Abstract
Highly pathogenic simian immunodeficiency virus/HIV chimeric viruses (SHIVs) cause extremely rapid, irreversible, and systemic depletions of CD4+ T lymphocytes in inoculated rhesus monkeys. In the absence of this T cell subset, virus production can be sustained for several months by tissue macrophage. During independent infections of seven animals with uncloned virus stocks, SHIV variants emerged bearing amino acid deletions that affected specific residues of the gp120 V2 loop. Some of these macrophage-phase SHIVs replicated to high levels in alveolar macrophage.
The capacity to infect macrophage is a property shared by all lentiviruses. Visna virus principally targets macrophage in the brains and lungs of infected sheep (1). The more promiscuous feline immunodeficiency virus infects both T and B lymphocytes in addition to macrophage (2). Although primate lentiviruses, such as HIV-1 and SIV, are recognized for their ability to infect and deplete CD4+ T lymphocytes, their capacity to infect macrophage was appreciated early in the AIDS epidemic when virus-producing microglia were detected in the brains of seropositive individuals (3). Most current knowledge concerning HIV-1 replication in macrophage derives from blood monocyte-derived macrophage (MDM), induced to differentiate by adherence to tissue-culture flasks or in the presence of specific cytokines. Virus-infected MDM have been reported to be refractory to the cytopathic effects of HIV-1 and resistant to the antiviral effects of reverse transcriptase and protease inhibitors (4, 5).
HIV-1-infected macrophage have been identified during all phases of the in vivo infection, becoming a more prolific source of progeny virions when associated with opportunistic infections (6). Although very little is known about the dynamics of virus production and spread within tissue macrophage, considerable effort has been expended to identify HIV-1 determinants conferring tropism for macrophage in the CNS and/or specific tissue compartments (7, 8).
We have reported (9–11) that highly pathogenic simian immunodeficiency virus/HIV chimeric viruses (SHIVs) can induce a rapid, systemic, and irreversible depletion of CD4+ T cells in rhesus monkeys within 2–4 weeks of i.v. inoculation. Infected animals survive an additional 4–6 months, and high levels of viremia are sustained in the absence of CD4+ T lymphocytes by macrophage in lymphoid tissues and the gastrointestinal tract (12). In the present study, we have investigated whether specific changes in the viral genome accompany the transition from the T cell to the macrophage phase of SHIV-infected macaques. By using two different, but related, uncloned virus stocks to inoculate seven rhesus monkeys, we observed specific amino acid changes (deletions and substitutions) affecting the V2 region of gp120 during independent in vivo infections. A subset of the late-phase SHIV variants replicated to high titers in cultured alveolar macrophage (AM).
Materials and Methods
Virus, Animal Inoculations, and Virus Load Measurements.
The tissue culture-derived SHIVDH12R stock has been described (10). Animal inoculations, lymphocyte subset analysis, and plasma viral load determinations were performed as reported (9).
Long env Gene PCR.
Viral RNA extraction from plasma and reverse transcription was conducted as described with minor modifications (13) by using 9493(−) 5′-GCGAGTATCCATCTTCCACCTCTC-3′ as cDNA primer. A 3-kbp fragment, encompassing the entire HIV-1DH12 Env coding region, was PCR-amplified as described (10) by using the following set of nested primers: 6428(+) 5′-GAGCCAGTAGATCCTAGACTAGAGCCC-3′ and 9493(−) as the outer primers, and the previously designed primers 6317(+) and 9190(−) (10) as the inner primers.
V2 Length Analysis.
A 181-bp fragment, encompassing the V2 region of gp120, was amplified by nested PCR as described with minor modifications (14). One primer in the second round of PCR was labeled with a fluorophore, 6-carboxy-fluorescein (6-FAM), at the 5′ end. The fluorescently labeled PCR-amplified products were separated on an ABI 377 DNA sequencer (Applied Biosystems) and sized with the GENESCAN 3.1 software (Applied Biosystems). Amino acid length of the V2-loop was calculated by subtracting 61 bp, representing the amplified constant regions outside of the V2-loop, from the measured nucleotide fragment length obtained at gel-run analysis.
Recombinant Viruses.
The specific V2 changes were introduced into pSHIVDH12R-CL-7 (R. Sadjadpour, T. Theodore, T.I., and M.A.M., unpublished work) by site-directed mutagenesis by using the QuickChange protocol (Stratagene) as described (15). Virus stocks were prepared by transfecting respective plasmids into RD cells as reported (15) with minor modifications.
Virus Replication in Monkey Peripheral Blood Mononuclear Cell (PBMC) and AM.
