Abstract
Subneutralizing concentrations of antibody may enhance virus infection by bringing the virus–antibody complex into contact with the cell surface Fc receptors; this interaction facilitates entry of virus into the cell and is referred to as antibody-dependent enhancement (ADE) of infection. Northern analysis of macrophage RNA demonstrated that ADE infection by the indigenous Australian alphavirus Ross River (RRV-ADE) ablated or diminished message for tumor necrosis factor α (TNF-α), nitric-oxide synthase 2 (NOS2), and IFN regulatory factor 1 (IRF-1), as well as for IFN-inducible protein 10 (IP-10) and IFN-β; the transcription of a control gene was unaffected. Additionally, electrophoretic mobility-shift assay (EMSA) studies showed that transcription factor IFN-α-activated factor (AAF), IFN-stimulated gene factor 3 (ISGF3), and nuclear factor-κB (NF-κB) complex formation in macrophage nuclear extracts were specifically suppressed post-RRV-ADE infection, emphasizing the capacity for ADE infections to compromise antiviral responses at the transcriptional level. The suppression of antiviral transcription factor complexes was shown to depend on replicating virus and was not simply a result of general antibody–Fc–receptor interaction. Although only a minority of cells (≈15%) were shown to be positive for RRV by immunostaining techniques post ADE, molecular (RT-PCR) analysis showed that unstained cells carried RRV-RNA, indicating a higher level of viral infectivity than previously suspected. Electron microscopy studies confirmed this observation. Furthermore, levels of cellular IL-10 protein were dramatically elevated in RRV-ADE cultures. This evidence demonstrates that RRV can potently disrupt the activation of specific antiviral pathways via ADE infection pathways, and may suggest a significant mechanism in the infection and pathogenesis of other ADE viruses.
Ross River virus (RRV) is a mosquito-borne alphavirus endemic to Australia and New Guinea that is responsible for outbreaks of epidemic polyarthritis (EPA) in the Australian community each year. The incidence of RRV infection, reflected by the consequent sequelae of RRV disease, including polyarthritis, appears to be increasing, with up to 7,000 cases reported annually in Australia (1). In 1979/1980, an explosive epidemic swept through several islands of the South Pacific, resulting in tens of thousands of disease cases (1). The predominant symptoms are arthritis/arthralgia, myalgia, rash, and lethargy, which in some patients can persist for extended periods (months to years). Synovial effusions taken from EPA patients predominantly contain monocytes, macrophages, T cells, and B cells, and recent studies in a mouse model of RRV disease have shown macrophages to be the cellular agent of disease, as well as the cell mediating severe muscle pathology (2).
In vitro enhancement of arbovirus infection by antibodies was originally demonstrated by Hawkes in the early 1960s (3). Antibody-dependent enhancement (ADE) of infection, which involves the entry of virus–antibody complexes into monocytic cells via Fc-γ-receptors (Fc-γRs), results in significantly enhanced virus titers and is recognized for several viruses, including influenza, dengue, and HIV (4–8). For dengue, ADE mechanisms associated with cross-reactive antibody responses to one of the four virus serotypes are considered a risk factor in the development of the life-threatening conditions dengue hemorrhagic fever and dengue shock syndrome post-secondary infection (4, 9).
Lipopolysaccharide (LPS) is the major component of the outer membrane of Gram-negative bacteria and induces innate immune responses and expression of cytokine genes, which include IL-1, IL-10, IFN-β, and tumor necrosis factor-α (TNF-α) (10). Interaction of LPS with its receptors on monocytic/macrophage cells results in the activation of a number of tyrosine and serine kinases, with consequent activation of nuclear transcription factors such as nuclear factor-κB (NF-κB), signal transducer and activator of transcription 1 (STAT-1), STAT-3, and Sp-1 (11). The activation of transcription factor STAT-1, NF-κB, and Sp-1 plays a major role in the transcriptional activation of type I IFNs (IFN-α/β), TNF-α, and IL-10, respectively (11). LPS can directly induce NF-κB activation whereas the activation of STAT-1 is mediated by LPS-induced type I IFN expression. LPS-induced type I IFN induced both IFN-stimulated gene factor 3 (ISGF3; STAT-1, Stat-2, and p48 heterodimers) and IFN-α activated factor (AAF; STAT-1 homodimers; ref. 11). AAF and ISGF3 complexes translocate to the nucleus and bind to IFN-γ-activated sequence elements and IFN-stimulated response elements, respectively. These different forms of STAT-1 complexes could cooperate with NF-κB to induce full transcriptional activation of many antiviral genes that are detrimental to the survival of the virus.
