Abstract
The low-shear environment of optimized rotation suspension culture allows both eukaryotic and prokaryotic cells to assume physiologically relevant phenotypes that have led to significant advances in fundamental investigations of medical and biological importance. This culture environment has also been used to model microgravity for ground-based studies regarding the impact of space flight on eukaryotic and prokaryotic physiology. We have previously demonstrated that low-shear modeled microgravity (LSMMG) under optimized rotation suspension culture is a novel environmental signal that regulates the virulence, stress resistance, and protein expression levels of Salmonella enterica serovar Typhimurium. However, the mechanisms used by the cells of any species, including Salmonella, to sense and respond to LSMMG and identities of the genes involved are unknown. In this study, we used DNA microarrays to elucidate the global transcriptional response of Salmonella to LSMMG. When compared with identical growth conditions under normal gravity (1 × g), LSMMG differentially regulated the expression of 163 genes distributed throughout the chromosome, representing functionally diverse groups including transcriptional regulators, virulence factors, lipopolysaccharide biosynthetic enzymes, iron-utilization enzymes, and proteins of unknown function. Many of the LSMMG-regulated genes were organized in clusters or operons. The microarray results were further validated by RT-PCR and phenotypic analyses, and they indicate that the ferric uptake regulator is involved in the LSMMG response. The results provide important insight about the Salmonella LSMMG response and could provide clues for the functioning of known Salmonella virulence systems or the identification of uncharacterized bacterial virulence strategies.
Optimized suspension culture using specially designed rotating bioreactors, called high-aspect-ratio rotating wall vessel bioreactors (HARVs), provides a low-shear growth environment that allows both prokaryotic and eukaryotic cells to assume medically and biologically important phenotypes that cannot be observed by using conventional culture methods. These phenotypes include the formation of three-dimensional tissue aggregates that morphologically and physiologically resemble in vivo tissues and are currently being engineered for use in tissue transplantation and infectious disease research (1–5). The growth environment achieved through optimized suspension culture is thought to provide low-shear growth cues [<1 dyne/cm2 (<10 μN/cm2)] similar to those encountered during normal in vivo fetal development and in certain low-shear areas of the body, such as between the brush border microvilli of epithelial cells (6–8). The latter environment is relevant to that encountered by numerous bacterial pathogens during the natural course of infection of the gastrointestinal, respiratory, and urogenital tracts. In addition, because several cellular responses observed under low-shear optimized suspension culture mimic those observed during cell culture in space, this culture environment is also used for ground-based studies of the effects of modeled microgravity on both eukaryotic and prokaryotic cellular physiology (1, 9). We have previously shown that growth conditions of low-shear modeled microgravity (LSMMG) increase the virulence potential of the invasive enteric pathogen Salmonella enterica serovar Typhimurium (9). Salmonella grown under LSMMG, as compared with identical growth conditions at normal gravity (1 × g), displayed a decreased LD50, shortened host time-to-death, and increased colonization of liver and spleen in the murine model of infection. LSMMG-grown Salmonella also displayed increased resistance to environmental stresses (acid, thermal, and osmotic), increased ability to survive within macrophages, and alterations in protein expression levels (9, 10). This global change in phenotype and protein expression by Salmonella in response to LSMMG indicates the existence of a regulatory network we term the “LSMMG regulon.” Our previous work indicates that the Salmonella LSMMG response does not require RpoS, the major regulator of environmental signal responses in both Escherichia coli and Salmonella (10). However, the mechanisms used by Salmonella, or the cells of any species, to sense and respond to LSMMG and the identities of the genes involved are not known.
Completion of the S. enterica serovar Typhimurium genome sequence (11) and the availability of DNA microarrays have provided an opportunity to study the wide-scale effects of LSMMG on this pathogen at the genomic and transcriptional level. Here we present the use of microarrays to elucidate the global transcriptional response of Salmonella to LSMMG as compared with identical conditions under 1 × g. This represents, to our knowledge, the first use of whole-genome analysis to identify genes that change expression in response to LSMMG in a prokaryotic organism and the first microarray-based global transcriptional analysis of Salmonella to any environmental signal. We report the identities of 163 genes differentially regulated by LSMMG; these genes represent numerous and diverse functional groups, indicating an overall reprogramming of Salmonella physiology in response to the LSMMG signal. The LSMMG gene groups include transcriptional regulators, virulence factors, lipopolysaccharide (LPS) biosynthetic enzymes, iron-utilization enzymes, and proteins of unknown function. Several of the genes are located in the same operon or in physically linked clusters. RT-PCR and phenotypic analysis further validated the results obtained with the microarrays. The information gained from the microarray analysis led to the discovery that the ferric uptake regulator (Fur) is involved in the Salmonella acid stress resistance that is induced by LSMMG. We also show that Salmonella LPS levels change in response to LSMMG, as predicted by the microarray results. Our findings indicate that LSMMG is a major global regulatory signal in Salmonella and help to direct future research that could provide clues to the functioning of known Salmonella virulence systems or to the identification of uncharacterized bacterial virulence and regulatory strategies.