Rhesus macaque PBMC, prepared from eight different animals, were suspended in RPMI medium 1640 supplemented with 10% (vol//vol) FBS and stimulated for 1 day with 25 μg/ml Con A (Amersham Pharmacia) followed by an additional 2-day cultivation with 20 units per ml of IL-2 (Roche Molecular Biochemicals). The cells were inoculated by spinoculation (16) for 60 min using 4.5 × 10−6 ng of p27 Gag per cell. Supernatants were collected daily, and p27 Gag production was measured by Retro-Tek p27 antigen ELISA kit (ZeptoMetrox Corporation, Buffalo, NY). AM were collected from uninfected rhesus macaques by bronchoalveolar lavage and cultivated as described (17). AM (≈4 × 105) were resuspended in Dulbecco's minimal essential medium supplemented with 10% human serum Type AB and 5% (vol/vol) FBS (400 μl) and plated in LabTek II glass chamber slides (Nalge Nunc International, Naperville, IL). After medium changes on days 1 and 2, the adherent cells were immediately infected with virus (normalized either by infectivity [8 × 103 TCID50] or reverse transcriptase (RT) activity [1 × 107 32P cpm]), as indicated in the text. In each case, the infected cultures were monitored for up to 25 days, with complete medium changes every other day. Progeny virus production was assessed by the RT activity in the culture supernatants, as described (10).
Results
Uncloned SHIV Stocks Consistently Induce High Virus Loads and Rapid Irreversible Depletions of CD4+ T Lymphocytes.
Two different uncloned highly pathogenic SHIV stocks were used to ascertain whether viral envelope glycoprotein changes accompanied the transition from the T cell phase of in vivo infections (during which rapid and irreversible CD4+ T lymphocyte depletion occurs) to the late stage of infection (when virus production is sustained by tissue macrophage; ref. 12). One of these viruses, SHIVDH12R, was isolated at the time of necropsy (week 68) from rhesus monkey 565Z, treated with an anti-human CD8 monoclonal antibody at the time of its primary infection with nonpathogenic SHIVDH12 (10). Virus isolated at week 52 from animal 565Z also induced an irreversible and extremely rapid depletion of CD4+ T lymphocytes after i.v. inoculation of rhesus monkey PS1 and was designated SHIVDH12R-PS1 (R.W., T.I., Y.E., and M.A.M., unpublished work). A tissue-culture stock of SHIVDH12R-PS1 was prepared by co-cultivating a mixture of PBMC and a lymphoid tissue suspension, recovered at the time of necropsy of macaque PS1, with mitogen-activated PBMC from a naïve rhesus monkey.
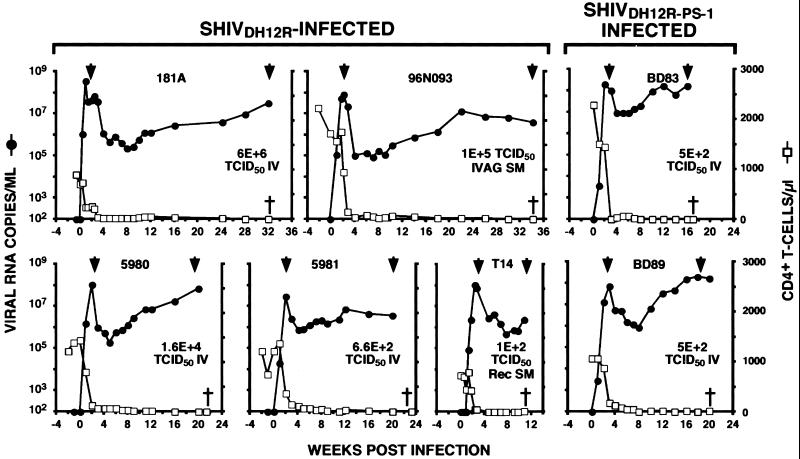
In the experiments to be described, five rhesus monkeys were inoculated with SHIVDH12R, and two received SHIVDH12R-PS1. All seven macaques exhibited a rapid and irreversible loss of CD4+ T lymphocytes within 3 weeks of infection (Fig. 1). The total number of CD4+ T cells declined to 10–70 cells per μl of blood within 3 to 5 weeks of infection (during the T cell phase of infection) and continued to fall to <10 cells per μl of blood over the next 1–8 months (during the macrophage phase of infection; ref. 12). In these seven SHIVDH12R/SHIVDH12R-PS1-infected animals, plasma viremia peaked at 107-108 viral RNA copies per ml of plasma by 2 weeks after infection, fell 50- to 1,000-fold over the next 3–5 weeks, and then gradually increased over the next 1–8 months (Fig. 1). Despite the absence of significant numbers of circulating CD4+ T cells, the SHIVDH12R/SHIVDH12R-PS1-infected monkeys continued to produce high plasma virus loads (105–108 viral RNA copies per ml) until they were euthanized because of their deteriorating clinical state.