Recently, in vitro studies on RRV-ADE infection of macrophages have provided vital clues to the mechanism of enhanced virus replication post Fc-γR-mediated infection (12). RRV-ADE infection specifically ablated the production of both TNF-α and nitric oxide (NO) in response to LPS (12). The specificity of gene/protein ablation was determined via the analysis of multiple constitutively expressed housekeeping genes, as well as studies on total cellular de novo protein synthesis post-RRV-ADE infection. These studies found that housekeeping control gene expression was unaffected by infection and general de novo protein synthesis for infected cells was not significantly perturbed (12). The ablation of antiviral protein production by RRV-ADE infection, which was concomitant with significantly increased RRV titers, was found in LPS-treated macrophage cultures. In contrast, uninfected (control) LPS-treated macrophage cultures were found to have enhanced TNF-α and NO production (12). Additionally, for non-ADE-infected macrophages (RRV alone or RRV + control antibody), LPS treatment significantly restricted RRV growth (12). This evidence demonstrates that the ADE infection pathway allows RRV to establish an enhanced infection in macrophages under conditions of substantial inflammatory activity where the virus would not normally survive.
This study describes investigations focused on the activity of the downstream transcription factor protein complexes ISGF3, AAF, and NF-κB in macrophages post-RRV-ADE-infection. RRV-ADE infection was found to significantly suppress the activity of these transcription factor protein complexes in macrophages, with replicating virus found to be crucial to this outcome. To address whether this suppression correlated with levels of infectivity, we investigated the degree of RRV infectivity in macrophage cultures. It was found that immunofluorescent antibody (IFA) techniques did not detect the full extent of infection, with IFA negative cells shown to contain RRV-RNA (confirmed quantitatively by electron microscopy). Therefore, RRV infectivity post-ADE infection was greater than previously suggested (12). In addition, IL-10 was prominently expressed in RRV-ADE-infected macrophages compared with controls. Together, these findings explain the global nature of antiviral gene suppression in infected cells post-RRV-ADE.
Methods
Cells and Virus.
RAW 264.7 mouse macrophages (ATCC TIB-71) were maintained, cultured, LPS treated, and RRV infected exactly as described previously (12). Details can be found in additional text, which is published as supporting information on the PNAS web site, www.pnas.org.
In addition to RRV-ADE infection, the following control infections/treatments were performed: PBS + 1% FCS alone, RRV alone, anti-RRV antibody alone, RRV + mouse anti-Barmah Forest virus (BFV) antibodies (titer = 1/5,120, 10−3 dilution), opsinized zymosan (a gift from W. Cowden, John Curtin School of Medical Research, Canberra, Australia), and UV-irradiated RRV (5 min, 55 cm from a 30-W source; irradiation before incubating with anti-RRV antibody). All virus and control preparations were diluted in PBS + 1% FCS.
Northern Hybridization Analysis of Macrophage RNA.
RAW 264.7 cells were infected for 24 h, after which the cells were trypsinized from the wells and washed twice in calcium/magnesium-free PBS (TRACE, Melbourne). Total RNA was isolated from cells with RNAzol B (Biotech Laboratories, Houston, TX) as recommended by the manufacturer. Details can be found in additional text in the supporting information.
RT-PCR and ELISA for IL-10.
RT-PCR was performed to determine relative quantities of mRNA for IL-10 as described (13). Details can be found in the supporting information.
IL-10 protein concentrations from macrophage culture supernatants were determined by ELISA as described previously (14). Recombinant IL-10 used in the assay as a standard was obtained from PharMingen. The sensitivity of the IL-10 ELISA was 15 pg/ml.