Materials and Methods
Bacterial Strains and Growth Conditions.
Studies were performed with wild-type S. enterica serovar Typhimurium χ3339 (12) (an animal-passaged isolate of SL1344) and an isogenic fur mutant derivative χ4659 (kindly provided by Roy Curtiss III, Washington University, St. Louis, MO). All strains were grown in Lennox broth (13) in the HARV (Synthecon, Houston) at 37°C under both 1 × g and LSMMG by using a 50-ml culture volume and 25 rpm rotational speed, as described previously (9). The cultivation conditions of LSMMG and 1 × g were achieved by changing the physical orientation of the HARV as shown in Fig. 5, which is published as supporting information on the PNAS web site. When the axis of the bioreactor's rotation is perpendicular to the gravitational vector, a condition of LSMMG is achieved. Likewise, positioning of the bioreactor axis of rotation parallel to the gravitational vector facilitates a 1 × g condition. All experiments were performed with LSMMG and 1 × g cultures simultaneously. The HARV cultures were harvested after 10 h of growth, at which time both the LSMMG and 1 × g cultures were at similar culture densities (5 × 108 to 1 × 109 cells), corresponding to mid-late log phase.
Preparation of Whole Genome Microarrays.
S. enterica serovar Typhimurium DNA microarrays were prepared by printing PCR amplicons representing ≈99% of the genome onto aminosilane-coated slides by using GeneMachine OmniGrid Array Maker (Genomic Instrumentation Services, San Carlos, CA), as described previously (14). Each DNA sample was printed in triplicate on each slide.
Preparation of Fluorescent Probes.
Total cellular RNA was obtained from Salmonella χ3339 grown under 1 × g and LSMMG by using the Qiagen RNeasy kit (Qiagen, Valencia, CA). Cells were harvested by centrifugation at 4°C, immediately resuspended in Qiagen RLT buffer, and lysed by agitation in the presence of glass beads. The RNA was then purified according to the manufacturer's instructions (Qiagen). Twenty micrograms of DNase-treated (Ambion, Austin, TX) total RNA was converted to fluorescently labeled cDNA by using Fluorolink Cy3- or Cy5-dUTP (Amersham Pharmacia), as described by P. Brown (http://cmgm.stanford.edu/pbrown/protocols/4_Ecoli_RNA.txt). To control for labeling differences, duplicate reactions were carried out where the Cy3 and Cy5 labels were switched during synthesis. Subsequent analysis of the two different labeling reactions was performed identically, as described below, by using the corresponding scan wavelength for each label during image acquisition. The labeled probes were then purified with a PCR purification kit according to the manufacturer's instructions (Qiagen), vacuum-dried, and stored at −20°C.
Probe Hybridization.
Microarrays were blocked according to the methodology of P. Brown (http://cmgm.stanford.edu/pbrown/protocols/3_post_process.html). Immediately before use, the probes were resuspended in 50 μl of hybridization buffer (5× SSC/0.1% SDS/0.2 mg/ml BSA). Microarrays were probed by cohybridizing the fluorescently labeled 1 × g and LSMMG cDNAs to the same microarray by using a Genomic Solutions automated hybridization chamber (Genomic Solutions, Ann Arbor, MI). Denatured probes were hybridized to slides for 18 h (3 h at 65°C, 3 h at 55°C, and 12 h at 50°C). The slides were then washed twice with 2× SSC/0.1% SDS at 50°C/four times with 1× SSC at 42°C/four times with 0.2× SSC at 42°C.
Image Acquisition and Data Analysis.
The microarrays were scanned for the Cy3 and Cy5 fluorescent signals by using ScanArray 3000 from GSI Lumonics (General Scanning, Watertown, MA), and the stored images were later analyzed by using imagene analysis software (Biodiscovery, Los Angeles) and gene spring software (Silicon Genetics, Palo Alto, CA). For both the Cy3 and Cy5 images, the intensity of each spot was defined as the summed intensities of each pixel within a circle that was precisely positioned over the spot. Background was defined as the summed intensity of an identical number of pixels directly surrounding the spot. The Cy3 and Cy5 values for each spot were normalized to account for any difference in total intensity between the two scanned images. The background-subtracted normalized LSMMG and 1 × g signal intensity values at each spot were used for calculating the ratio of gene expression of LSMMG to 1 × g. Finally, all replicated data were combined and the mean expression ratio and standard deviations calculated for ORFs showing >2-fold change. All mean expression ratios were calculated from experimental results obtained from at least three independent HARV cultures.