Figure 1.
Plasma viral load and CD4+ T cell counts. Rhesus monkeys were inoculated with SHIVDH12R or SHIVDH12R-PS1 through the indicated routes: IV, i.v. injection; IVAG SM, intravaginal submucosal injection; Rec SM, rectal submucosal injection. †, when animals were euthanized; ▾, when plasma samples were collected for RT-PCR and sequencing analyses.
Common gp120 V2 Changes Emerge During the Transition from the T Cell to the Macrophage Phase of Infection in Infected Rhesus Macaques.
To assess changes introduced into the SHIV-envelope glycoprotein during in vivo infections, 3-kbp segments were PCR-amplified from the starting virus stocks and the plasma viral RNAs at week 2 (T cell phase) or following CD4+ T cell depletion (macrophage phase). Compared with the gp120 present in the parental nonpathogenic, molecularly cloned SHIVDH12 (18), sequence analyses (21 independent PCR clones) revealed that the starting, uncloned SHIVDH12R virus stock was genetically heterogeneous, especially in the V2 region of gp120 (Fig. 2 Top). However, by week 2 after infection, only two classes of gp120 V2 variants were detected in the five SHIVDH12R infected monkeys (7–10 independent RT-PCR clones of plasma viral RNA for each). One population had D185G plus R186K changes, and the second had I165R, R186M, N187S substitutions (Fig. 2 Middle). The first variant comprised 25% of the original SHIVDH12R stock. The overall homogenization of the V2 quasi-species in virus circulating during the T cell phase of infection plus the presence of similar V2 variants in both the virus inoculum and plasma virus 2 weeks after infection suggested that they arose as a result of outgrowth/selection of preexisting virions in the uncloned SHIVDH12R stock.
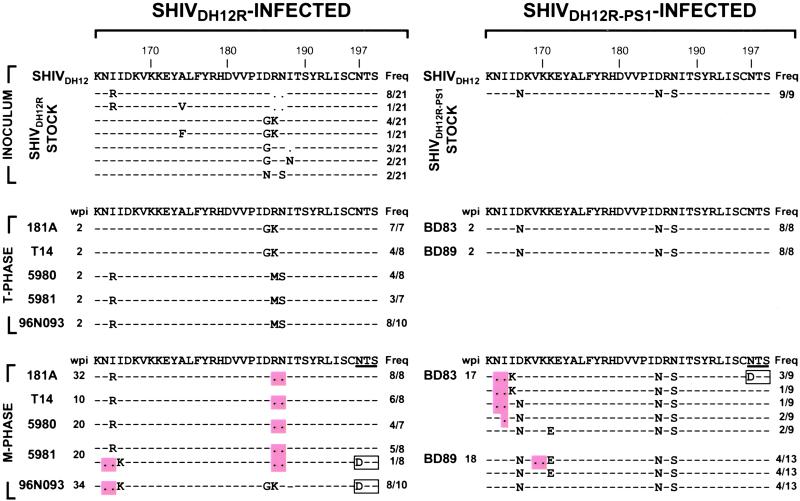
Figure 2.
gp120 sequences from the V2 region of SHIVDH12R/SHIVDH12R-PS1-infected monkeys. The deduced amino sequences of V2 regions in the inocula (INOCULUM) and the plasma viral RNA during the T cell phase (T-PHASE) and the macrophage phase (M-PHASE) of infection were aligned with V2 sequences present in the parental SHIVDH12 at the top. The predominant variants detected during the macrophage phase are shown. —, identical sequence; ●, gap; wpi, weeks postinfection; Freq, frequency of env PCR clone. Amino acid deletions are highlighted; an amino acid change resulting in a loss of an N-glycosylation site is boxed.
A similar RT-PCR analysis of plasma viral RNA was conducted on samples recovered from the five SHIVDH12R-infected rhesus monkeys during the macrophage phase of infection. Remarkably, the only consistent changes observed within the entire gp120-coding region were deletions mapping to the V2 region (Fig. 2 Bottom). The major gp120 variant contained a double amino acid deletion affecting residues 186 and 187 that accompanied an I165R substitution (I165R+Δ186–187). This variant arose independently in four of five SHIVDH12R-infected monkeys (181A, T14, 5980, and 5981) and was present in relatively high frequencies (57–100% of all of the macrophage phase-derived gp120 sequences detected in each animal). The I165R+Δ186–187 variant comprised approximately one-third of the starting uncloned SHIVDH12R stock (Fig. 2 Top), yet was never amplified during the T cell phase. Nonetheless, this gp120 variant re-emerged and became predominant during the macrophage phase of infection. A second variant, observed during the macrophage phase of infection, contained a different double amino acid deletion (affecting residues 164 and 165) and was associated with I166K, D185G, and R186K amino acid substitutions (Δ164–165+I166K+D185G+R186K). A low-frequency variant also was detected during the macrophage phase in monkey 5981, which contained both double amino acid deletions (affecting residues 164, 165, 186, and 187) and was associated with the I166K amino acid substitution (Δ164–165+I166K+Δ186–187) of the gp120 V2. The V2 deletions at residues 164 and 165 in these latter variants were invariably accompanied by a loss of the N-glycosylation site (gs) at residue 197. Random substitutions were observed in the remainder of gp120; none were consistently detected during the macrophage phase of infection.