Electrophoretic Mobility-Shift Assay (EMSA) Studies.
EMSAs were done to determine the activation of AAF, ISGF3, and NF-κB complexes in RRV-ADE-infected macrophages by previously described methods (15, 16). Details can be found in the supporting information.
Determination of RRV Infectivity by IFA.
RAW 264.7 cultures were infected with RRV exactly as described earlier. The percentage of infected cells was determined by IFA at 4, 7, 12 and 24 h postinfection (p.i.) and is described in the supporting information.
Fluorescence-Activated Cell Sorter (FACS)/RT-PCR Analysis of RRV Infectivity.
Refer to the supporting information.
Electron Microscopy.
Refer to the supporting information.
Statistical Analysis.
Refer to the supporting information.
Results
Kinetic Analysis of RRV-ADE Infectivity.
The kinetics of RRV infectivity were determined over the first 24 h of infection for RRV-ADE and RRV-normal mouse serum (NMS) cultures (Table 1). We observed at 4 h p.i. that there was no difference in the percentage of cells positive for virus after RRV-ADE or RRV-NMS infection. At 7 h p.i., there was increased infectivity for RRV-ADE-treated cells, but this finding was not statistically significant compared with the control (P = 0.0824; n = 3). As the data show, a clear and significant increase in infectivity was first seen at 12 h p.i., with a further substantial increase in infectivity seen for RRV-ADE at 24 h p.i.
Table 1.
Comparison of percent RRV infectivity in RAW 264.7 macrophages at 4–24 h post-ADE or control (NMS) infection
| RAW 264.7 treatment | Mean (± SEM) % infectivity
|
|||
|---|---|---|---|---|
| 4 h | 7 h | 12 h | 24 h | |
| RRV-ADE | 0.13 ± 0.01 | 0.20 ± 0.03* | 5.2 ± 0.3† | 12.6 ± 0.3† |
| RRV-NMS | 0.11 ± 0.01 | 0.14 ± 0.02 | 0.16 ± 0.03 | 0.22 ± 0.02 |
Considered not significant when compared with RRV-NMS (P = 0.0824).
Considered significant when compared with RRV-NMS (P < 0.05).
Therefore, initial RRV infectivity was identical for ADE vs. non-ADE infection of macrophages, with obvious enhancement of ADE-mediated infection seen only after 12 h of viral replication.
Expression of Antiviral Genes in RRV-ADE-Infected Macrophages.
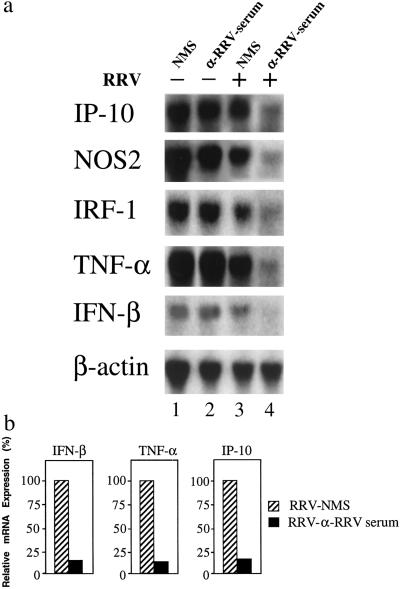
We first examined the expression of selected antiviral genes post ADE infection. Fig. 1a shows Northern blot analysis of mRNA for IFN-inducible protein 10 (IP-10), nitric-oxide synthase 2 (NOS2), IRF-1, TNF-α, and IFN-β from RRV-ADE-infected macrophages cultured with LPS (250 ng/ml). Message for IP-10, NOS2, IRF-1, TNF-α, and IFN-β was only marginally detectable for RRV-ADE-infected macrophages (Fig. 1a, lane 4), whereas, for noninfected LPS-stimulated cells, specific mRNA was strongly expressed for IP-10, NOS2, IRF-1, and TNF-α (IFN-β mRNA expression was weaker but clearly detectable; lanes 1 and 2). Message for IP-10, NOS2, IRF-1, TNF-α, and IFN-β was also clearly detected in RRV (+ NMS)-infected macrophages. Expression of these factors in RRV (+ NMS, lane 1)-infected macrophages was similar to that observed in infected LPS-treated macrophages in the absence of NMS (data not shown). No message was detected for any of these factors in macrophages in the absence of LPS treatment (data not shown). Exposure of macrophages to anti-RRV antibody alone had no effect on mRNA levels for any of the genes examined (Fig. 1a, lane 2). Although the ablation of mRNA for IP-10, NOS2, IRF-1, TNF-α, and IFN-β was found for RRV-ADE infection, β-actin transcription was not affected (Fig. 1a). Quantitative analysis showed that the levels of IP-10, TNF-α, and IFN-β expression in RRV-ADE-infected macrophages were only between 10–15% of those in RRV (+ NMS)-infected macrophages (Fig. 1b).