RT-PCR.
DNase-treated LSMMG and 1 × g total RNA were reverse transcribed with SuperScriptII (Life Technologies, Gaithersburg, MD) by using random primers (Invitrogen) as described by the manufacturers. A subset of the genes or operons that showed >2-fold change in expression as determined from microarray analysis was then amplified with gene-specific primers. Reactions without SuperScript II were done in parallel to rule out amplification from contaminating DNA. PCR products were run on agarose gels, quantitated by using alphaease fc (Alpha Innotech, San Leandro, CA), and normalized by using ribosomal RNA content. The nucleotide sequences of the primers used in this study are in Table 2, which is published as supporting information on the PNAS web site.
LPS Isolation and Quantitation.
To isolate LPS, bacterial lysates were subjected to Proteinase K digestion as described previously (15). Preparations were subjected to SDS/PAGE, and the LPS was visualized by silver staining, as described previously (16). The digital gel image was captured and quantitated by using alphaease fc software. Total protein content of the bacterial lysates was quantitated by using a protein assay kit as described by the manufacturer (Bio-Rad). Normalization of the quantitated LPS values was achieved by dividing the LSMMG to 1 × g LPS ratio by the LSMMG to 1 × g total protein ratio.
Acid Stress Assays.
Bacteria grown under LSMMG and 1 × g were subjected to acid stress of pH 3.5, as described previously (9). The average percent survival of three independent trials was calculated as the ratio of colony-forming units (cfu) at each time point to cfu at time 0.
Results
Microarray Analysis of Salmonella Response to LSMMG.
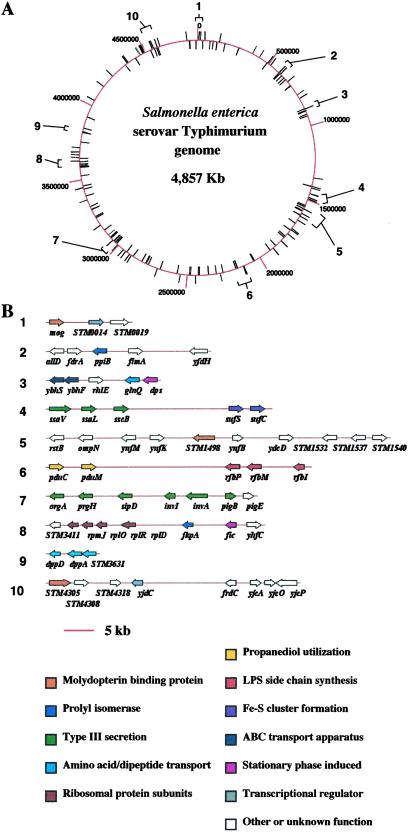
To characterize the global expression changes of Salmonella in response to LSMMG, we used DNA microarrays representing ≈99% of the ORFs of Salmonella to examine the genes that were up- or down-regulated by this organism in response to LSMMG, as compared with identical growth conditions under 1 × g. Total RNA from LSMMG- and 1 × g-grown Salmonella χ3339 was converted to fluorescently labeled cDNA and cohybridized to the microarrays. Of the ≈4,600 genes on the array, 163 were identified with statistically significant changes in the level of expression—at least a 2-fold change—and are described by using an expression ratio (LSMMG/1 × g). Genes with expression ratios <0.5 were considered down-regulated and those with >2, up-regulated. Mapping of all of the LSMMG-regulated genes showed them to be distributed across the entire Salmonella chromosome as shown in Fig. 1A. Many of these genes were organized into clusters and represented members of the same operon, which indicates that, although the LSMMG regulon is globally distributed on the chromosome, certain genetic loci may be targeted by the LSMMG response. On the basis of their distribution, at least 10 such clusters comprised of genes belonging to various functional categories could be identified (Fig. 1B). The complete list of both up- and down-regulated genes identified by the microarray analysis is published as supporting information on the PNAS web site, www.pnas.org, and shows that the number of down-regulated genes is slightly larger than the number of up-regulated (97 vs. 68, respectively). This observation is consistent with previous results that showed the number of proteins down-regulated by LSMMG was greater than the number of up-regulated proteins as detected by two-dimensional gel analysis (9). The listed genes are broadly categorized by function or location in the cell envelope if predicted to be a membrane protein. The genes belong to diverse functional groups, including transcriptional regulators, type III secretion components, LPS O-antigen biosynthetic enzymes, ribosomal subunits, iron-utilization enzymes, and membrane transporters. The latter two groups, along with the transcriptional regulators, have genes that are both up- and down-regulated, indicating a differential regulation of these functions by LSMMG. Numerous genes were not easily categorized into functional groups and represent a wide range of cellular metabolic processes. Some genes encode cytoplasmic and membrane proteins with no detectable homology to other proteins and were not assigned a function. This result indicates that LSMMG is regulating genes whose functions are yet to be discovered and may be involved in uncharacterized cellular processes. The microarray results demonstrate that LSMMG is a growth signal that affects a large number of cell functions and most likely serves to globally reprogram Salmonella physiology to allow the bacteria to adapt to this environment.