Nine independent clones were amplified from the uncloned SHIVDH12R-PS1 stock used to inoculate two of the rhesus monkeys. In contrast to the SHIVDH12R inoculum, the SHIVDH12R-PS1 stock was quite homogeneous, containing the D167N+D185N+N187S changes compared with the nonpathogenic SHIVDH12 (Fig. 2 Top). This species was also the only gp120 species detected at the peak of the T cell phase of infection in these two infected macaques (Fig. 2 Middle). Nonetheless, during the transition to the macrophage phase of infection, V2 sequences did change markedly and included 1–2 amino acids deletions at two locations within V2 (residues 164–165 or at 169–170). In macaque BD83, 70% of the independent PCR clones contained either a two-amino acid deletion (Δ164–165) or a one-amino acid deletion (Δ165); some were present in association with a I166K change and a loss of a gs at residue 197. In animal BD89, ≈30% of the variants present during the macrophage phase contained V2 deletions at a different location (at residues 169–170) in association with K171E change (Δ169–170+K171E). The K171E change also was detected alone in 30% of SHIV variants derived circulating in this animal. Similar to the env sequences derived from SHIVDH12R infected monkeys, no other deletions were found in the remainder of the gp120 coding sequences. However, in contrast to the SHIVDH12R-infected macaques, none of the V2 changes present in the macrophage phase-derived variants were detected in either the SHIVDH12R-PS1 virus stock or during the T cell phase of the SHIVDH12-PS1 infection, implying that they arose de novo during the in vivo infections rather than as an outgrowth of a preexisting quasi-species. Taken together, these results suggest that variants containing V2 deletions emerge during the transition from the T cell phase to the macrophage phase of infection in SHIVDH12R/SHIVDH12R-PS1-infected monkeys. These deletions were invariably present at three specific locations within the V2 region of gp120: at residues 164–165, 169–170, and 186–187.
The Use of a Fluorescent-Amplified Fragment-Length Polymorphism Assay to Detect Amino Acid Deletions in the gp120 V2 Region of Plasma Virus Populations.
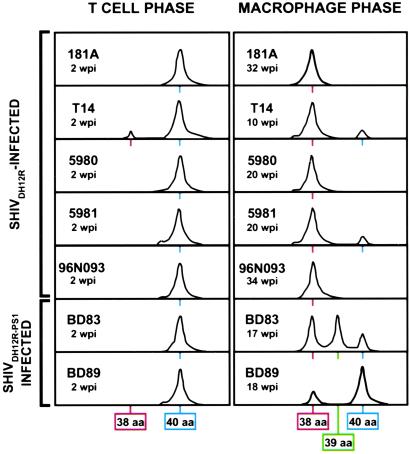
To confirm independently that SHIVDH12R/SHIVDH12R-PS1 variants containing V2 deletions were being generated during the macrophage phase of pathogenic SHIV infections, a fluorescent-amplified fragment-length polymorphism assay, capable of sampling the entire virus population and detecting minor gp120 variants present at frequencies ≥5%, was used. Fig. 3 depicts the gp120 V2 region-length polymorphism analysis of virus populations present during the T cell and macrophage phases of infection in SHIVDH12R and SHIVDH12R-PS1 inoculated monkeys. A single peak, corresponding to the 40-aa V2 loop, was detected for plasma virus from six of the seven SHIV-infected macaques during the T cell phase of infection, indicating that virtually all of the virus circulating in these animals 2 weeks after inoculation carried a full-length V2-loop.
Figure 3.
The length profiles of the V2 region of gp120. The V2 length polymorphism was evaluated for plasma viruses circulating during the T cell and the macrophage phase of infection. The amino acid lengths of the V2 loop are indicated at the bottom.