Figure 1.
Suppression of antiviral genes in RRV-ADE-infected macrophages. (a) Northern blot analysis of messenger RNA expression specific for IP-10, NOS2, IRF-1, TNF-α, and IFN-β in LPS-stimulated (250 ng/ml) RAW 264.7 cells 24 h post-RRV-ADE infection (RRV + 10−3 anti-RRV serum). Control mRNA expression is represented by β-actin. (b) Northern blots were quantified by PhosphorImager (Molecular Dynamics) analysis, and relative mRNA levels (IFN-β, TNF-α, and IP-10) are presented as the percentage of expression in RRV-non-ADE (RRV + NMS) control-infected cultures.
At the time of mRNA analysis (24 h p.i.), viral titers in macrophage cultures were: RRV-NMS = 5.56 ± 0.08 plaque-forming units (pfu)/ml; RRV-NMS + 250 ng/ml LPS = 3.74 ± 0.07 pfu/ml; RRV-ADE + 250 ng/ml LPS = 6.66 ± 0.19 pfu/ml. For a full analysis of RRV-ADE titers in RAW 264.7 macrophage cultures by plaque assay, see ref. 12.
EMSA Analysis of Antiviral-Associated Transcription Factor Activity in RRV-ADE-Infected Macrophages.
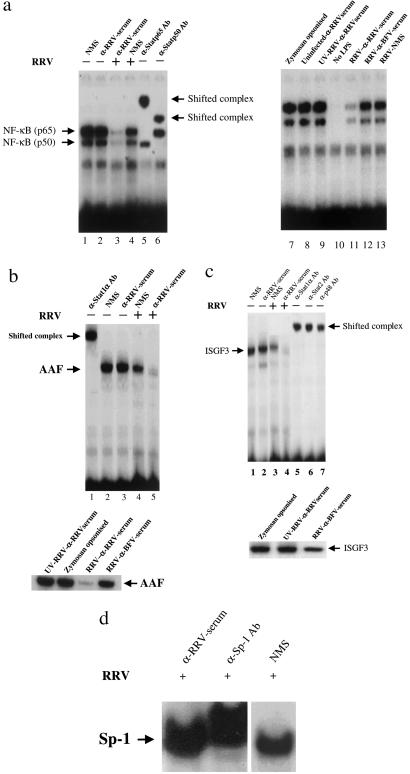
Results from the preceding section clearly show that there was a down-regulation of antiviral gene transcription in RRV-ADE cultures. Therefore, it was of interest to determine whether this suppression was associated with the disruption of specific transcription factor complexes. Greatly reduced NF-κB protein activity was detected in the nuclei of RRV-ADE-infected macrophages (Fig. 2a, lane 3), but RRV (+ NMS)-infected (lane 4) and noninfected controls showed abundant NF-κB activity in LPS-treated (250 ng/ml) cultures. Fig. 2 b and c shows the results of the AAF and ISGF3 protein complex formation in RRV-infected macrophages (+250 ng/ml LPS) by EMSA. AAF activity (Fig. 2b) was significantly reduced for RRV-ADE-infected macrophages (lane 5), but was clearly detected for RRV (+ NMS)-infected (lane 4) and noninfected controls cultured with NMS or anti-RRV alone. Results identical to that detected for AAF were found for ISGF3 (Fig. 2c), with RRV-ADE ablating this protein (lane 4). Interestingly, there appears to be a mild down-regulation of NF-κB, AAF, ISGF3 complex formation in non-ADE-infected (RRV + NMS) macrophages, suggesting that RRV infection alone may have a modest inhibitory effect. However, the nuclear activity of a transcription factor protein complex not associated with antiviral pathways, Sp-1, was not suppressed by RRV-ADE infection (Fig. 2d). A mild down-regulation of Sp-1 complex formation in non-ADE-infected (RRV + NMS) macrophages was noted.