Figure 1.
Chromosomal organization of the S. enterica serovar Typhimurium LSMMG regulon. (A) The circular Salmonella chromosome is schematically depicted with kilobase coordinates, noted as red lines extending from the chromosome. LSMMG-regulated genes as identified by microarray analysis are noted as black lines extending from the chromosome. Numbered brackets indicate clusters of genes that are physically linked (within 50 kb) or organized into the same operon and are shown in greater detail in B immediately below. (B) Clusters of LSMMG-regulated genes are schematically depicted. The numbers next to each cluster correspond to the numbered brackets in A. The LSMMG-regulated genes are depicted as arrows and are colored according to a common function. Only those genes that are LSMMG-regulated are shown for each cluster. Both up- and down-regulated genes are sometimes present in the same cluster. The regulatory profiles of the genes are found in Table 1, which is published as supporting information on the PNAS web site.
RT-PCR of Specific LSMMG-Regulated Genes.
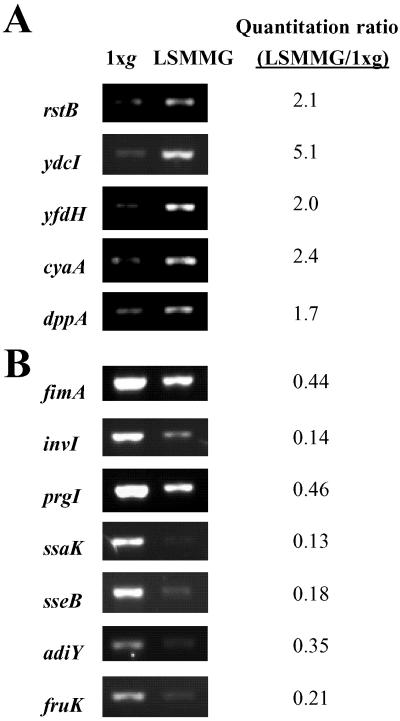
To confirm our microarray expression data, we assessed the levels of specific 1 × g and LSMMG Salmonella mRNAs by RT-PCR. Oligonucleotide primers were designed for a subset of genes identified by the microarrays as differentially expressed in response to LSMMG. These genes represent diverse functional categories and are widely distributed on the Salmonella chromosome. Fig. 2 shows the results of RT-PCR analysis of 12 genes from 1 × g- and LSMMG-grown Salmonella. The prgI and ssaK genes, although not identified by microarray analysis, are immediately adjacent to genes identified as LSMMG-regulated in their respective operons, indicating that those entire transcriptional units are LSMMG responsive. The RT-PCR results are in complete agreement with those obtained by using the microarrays and confirm our data obtained by using the whole-genome analysis.
Figure 2.
RT-PCR analysis of Salmonella LSMMG-regulated genes. The expression of Salmonella genes identified by microarray analysis was analyzed using RT-PCR with total RNA from LSMMG and 1 × g HARV cultures as described in Materials and Methods. The ratio of the normalized quantitated signals is presented as the LSMMG value divided by 1 × g value. The DNA sequences of the primers used are published as supporting information on the PNAS web site. The prgI and ssaK genes are adjacent to other LSMMG-regulated genes in their respective operons. (A) LSMMG up-regulated genes. (B) LSMMG down-regulated genes.
LPS Profile of 1 × g- and LSMMG-Grown Salmonella.