In contrast, the SHIVs circulating in the blood during the macrophage phase carried gp120s with deleted and/or highly divergent V2 loop lengths (Fig. 3). In three of the monkeys (181A, 5980, and 96N093), the only variant detected contained a shortened 38-aa V2 region. Two populations of gp120 variants were present in animals T14 and 5981: a single predominant species and a minor component with V2 loop length corresponding to 38 and 40 aa, respectively. A mixture of three virus populations, with V2 regions of 38, 39, and 40 aa, respectively, was observed in animal BD83. Taken together, these data confirm the emergence of SHIVDH12R variants containing V2 loop deletions during the transition from the T cell phase to the macrophage phase of infection.
Construction of Full-Length SHIV Molecular Clones Containing Variant gp120 V2 Regions.
We have recently obtained an infectious molecular clone, designated pSHIVDH12R-CL-7, which directs production of virus that consistently induces rapid, irreversible depletion of CD4+ T lymphocytes after i.v. inoculation of rhesus monkeys (R. Sadjadpour, T. Theodore, T.I., and M.A.M., unpublished work). pSHIVDH12R-CL-7 was isolated from rhesus macaque PBMC, infected with SHIVDH12R-PS1 by using a λ-phage vector to clone unintegrated viral DNA. Not unexpectedly, pSHIVDH12R-CL-7 contains the “signature” SHIVDH12R-PS1 V2 region (shown at the top of Fig. 4).
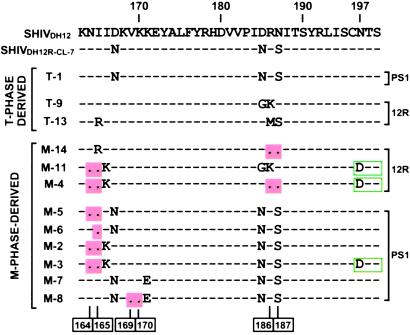
Figure 4.
SHIVDH12R envelope variants. Site-specific mutagenesis was used to generate SHIVs bearing V2 changes detected in infected monkeys. The positions of deleted amino acids are indicated at the bottom. Amino acid changes resulting in a loss of the N-glycosylation site at residue 197 are boxed. 12R, SHIVDH12R-infected; PS1, SHIVDH12R-PS1-infected animal-derived.
The two V2 loop variants detected during the T cell phase of the SHIVDH12R infection were introduced into pSHIVDH12R-CL-7 by site-directed mutagenesis. The single variant identified at week 2 after SHIVDH12R-PS1 infection possessed a V2 region identical to SHIVDH12R-CL-7. These full-length viral DNA constructs are designated T-1, T-9, and T-13 in Fig. 4. The nine macrophage phase gp120 V2 variant regions were similarly introduced into pSHIVDH12R-CL-7. Three of these (M-14, M-11, and M-4) were identified in SHIVDH12R-infected animals, and six (M-7, M-8, M-5, M-6, M-2, and M3) were present in macaques inoculated with SHIVDH12R-PS1 (Fig. 4). A majority of these macrophage-phase SHIV clones contained V2 regions with 1, 2, or 4 aa deletions.
Replicative Properties of SHIVs Containing gp120 V2 Variants in Rhesus Monkey PBMCs.
The three SHIVs (T-1, T-9, and T-13) bearing T cell phase V2 loop changes replicated robustly and to similar levels (207–346 ng of p27 per ml on day 5 or 6 post infection) in macaque PBMC (Fig. 5). In contrast, the nine SHIVs carrying macrophage phase gp120 V2 variants exhibited variable replicative phenotypes. The infection kinetics and peak virus production of three of these (M-14, M-7, and M-8) were comparable to those observed with the T cell phase gp120 V2 viruses (206–281 ng of p27 per ml). Of the remaining six macrophage phase-derived variants, two (M-11 and M-4) replicated with markedly delayed infection kinetics and produced relatively low amounts of progeny virions (41 and 54 ng of p27 per ml on day 8 pi). In general, double amino acid deletions affecting residues 169–170 or 186–187 of gp120 had no deleterious effect on infectivity, whereas a similar deletion at positions 164–165 suppressed progeny virion production to varying extents. The loss of the potential gs at residues 197–199 had no demonstrable effect on replicative capacity in macaque PBMC (compare M-2 and M-3 in Fig. 5), but the presence of two sets of double amino acid deletions in V2 (variant M-4) severely compromised infectivity.
Figure 5.
Replication of SHIVDH12R V2 variants in rhesus monkey PBMC. Representative results of two independent experiments are shown. V2 changes introduced in each SHIVDH12R variant are indicated. T-1, T-9, and T-13, T cell phase-derived; M-14, M-11, M-4, M-5, M-6, M-2, M-3, M-7, and M-8, macrophage phase-derived.
Some Late Stage gp120 V2 Variants Exhibit Macrophage Tropism.