Figure 2.
EMSA analysis of (a) NF-κB complex (p50 + p65), (b) AAF, (c) ISGF3, and (d) Sp-1. Various transcription factor activity was investigated in LPS-stimulated (250 ng/ml) RAW 264.7 cells 24 h post-RRV-ADE infection. A number of different controls were included for comparisons. Five micrograms of each nuclear extract were analyzed for binding activity by EMSA, by using radiolabeled oligonucleotides containing the appropriate sequence motif. Confirmation of protein identity is confirmed by reactivity with specific antisera, shown as shifted complexes.
Specific antibody probes confirmed the identities of the examined transcription factor protein complexes.
Fig. 2 also describes a number of control infection and treatment regimes designed to examine the extent to which antigen–antibody complex interaction with cellular Fc-γRs (17) featured in the ablation of antiviral transcription factors for LPS-treated cells. ADE infection of RAW 264.7 macrophages with UV-inactivated RRV did not result in the diminution of NF-κB (Fig. 2a), AAF (Fig. 2b), and ISGF3 (Fig. 2c) complex formation, emphasizing the need for functional virus to perturb antiviral pathways. UV-inactivated virus could be detected by immunostaining, suggesting that while there was no infection, there was viral attachment to cells and viral antigens were intact (data not shown). In addition, polyclonal antiserum to the serologically related alphavirus Barmah Forest induced only a mild suppression of transcription factor complex formation post-RRV-ADE infection (Fig. 2). Macrophages were also incubated with zymosan–antibody complexes (opsinized zymosan). At 24 h posttreatment, high levels of antiviral transcription factor complex formation were observed, suggesting that nonviral antibody complexes do not specifically ablate antiviral transcription factor activation in LPS-treated cultures.
Quantitative Analysis of RRV-ADE Infectivity.
We have previously shown by IFA techniques that 12–15% of RAW 264.7 cells were infected at 24 h post-ADE infection (12). FACS studies showed that, at an identical time point, 14.2% of the total cell population stained FITC positive, indicating biological infection of these cells by RRV.
Flow cytometric sorting and RT-PCR detection of viral RNA.
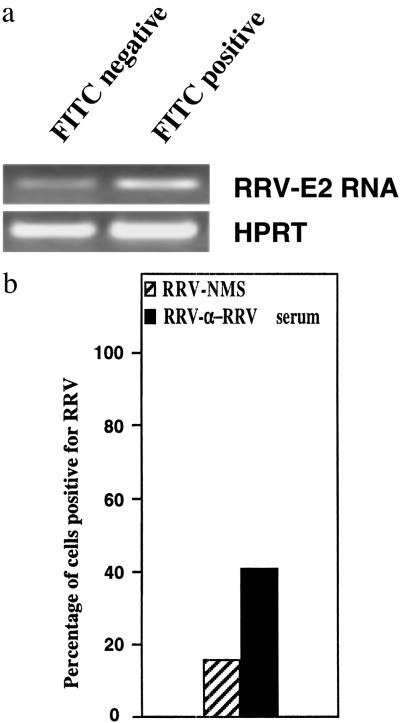
To fully assess the extent of RRV infectivity in RAW 264.7 macrophages post-ADE infection, RRV (FITC)-negative cells were purified by FACS and subjected to RT-PCR analysis. Whereas IFA and FACS data indicated that only a minority of cells were RRV infected, the RT-PCR data demonstrated that the unstained RAW 264.7 population also harbored virus, but at levels not detectable by immunostaining techniques. RT-PCR analysis of the purified unstained (FITC-negative) macrophage population showed clearly that RRV-RNA was present in these cells (Fig. 3a).