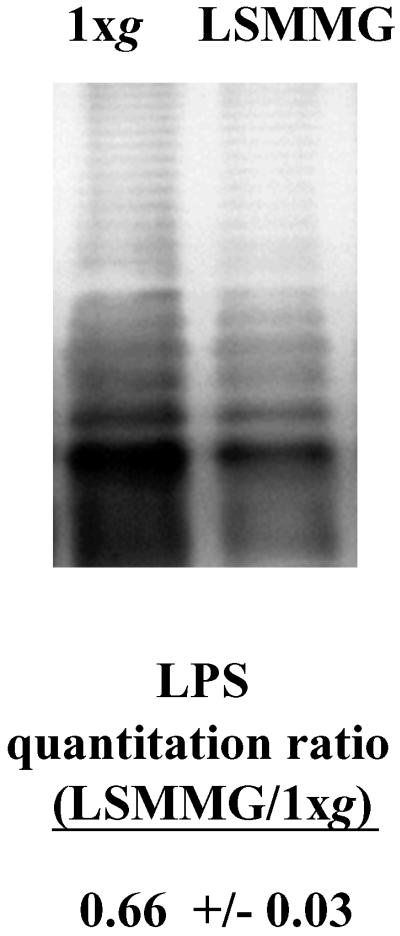
Three genes detected by our microarray analysis, rfbP, rfbM, and rfbI, are expressed from the same large operon involved in LPS O-antigen biosynthesis and were found to be down-regulated by LSMMG. The surface expression of LPS can affect the virulence of bacterial pathogens, including Salmonella, and could be related to the increase in Salmonella virulence observed under LSMMG. Therefore, we compared the LPS O-antigen profiles of 1 × g- and LSMMG-grown Salmonella χ3339 (Fig. 3). Although the O-antigen profile of both the 1 × g and LSMMG cultures showed similar banding patterns, a distinct difference in the intensity of the pattern was observed and indicated that less LPS appeared to be recovered from the LSMMG cultures. This difference was confirmed by quantitation by using densitometric analysis with the values being normalized based on total protein content of the samples. The amount of LPS isolated from the LSMMG-grown χ3339 was nearly half that of the 1 × g culture. This finding is consistent with the microarray results, which indicated the down-regulation of genes involved in LPS O-antigen biosynthesis.
Figure 3.
The LPS profiles of LSMMG- and 1 × g-grown Salmonella. The Salmonella LPS from LSMMG and 1 × g HARV cultures was isolated and visualized as described in Materials and Methods. The ratio of the quantitated LPS signal is presented as the LSMMG value divided by 1 × g value. This ratio was normalized by comparing the quantitated amounts of total protein present in the LSMMG- and 1 × g-grown cells.
Involvement of fur in the Salmonella LSMMG Response.
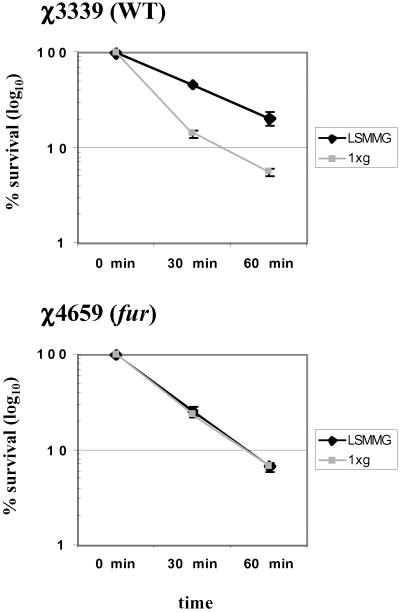
Our microarray analysis indicated that several genes involved in iron metabolism (fepD, STM1537, hscB, feoB, yliG, sufC, sufS) are regulated by LSMMG. Several ORFs encoding putative hydrogenases, dehydrogenases, and reductases that could potentially use iron for normal function were identified as well (see supporting information on the PNAS web site). In addition, a number of potential Fur-binding sites were found to be located upstream of several different LSMMG regulated genes (data not shown; unpublished results). At six of these genes, two potential Fur-binding sites were identified, and in five of those cases, the sites are within 300 bp of each other. This is a Fur-binding site arrangement that has been previously observed (17). The Fur-binding site-associated genes included those known to be regulated by Fur (fepD, sufC, sufS, feoB) and those not previously characterized as Fur regulated. Finally, Fur has previously been shown to regulate a number of other processes besides iron metabolism in E. coli and Salmonella, including acid resistance and succinate utilization (18, 19). These observations prompted us to examine the LSMMG response in a fur mutant strain. We examined the acid survival profile of a fur mutant, χ4659, as compared with its isogenic wild-type parent χ3339 under conditions of both 1 × g and LSMMG (Fig. 4). We have previously shown that LSMMG induces acid resistance in Salmonella χ3339 (9). Using conventional growth conditions in a shaker culture flask, previous studies have shown that χ3339 and χ4659 have very similar acid resistance profiles from 0 to 60 min (20, 21). However, comparison of the acid survival profiles of both strains cultured at LSMMG and 1 × g indicates that, whereas χ3339 displays LSMMG-induced acid resistance as expected, the fur mutant χ4659 does not show any detectable acid resistance to be induced by LSMMG. This result indicates that Fur is required for LSMMG-induced acid resistance in Salmonella, a finding that is consistent with the involvement of Fur in transmitting the LSMMG signal as suggested by our microarray results.
Figure 4.
Involvement of fur in the Salmonella LSMMG response. Salmonella strains χ3339 (wild type) and χ4659 (fur) grown at both LSMMG and 1 × g were subjected to acid stress (pH 3.5) as described in Materials and Methods. Percent survival is plotted as the number of viable colony-forming units (cfu) at the indicated time divided by the viable cfu at time 0. At least three independent trials of each experiment were performed, and the error bars represent the standard error of the mean.