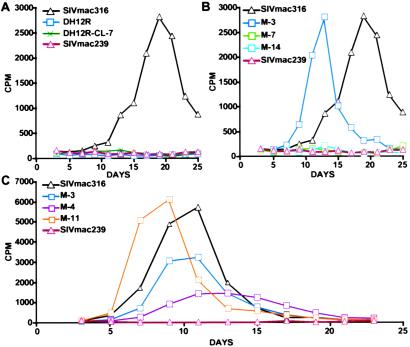
The possibility that the SHIV variants emerging during the macrophage phase of infection had acquired tropism for tissue macrophage was examined by using AM that was obtained by bronchoalveolar lavage. As shown in Fig. 6A, SIVmac316, previously propagated and titered in rhesus PBMC, readily replicated in AM, reaching peak levels of virus production on day 19 pi. In contrast, SIVmac239 and the two highly pathogenic SHIV stocks, SHIVDH12R and SHIVDH12R-CL-7, failed to produce measurable amounts of progeny virions.
Figure 6.
Replication of SHIVDH12R V2 variants in rhesus monkey AM. Virus inocula (8 × 103 TCID50), prepared in rhesus PBMC, were used to infect rhesus macaque AM (A and B). (C) Inocula used (normalized by RT activity [1 × 107 32P cpm]) was harvested from the supernatants of transfected HeLa cells. Virus replication was assessed by RT activity released into the culture medium.
In the initial experiment to assess the tropic properties of macrophage-phase SHIVs, three representative variants (M-3, M-7, and M-14), exhibiting robust replicative properties in rhesus PBMC and containing deletions/insertions frequently observed in late stage plasma virus, were examined (Fig. 6B). Of the three, only M-3 was able to replicate in AM. In the experiment shown (normalized for input infectivity), SHIV M-3 replicated with faster kinetics and to similar levels as the macrophage tropic SIVmac316. As shown in Fig. 4, the frequently observed double amino acid deletions affecting residues 164/165 of V2 distinguishes M-3 from both M-7 and M-14. To assess whether this V2 deletion was associated with macrophage tropism, two other late-stage variants (M-4 and M-11), bearing the same deletion, were evaluated in the cultured AM. In this case, untitered supernatants from transfected HeLa cells were used to infect AM after normalizing for RT activity (1 × 107 32P RT cpm). As shown in Fig. 6C, three of the late-stage SHIVs (M-3, M-4, and M-11) carrying the 164/165 deletion replicated in AM. These results indicate that some of the SHIV variants, emerging in the absence of CD4+ T cells, acquired the capacity to infect tissue macrophage.
Discussion
The selective factors responsible for the emergence of viral variants containing deleted gp120 V2 regions during independent SHIVDH12R infections of rhesus monkeys are presently unknown. Because tissue macrophage represents the only remaining cell target capable of sustaining the virus infection in animals depleted of CD4+ T lymphocytes, the capacity to enter and replicate efficiently in this cell lineage is very likely strongly selected for. The V1 and V2 variable loops of HIV-1 gp120 (also present on the SHIVDH12R gp120) are exposed on the surface of the envelope glycoprotein complex and are thought to partially occlude the binding sites for CD4 and coreceptor (19). During virus entry, binding of HIV-1 to CD4 induces a conformational change in V1/V2 and a repositioning of the V1/V2 stem, an alteration that contributes to the formation of the bridging sheet, a gp120 element containing high-affinity binding sites for the CCR5 coreceptor (19–23). Taken at face value, the reduction in the size of the V2 loop observed in macrophage phase SHIVs might be expected to result in greater exposure of the CD4 and coreceptor binding sites.
It is now appreciated that cell-type-specific expression of the CD4 receptor and/or chemokine coreceptor can be influenced by the state of differentiation or activation and can significantly affect the apparent tropism of a primate lentivirus (24–27). For example, compared with activated human PBMC, the levels of cell surface CD4, CCR5, and CXCR4 have been reported to be very low in human AM (28). Similar studies have shown that rhesus monkey MDM and AM express extremely low to undetectable levels of CD4 and can only support the replication of SIV strains containing specific changes in gp120 (29–31). For SIVmac239 and its macrophage tropic derivative, SIVmac316, this change in tropism is not due to altered use of the CCR5 coreceptor but to the capacity of SIVmac316, not SIVmac239, to infect MDM with low densities of surface CD4. It also has been recognized that macrophage-tropic SIV strains exhibit varying degrees of CD4 independence, being able to infect CD4−, CCR5+ cells (32, 33). In general, such SIV strains also are more sensitive to neutralizing antibodies and are less pathogenic than T cell tropic SIVs (34).