Figure 3.
(a) RT-PCR analysis of RRV-ADE infectivity in RAW 267.4 macrophages at 24 h p.i. RNA (RRV-ADE cultures) was extracted from equal numbers of FACS-purified RRV (FITC) positive and RRV (FITC) negative macrophage populations, and first strand cDNA was prepared as described in Methods. PCR was performed with primers specific for the viral E2 gene, with hypoxanthine phosphoribosyltransferase used as a housekeeping control. (b) Transmission electron microscopy analysis of RRV infectivity in RAW 264.7 macrophages at 24 h p.i. A total of 100 fields (each representing a single cell) were examined randomly to detect RRV in ADE vs. non-ADE control cultures. The number of high powered fields found to contain virus was determined, and the data were presented as a percentage of cells found to contain virus.
Electron microscopy.
A subsequent study to accurately quantitate the number of macrophages that harbor RRV post-ADE or control infection was performed by electron microscopy. A total of 100 fields were analyzed randomly, and the presence of virus was noted for each field (data presented as a percentage of cells associated with virus). Fig. 3b shows that, for ADE infection, 42% of macrophages harbored RRV, compared with only 15% of cells for RRV-NMS-infected control macrophage cultures.
Levels of IL-10 in RRV-ADE-Infected Macrophages.
In the preceding section, we showed that Sp-1 was not suppressed by RRV-ADE infection. Sp-1 has been shown to play a prominent role during LPS-mediated induction of the IL-10 gene (14). In addition, IL-10 is known to potently inhibit NF-κB and IFN-induced STAT-1 activation (18). We therefore investigated the activity of IL-10 in this system.
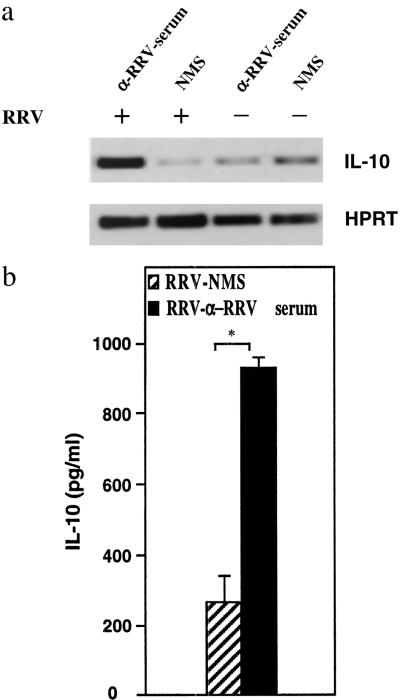
RT-PCR analysis revealed high levels of IL-10 mRNA expression (10-fold) in RRV-ADE cultures when compared with controls (Fig. 4a). To address whether this observation was consistent at the protein level, supernatants were then analyzed for IL-10 protein by ELISA. Supernatants taken from RRV-ADE-infected macrophage cultures contained significantly (P < 0.05) higher levels (4-fold) of IL-10 protein when compared with control (RRV-NMS) culture supernatants (Fig. 4b).
Figure 4.
Induction of IL-10 mRNA and protein in macrophages. (a) Total RNA was prepared from macrophage cultures, and IL-10 mRNA was amplified by RT-PCR. The amplification products were blotted onto nylon membranes and hybridized to fluorescein-labeled oligonucleotide probes specific for the PCR product. The reference gene, hypoxanthine phosphoribosyltransferase, was used to assess variation between RNA and cDNA loading. Non-ADE-RRV-infected macrophages and LPS alone treated macrophages served as controls. (b) Supernatants from various groups were collected and analyzed for IL-10 protein by ELISA. Data are presented as means of triplicate cultures ± SEM and are representative of two experiments. Asterisk indicates statistically significant differences between groups (P < 0.05).
Discussion
The in vitro enhancement of virus infection in macrophage cell lines by virus-specific antibodies at subneutralizing concentrations has been reported for several viruses (4–8). The initial stages of virus enhancement require the formation of a virus–antibody complex that then attaches, via the Fc portion of the antibody, to the macrophage Fc receptor; this complex facilitates the entry of the virus, as compared with virus infection in the absence of antibody.