Discussion
Optimized suspension culture using rotating HARV bioreactors offers enormous potential to help us understand the low-shear growth signals present in vivo and in space and has already led to major advances in several fields, including tissue engineering, infectious disease, and microgravity research (1–5, 10). The LSMMG growth signal provided by the HARV bioreactors induces a genetic regulon in Salmonella and results in increased bacterial virulence and stress resistance and alteration in protein expression levels (9, 10). Here, using whole genome DNA microarray analysis, we present the identification of 163 Salmonella genes whose expression is altered in response to LSMMG and which therefore belong to the LSMMG regulon. The global distribution of these genes across the Salmonella chromosome and the diverse functions represented by the LSMMG gene groups indicate that a wide-scale reprogramming of Salmonella physiology occurs under LSMMG growth conditions. These results provide an important step in understanding the response to LSMMG growth signals and form a potential “road map” for future studies involving the molecular response to LSMMG. These results are also remarkably consistent with our previous 2D protein gel analysis of LSMMG gene expression in two ways. First, the majority of the detected changes in both the 2D gel and microarray analyses were decreases in expression in response to LSMMG (9). Second, although the 2D gel analysis did not allow identification of the LSMMG-responsive proteins, several of those proteins ran at positions extremely similar to those of iron-binding, Fur-regulated proteins (unpublished observation).
We have identified LSMMG regulon gene groups involved in transcriptional regulation, type III secretion, LPS, and cell wall synthesis, iron metabolism, ribosomal structure, and membrane transport. We also detected several genes with no homology to other genes in the current databases, suggesting that novel functions may be regulated by LSMMG. Several of the genes identified by the microarray analysis are found in the same operon or within a physically linked cluster of genes, indicating that certain areas of the chromosome are specifically targeted by the LSMMG signaling pathway. Those LSMMG regulon genes found in the same operon are presumably expressed from the same LSMMG-regulated promoter. The clustered genes not expressed in the same operon would likely have to be regulated by a different means. Coregulation of genes found in the same physical location but not expressed from the same promoter frequently involves changes to the local DNA–protein architecture that can affect the expression of genes spread across several kilobases (22). This type of gene regulation may be occurring under LSMMG, and two genes encoding modulators of DNA topology (himA and dps) were identified in our microarray analysis. The IHF (composed of HimA and HimB subunits) and DPS proteins, or other chromatin-associated proteins, may be involved in coregulation of the physically linked clusters of genes found to be responsive to LSMMG.
The results indicate that the Fur protein is involved in the Salmonella LSMMG response on the basis of a number of observations. First, our microarray analysis revealed several genes involved, or potentially involved, in iron metabolism to be regulated by LSMMG. Second, a number of potential Fur-binding sites were found to be located upstream of several different LSMMG regulon genes. Third, LSMMG-induced acid resistance is not observed in a Salmonella fur mutant strain. Thus, LSMMG may be a novel example of an environmental signal that utilizes Fur for cellular transmission and opens up the possibility of identifying several new Fur-regulated genes. Whether Fur is responding to changes in iron concentration or some other coinducer under the LSMMG growth conditions is not known at this time. However, results suggest that Fur could be regulating a wide array of genes affecting multiple aspects of cellular metabolism in response to LSMMG. The role of Fur as a global regulator of a variety of cellular functions in response to an environmental signal (in addition to iron-related functions) has been previously suggested (17).
Fourteen genes encoding transcriptional regulators were found to be regulated by LSMMG (see supporting information on the PNAS web site). Eight of these genes were up-regulated, and six were down-regulated. These regulators may be involved in eliciting changes in gene expression that affect the reprogramming of Salmonella physiology in response to LSMMG. Seven of the eight up-regulated transcriptional regulators are encoded by genes that are previously uncharacterized. Half (7 of 14) of the LSMMG-regulated transcriptional factors are predicted to belong to the LysR family of regulators that respond to different coinducers and regulate a variety of genes with different functions. This family of transcriptional regulators may be important for responding to LSMMG in Salmonella. In addition to the transcription factors, a putative membrane sensory kinase (encoded by rstB) that functions as part of a two-component system was identified in our microarray analysis and may function in the transmission of the LSMMG signal to the cell. The roles of these genes in the Salmonella LSMMG response are currently being investigated in our laboratory.