The acquisition of macrophage tropism by some of the late-stage SHIV variants comes as no surprise, given the paucity of CD4+ T cell targets systemically (11). Unlike the highly pathogenic parental viruses (SHIVDH12R and SHIVDH12R-PS1), these variants exhibited replicative properties similar to SIVmac316, a prototypic macrophage-tropic SIV strain (30). In addition to the double 164-/165-aa deletion in V2, macrophage-tropic SHIV variants M-3, M-4, and M-11 have all lost the glycosylation site at position 197 (Fig. 4). It is of interest that elimination of this same gs conferred CD4 independence in an HIV-1 strain-specific fashion by augmenting both the binding and entry of HIV-1ADA into cells lacking CD4 (35, 36). The contribution of the two different V2 alterations on the capacity to infect AM can be evaluated by testing additional late-stage SHIVs such as M-2 and M-5 (see Fig. 4). It also is worth noting that SHIV variant M-11, which produced quite low amounts of progeny virus in rhesus PBMC (Fig. 5), exhibited robust replication kinetics in AM (Fig. 6).
Very little is currently known about the dynamics of HIV-1 production and spread within tissue macrophage in infected individuals. It has been long debated whether sequestration of HIV-1 occurs within specific body compartments and whether virus strains, within the CNS for example, evolve independently from virus present in other tissues (7, 8). Compartmentalization of HIV-1 has been exceedingly difficult to demonstrate in infected persons for several reasons, including intrapatient viral genetic heterogeneity and the logistical problems attending the longitudinal sampling of relevant tissues. In the pathogenic SHIV/macaque system described, the emergence of viruses bearing specific V2 alterations after independent in vivo infections with uncloned virus stocks would appear to be biologically relevant. The properties of some of these macrophage-tropic SHIV variants, particularly those recovered directly from tissue, should provide important information pertaining to the compartmentalization question as well as to the immunodeficiency induced by the primate lentiviruses.
Acknowledgments
We thank A. Buckler-White and R. J. Plishka for the monkey plasma viral load assessments, Dusty Rhodes for the bronchoalveolar lavage protocol, and Ronald C. Desrosiers and Robert E. Means for helpful suggestions on AM culture. A part of this work was supported by the National Institute of Allergy and Infectious Diseases under contract N01-CO-12400 with Science Applications International Corporation–Frederick, Inc.
Abbreviations
- SHIV
simian immunodeficiency virus/HIV chimeric virus
- AM
alveolar macrophage
- PBMC
peripheral blood mononuclear cell
- RT
reverse transcriptase
References
- 1.Gendelman H E, Narayan O, Molineaux S, Clements J E, Ghotbi Z. Proc Natl Acad Sci USA. 1985;82:7086–7090. doi: 10.1073/pnas.82.20.7086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brown W C, Bissey L, Logan K S, Pedersen N C, Elder J H, Collisson E W. J Virol. 1991;65:3359–3364. doi: 10.1128/jvi.65.6.3359-3364.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koenig S, Gendelman H E, Orenstein J M, Dal Canto M C, Pezeshkpour G H, Yungbluth M, Janotta F, Aksamit A, Martin M A, Fauci A S. Science. 1986;233:1089–1093. doi: 10.1126/science.3016903. [DOI] [PubMed] [Google Scholar]
- 4.Meltzer M S, Nakamura M, Hansen B D, Turpin J A, Kalter D C, Gendelman H E. AIDS Res Hum Retroviruses. 1990;6:967–971. doi: 10.1089/aid.1990.6.967. [DOI] [PubMed] [Google Scholar]
- 5.Perno C F, Newcomb F M, Davis D A, Aquaro S, Humphrey R W, Calio R, Yarchoan R. J Infect Dis. 1998;178:413–422. doi: 10.1086/515642. [DOI] [PubMed] [Google Scholar]
- 6.Orenstein J M, Fox C, Wahl S M. Science. 1997;276:1857–1861. doi: 10.1126/science.276.5320.1857. [DOI] [PubMed] [Google Scholar]