As previously reported, ADE entry into macrophages afforded RRV with protection from LPS-induced antiviral activity in vitro due to the ablation of key antiviral gene expression and the associated suppression of antiviral proteins (12). In light of these previous results, this subsequent study focused on the activity of relevant transcription factor protein complexes in the nuclei of RRV-infected and LPS-stimulated macrophages. Here, we show that AAF, ISGF3, and NF-κB were significantly suppressed post-RRV-ADE infection. AAF is a STAT-1 homodimer that binds to IRF-1, IFN-β, and NOS2 promoters and positively regulates the transcriptional activation of these genes. RRV-ADE also ablated ISGF3 and NF-κB activity, with the downstream impact of this transcription factor disruption shown by the suppression of IP-10 and TNF-α, respectively.
The requirement for active virus in the suppression of antiviral responses by macrophages also needed exploration. Was the specific suppression of antiviral activity simply a result of Fc-γR cross-linking by antibody, regardless of the antigen involved? EMSA studies showed that nonviral immune complexes (zymosan–antibody complexes) did not induce the suppression of STAT-1 and NF-κB complexes in LPS-treated macrophages. Zymosan is a yeast capsular protein that is used extensively in phagocytosis studies (19). Also, RRV that was briefly UV irradiated before ADE infection did not suppress these complexes. It was viral replication post-antibody-enhanced entry, therefore, and not Fc-γR cross-linking that was responsible for the suppression/ablation of key antiviral responses.
Recent studies with dengue virus lend considerable support into observations of disruption to antiviral cytokine production post-ADE infection. Subneutralizing titers of immune sera against dengue-1 were found to enhance dengue-2 growth in monocytes, with an associated increase in cellular proliferation, but a decrease in the production of the potent antiviral cytokine IFN-γ (20, 21). This evidence led these workers to suggest that suppression of Th-1 immune responses may be linked to ADE associated with heterotypic dengue infection. However, the findings for monocytes/macrophages cannot be generalized to all cells. Another study found that, for antibody-enhanced dengue infection of a mast cell/basophil cell line, a significant increase in IL-1β production was observed and a “modest” increase for IL-6, but no increases were observed for granulocyte-macrophage colony-stimulating factor (22). These results lend some support to the concept of cytokine disruption post-ADE infection, but the enhanced production of IL-1β, an important inflammatory cytokine although not antiviral, is contrary to this position.
It should also be considered that a report has shown augmented serum TNF-α levels in patients suffering dengue hemorrhagic fever/dengue shock syndrome (DHF/DSS), compared with patients with primary dengue fever (23). While the serum TNF-α levels reported for DHF/DSS patients were a convincing correlate to the severity of these diseases, ex vivo monocyte/macrophage cultures from these patients were not examined directly for spontaneous or in vitro stimulated TNF-α production, leaving the question open as to the nature of the cells producing the excessive TNF-α concentrations in these patients. The timing of infection may also be of importance to the cytokine profile post-ADE infection. Whereas RRV-ADE in vitro infection of macrophages results in dramatic increases of virus titers initially, by day 5 p.i. the ADE titers are similar to non-ADE control (RRV) titers, with virus titers dropping to undetectable levels (plaque assay) more rapidly for ADE-infected cultures (data not shown). Initial ADE infection pathways ablate early antiviral responses in infected macrophages, allowing the establishment of infection, but this result may not guarantee that the expression of inflammatory/antiviral cytokines is suppressed for the duration of the infection or associated disease, particularly because the dynamic of the virus–host interaction may change greatly once an infection becomes prolonged or persistent.