Our microarray analysis indicated that the large rfb operon involved in LPS side chain synthesis was down-regulated by LSMMG. We compared the LPS profile of both 1 × g- and LSMMG-grown Salmonella and found the overall level of LPS to be decreased under LSMMG conditions. Other surface-exposed cellular functions are also targeted for regulation by LSMMG, including fimbrial subunits (fimA and sthE), type III secretion machinery (see below), and several other membrane proteins. This aspect of the Salmonella LSMMG response may be related to the increase in Salmonella virulence that we have previously observed. The alteration of Salmonella surface components (which involve decreases in LPS, FimA, and type III secretion machinery levels) that serve as targets for recognition by the host immune system may allow LSMMG-grown Salmonella to evade an initial immune response to a greater degree and enhance establishment of Salmonella in the host. This explanation would assume that a sufficient amount of these surface components is available to perform their functions related to the Salmonella infection process. The immune response to LSMMG-grown Salmonella is currently being investigated in our laboratory. The alteration of surface components likely reflects the global reprogramming of Salmonella physiology that is occurring under LSMMG. Of particular note is that several of the membrane proteins identified in our microarray analysis are previously uncharacterized and could even function together to form membrane complexes involved in the LSMMG response.
Our results indicate that the mRNA levels of genes located in the SPI-1 and -2 pathogenicity islands encoding the two Salmonella type III three secretion systems are down-regulated by LSMMG (orgA, prgH, sipD, invI, invA, pigB, sseB, ssaL, ssaV, and sseJ). It appears that the physiological reprogramming by LSMMG involves a pathway that decreases transcription of type III secretion genes, although we do not presently know the functional reasons for this phenomenon. Preliminary data in our laboratory suggest that the accumulation of Salmonella secreted proteins into the external media under LSMMG may be decreased (data not shown), which could be a reflection of the down-regulation of type III and/or other secretion systems. The resulting decrease in type III gene transcription due to LSMMG is curious because the SPI-1 and -2 type III secretion systems play roles in Salmonella virulence, and LSMMG causes an increase in Salmonella virulence as measured by mouse infection and macrophage survival assays (9), two assays that depend on the type III secretion systems. One possible explanation could be that the LSMMG signal serves to modulate type III secretion to the optimal level, which involves an overall decrease in transcription of the type III genes. Another explanation could be that LSMMG increases Salmonella virulence via a mechanism that does not involve type III secretion, and the decrease in type III gene expression is not involved in the LSMMG-induced increase in Salmonella virulence. The possibility that novel Salmonella virulence mechanisms are induced by LSMMG is discussed below. A third explanation for decrease in type III secretion and increase in virulence is that this may indicate that LSMMG is decreasing the expression of surface components that are recognized by the host immune system, as discussed above. Again, this explanation would assume that a sufficient amount of surface-exposed type III secretion machinery is available to perform its virulence-related functions. Experiments to help clarify some of these explanations are currently underway in our laboratory.
We do not see an induction of known virulence genes that could be responsible for the LSMMG-induced increase in Salmonella virulence. Likewise, none of the LSMMG up-regulated genes display any homology to known functions that have an obvious role in virulence, with the possible exception of the sthE gene that codes for a putative fimbrial subunit. This raises the exciting possibility that LSMMG is altering Salmonella virulence by a previously uncharacterized mechanism that could involve novel virulence functions. Another possibility is that the increase in Salmonella virulence due to LSMMG is the result of contributions of multiple genes of different functions that are regulated as part of the reprogramming of Salmonella under LSMMG. The additive result of this global change in multiple functions may be that Salmonella is better poised to initiate infection. In this case, the up-regulation of one or two classic virulence functions (previously characterized or unknown) may not explain the increase in virulence. Our results in this regard are remarkably similar to those found in a recent study demonstrating that Vibrio cholera freshly isolated from contaminated human stools is significantly more infectious in mice than are laboratory-grown strains (23). Microarray analysis of the transcriptional profiles of both strains revealed a number of genes to be differentially regulated in the stool strains, but not one previously characterized virulence gene of the ToxR/TcpP/ToxT regulon that is essential for infection in humans and mice was detected. These findings help to underscore the point that the interplay among many genes of different functional groups may have just as important a role in virulence as the expression of classic virulence factors. A third possible explanation for our lack of detectable LSMMG-induced Salmonella virulence genes is that certain known virulence functions could actually be up-regulated by LSMMG, but the microarray analysis is unable to detect these changes. The unraveling of the LSMMG signaling pathway in Salmonella will allow us to understand how cells respond to low-shear growth signals present in vivo and how this impacts multiple aspects of cellular physiology including bacterial virulence, gene expression, and cellular metabolism.
Supplementary Material
Acknowledgments
We thank Laurel Stewart for invaluable help with the genespring analysis software, Bill Halford for advice on RT-PCR, Roy Curtiss III for generously providing strain χ4659, and Brian Morrow for critical review of the manuscript. This work was supported by the National Aeronautics and Space Administration (NASA) Ames Grant NAG 2-1378, and by the Louisiana Board of Regents Support Foundation (LEQSF-1999-02-RD-A-41). S.P. and M.M. are supported by National Institutes of Health Grant R01 AI34829-14. T.H. and P.A. are supported by NASA Cooperative Agreement NCC2-1177.