- 7.Korber B T, Kunstman K J, Patterson B K, Furtado M, McEvilly M M, Levy R, Wolinsky S M. J Virol. 1994;68:7467–7481. doi: 10.1128/jvi.68.11.7467-7481.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong J K, Ignacio C C, Torriani F, Havlir D, Fitch N J, Richman D D. J Virol. 1997;71:2059–2071. doi: 10.1128/jvi.71.3.2059-2071.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endo Y, Igarashi T, Nishimura Y, Buckler C, Buckler-White A, Plishka R, Dimitrov D S, Martin M A. J Virol. 2000;74:6935–6945. doi: 10.1128/jvi.74.15.6935-6945.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Igarashi T, Endo Y, Englund G, Sadjadpour R, Matano T, Buckler C, Buckler-White A, Plishka R, Theodore T, Shibata R, Martin M. Proc Natl Acad Sci USA. 1999;96:14049–14054. doi: 10.1073/pnas.96.24.14049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Igarashi T, Brown C R, Byrum R A, Nishimura Y, Endo Y, Plishka R J, Buckler C, Buckler-White A, Miller G, Hirsch V M, Martin M A. J Virol. 2002;76:379–391. doi: 10.1128/JVI.76.1.379-391.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Igarashi T, Brown C R, Endo Y, Buckler-White A, Plishka R, Bischofberger N, Hirsch V, Martin M A. Proc Natl Acad Sci USA. 2001;98:658–663. doi: 10.1073/pnas.021551798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Imamichi H, Crandall K A, Natarajan V, Jiang M K, Dewar R L, Berg S, Gaddam A, Bosche M, Metcalf J A, Davey R T, Jr, Lane H C. J Infect Dis. 2001;183:36–50. doi: 10.1086/317641. [DOI] [PubMed] [Google Scholar]
- 14.Jansson M, Backstrom E, Scarlatti G, Bjorndal A, Matsuda S, Rossi P, Albert J, Wigzell H. AIDS Res Hum Retroviruses. 2001;17:1405–1414. doi: 10.1089/088922201753197079. [DOI] [PubMed] [Google Scholar]
- 15.Imamichi T, Berg S C, Imamichi H, Lopez J C, Metcalf J A, Falloon J, Lane H C. J Virol. 2000;74:10958–10964. doi: 10.1128/jvi.74.23.10958-10964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Doherty U, Swiggard W J, Malim M H. J Virol. 2000;74:10074–10080. doi: 10.1128/jvi.74.21.10074-10080.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Means R E, Matthews T, Hoxie J A, Malim M H, Kodama T, Desrosiers R C. J Virol. 2001;75:3903–3915. doi: 10.1128/JVI.75.8.3903-3915.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shibata R, Hoggan M D, Broscius C, Englund G, Theodore T S, Buckler-White A, Arthur L O, Israel Z, Schultz A, Lane H C, et al. J Virol. 1995;69:4453–4462. doi: 10.1128/jvi.69.7.4453-4462.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 22.Sattentau Q J, Moore J P, Vignaux F, Traincard F, Poignard P. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bleul C C, Wu L, Hoxie J A, Springer T A, Mackay C R. Proc Natl Acad Sci USA. 1997;94:1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Di Marzio P, Tse J, Landau N R. AIDS Res Hum Retroviruses. 1998;14:129–138. doi: 10.1089/aid.1998.14.129. [DOI] [PubMed] [Google Scholar]
- 26.Platt E J, Wehrly K, Kuhmann S E, Chesebro B, Kabat D. J Virol. 1998;72:2855–2864. doi: 10.1128/jvi.72.4.2855-2864.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yi Y, Rana S, Turner J D, Gaddis N, Collman R G. J Virol. 1998;72:772–777. doi: 10.1128/jvi.72.1.772-777.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Worgall S, Connor R, Kaner R J, Fenamore E, Sheridan K, Singh R, Crystal R G. J Virol. 1999;73:5865–5874. doi: 10.1128/jvi.73.7.5865-5874.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bannert N, Schenten D, Craig S, Sodroski J. J Virol. 2000;74:10984–10993. doi: 10.1128/jvi.74.23.10984-10993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mori K, Ringler D J, Kodama T, Desrosiers R C. J Virol. 1992;66:2067–2075. doi: 10.1128/jvi.66.4.2067-2075.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mori K, Rosenzweig M, Desrosiers R C. J Virol. 2000;74:10852–10859. doi: 10.1128/jvi.74.22.10852-10859.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edinger A L, Mankowski J L, Doranz B J, Margulies B J, Lee B, Rucker J, Sharron M, Hoffman T L, Berson J F, Zink M C, et al. Proc Natl Acad Sci USA. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edinger A L, Blanpain C, Kunstman K J, Wolinsky S M, Parmentier M, Doms R W. J Virol. 1999;73:4062–4073. doi: 10.1128/jvi.73.5.4062-4073.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puffer B A, Pohlmann S, Edinger A L, Carlin D, Sanchez M D, Reitter J, Watry D D, Fox H S, Desrosiers R C, Doms R W. J Virol. 2002;76:2595–2605. doi: 10.1128/JVI.76.6.2595-2605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney L J, Choe H, Sodroski J. J Virol. 1999;73:8120–8126. doi: 10.1128/jvi.73.10.8120-8126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kolchinsky P, Kiprilov E, Bartley P, Rubinstein R, Sodroski J. J Virol. 2001;75:3435–3443. doi: 10.1128/JVI.75.7.3435-3443.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]