [PubMed] [Google Scholar]
- 55.Elekima O T, Mills C O, Ahmad A, Skinner G R, Ramsden D B, Bown J, Young T W, Elias E. Liver. 2000;20:45–50. doi: 10.1034/j.1600-0676.2000.020001045.x. [DOI] [PubMed] [Google Scholar]
- 56.Hobkirk R. Can J Biochem Cell Biol. 1985;63:1127–1144. doi: 10.1139/o85-141. [DOI] [PubMed] [Google Scholar]
- 57.Pasqualini J R, Varin C, Nguyen B L. Cancer Lett. 1992;66:55–60. doi: 10.1016/0304-3835(92)90280-9. [DOI] [PubMed] [Google Scholar]
- 58.Dotzlaw H, Leygue E, Watson P, Murphy L C. Clin Cancer Res. 1999;5:2103–2107. [PubMed] [Google Scholar]
- 59.Seo K W, Oh M H, Choung S Y, Kim S J, Kim H J. Biol Pharm Bull. 1999;22:261–264. doi: 10.1248/bpb.22.261. [DOI] [PubMed] [Google Scholar]
- 60.Rae J M, Johnson M D, Lippman M E, Flockhart D A. J Pharmacol Exp Ther. 2001;299:849–857. [PubMed] [Google Scholar]