Abbreviations
- LSMMG
low-shear modeled microgravity
- HARV
high-aspect-ratio rotating wall vessel bioreactor
- LPS
lipopolysaccharide
- Fur
ferric uptake regulator
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Freed L E, Langer R, Martin I, Pellis N R, Vunjak-Novakovic G. Proc Natl Acad Sci USA. 1997;94:13885–13890. doi: 10.1073/pnas.94.25.13885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Freed L E, Vunjak-Novakovic G. In Vitro Cell Dev Biol Anim. 1997;33:381–385. doi: 10.1007/s11626-997-0009-2. [DOI] [PubMed] [Google Scholar]
- 3.Hammond T G, Lewis F C, Goodwin T G, Lennehan R M, Wolf D A, Hire K P, Campbell W C, Benes E, O'Reilly K C, Globus R K, Kaysen J H. Nat Med. 1999;4:359. doi: 10.1038/7331. [DOI] [PubMed] [Google Scholar]
- 4.Nickerson C A, Goodwin T J, Terlonge J, Ott C M, Buchanan K L, Uicker W B, Emami K, Cedor C L, Ramamurthy R, Clarke M S, et al. Infect Immun. 2001;69:7106–7120. doi: 10.1128/IAI.69.11.7106-7120.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Unsworth B R, Lelkes P I. Nat Med. 1998;4:901–907. doi: 10.1038/nm0898-901. [DOI] [PubMed] [Google Scholar]
- 6.Goodwin T J, Prewett T L, Wolf D A, Spaulding G F. J Cell Biochem. 1993;51:301–311. doi: 10.1002/jcb.240510309. [DOI] [PubMed] [Google Scholar]
- 7.Guo P, Weinstein A M, Weinbaum S. Am J Physiol. 2000;279:F698–F712. doi: 10.1152/ajprenal.2000.279.4.F698. [DOI] [PubMed] [Google Scholar]
- 8.Stock U A, Vacanti J P. Tissue Eng. 2001;7:1–7. doi: 10.1089/107632701300003241. [DOI] [PubMed] [Google Scholar]
- 9.Nickerson C A, Ott C M, Mister S J, Morrow B J, Burns-Keliher L, Pierson D L. Infect Immun. 2000;68:3147–3152. doi: 10.1128/iai.68.6.3147-3152.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wilson, J., Ott, C. M., Ramamurthy, R., Porwollik, S., McClelland, M., Pierson, D. L. & Nickerson, C. A. (2002) Appl. Environ. Microbiol., in press. [DOI] [PMC free article] [PubMed]
- 11.McClelland M, Sanderson K E, Spieth J, Clifton S W, Latreille P, Courtney L, Porwollik S, Ali J, Dante M, Du F, et al. Nature. 2001;413:852–856. doi: 10.1038/35101614. [DOI] [PubMed] [Google Scholar]
- 12.Gulig P A, Curtiss R., III Infect Immun. 1987;55:2891–2901. doi: 10.1128/iai.55.12.2891-2901.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lennox E S. Virology. 1955;1:190–206. doi: 10.1016/0042-6822(55)90016-7. [DOI] [PubMed] [Google Scholar]
- 14.Porwollik S, Wong R, Sims S, Schaaper R, Demarini D, McClelland M. Mutat Res. 2001;483:1–11. doi: 10.1016/s0027-5107(01)00239-1. [DOI] [PubMed] [Google Scholar]
- 15.Hitchcock P J, Brown T M. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai C M, Frasch C E. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- 17.Escolar L, Perez-Martin J, de Lorenzo V. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hall H K, Foster J W. J Bacteriol. 1996;178:5683–5691. doi: 10.1128/jb.178.19.5683-5691.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hantke K. Mol Gen Genet. 1987;210:135–139. doi: 10.1007/BF00337769. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-del Portillo F, Foster J W, Finlay B B. Infect Immun. 1993;61:4489–4492. doi: 10.1128/iai.61.10.4489-4492.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Riesenberg-Wilmes M R, Bearson B, Foster J W, Curtiss R., III Infect Immun. 1996;64:1085–1092. doi: 10.1128/iai.64.4.1085-1092.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorman C J. Microbiology. 1995;141:1271–1280. doi: 10.1099/13500872-141-6-1271. [DOI] [PubMed] [Google Scholar]
- 23.Merrell D S, Butler S M, Qadri F, Dolganov N A, Alam A, Cohen M B, Calderwood S B, Schoolnik G K, Camilli A. Nature. 2002;417:642–645. doi: 10.1038/nature00778. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.