Although the ability of double-stranded DNA viruses to perturb antiviral activity has been well recognized (24–26), much evidence has recently emerged on the capacity of RNA viruses to disrupt cellular antiviral pathways via the action of viral genes. The sole nonstructural protein of influenza A virus, NS1, has been recognized as a key virulence factor for its ability to inhibit type 1 IFN responses in the infected host (27). The ability of NS1 to block IFN-α/β activation has been found to be associated with the perturbation of double-stranded-RNA-activated protein kinase (PKR; ref. 28), with further investigations finding that IRF-3 and NF-κB were also inhibited (29, 30). This property of type I IFN antagonism has also been identified for Ebola virus VP35 protein (31), suggesting dual and possibly multiple roles for proteins encoded by small genome RNA viruses in cell interactions and immune evasion. TNF-α expression has been shown to be sensitive to in vitro infection by RRV alone (12), and this finding has been confirmed by this study (see Fig. 1). Like influenza and Ebola, RRV appears to have an innate capacity to disrupt host gene expression, but ADE entry is required for the maximum level of broad antiviral suppression post-macrophage infection.
With RRV alone displaying the ability to mildly disrupt TNF-α expression, consideration of possible disparities in the initial infection conditions for RRV-ADE vs. RRV alone was required. An infectivity kinetics study (Table 1) definitively demonstrated that the initial infectivity for RRV-ADE was identical to that found for the RRV control at 4 h p.i. It was only after 12 h p.i. that significant increases in infectivity were observed for RRV-ADE. This study focused on the disruption of the transcriptional regulation of antiviral responses in macrophages, and the infection kinetics data show that the ADE-mediated suppression of antiviral expression at 24 h p.i. was not due simply to either increased initial virus dose, or an initial enhanced infectivity for ADE treatment.
Central to the role of viral genes and proteins in the ablation of antiviral responses by macrophages are questions concerning the true degree of cellular infectivity at 24 h p.i. Whereas previous work by us and others (12, 32) has shown that ADE enormously enhanced RRV infectivity for macrophages in vitro, rates of between 12 and 15% (12) of the total cell population infected posed questions on how a minority of infected cells could induce global suppression of cellular genes and proteins. To address such concerns, RT-PCR studies were performed to examine the RRV-RNA status of cells found to be IFA negative for viral antigen. Viral RNA was detected in the unstained macrophage population, indicating that true RRV infectivity exceeded the 12–15% levels previously reported (12). In light of this RT-PCR result for RRV-ADE-infected cells, further studies were performed to precisely quantitate RRV infectivity for both RRV-ADE-infected macrophages and RRV-NMS-infected controls via transmission electron microscopy. This analysis demonstrated a substantially higher percentage of infected cells for ADE-infected macrophage cultures (42%) compared with non-ADE control cultures (15%). Furthermore, a separate study has also detected low level RRV infection of RAW 264.7 macrophages by RT-PCR and electron microscopy in IFA-negative cells post-RRV-NMS infection (39). Such evidence clearly demonstrates that overt infection, whether detected as cytopathic changes to cells or antigen immunostaining, does not necessarily indicate the full extent of viral infectivity. Whereas many more macrophages were infected by RRV than initially determined by IFA techniques alone (12), our new evidence shows that 58% of cells remained uninfected post-RRV-ADE. This finding stimulated investigations of virally induced cellular proteins in the suppression of antiviral activity for uninfected macrophages.
Whereas direct viral intervention in molecular processes within infected cells may account for a significant proportion of specific antiviral suppression, cellular proteins/factors stimulated by infection can suppress the expression of antiviral genes. In this regard, IL-10 is a primary candidate because levels of this cytokine were elevated in RRV-ADE-infected macrophage cultures. Interestingly, IL-10 transcription can be potently regulated by transcription factor Sp-1 (14), and, indeed, Sp-1 complex formation was not suppressed in RRV-ADE-infected macrophages. Furthermore, IL-10 is a potent immunosuppressive molecule that can mediate the down-regulation of Th1 responses by inhibiting the production of IL-12, IFN-γ, and TNF-α (18). IL-10 is capable of inhibiting NF-κB activation in human monocytes as well as IFN-α- and IFN-γ-induced genes by suppressing tyrosine phosphorylation of STAT-1 (18, 33, 34). Studies with other alphaviruses have considered the expression of IL-10 p.i. The inflammatory response to nonfatal Sindbis virus infection in SJL mice has shown that lymphocytes isolated from the CNS produced high levels of IL-10 (35). Venezuelan equine encephalitis virus infection of C57BL/6 mice resulted in the enhanced gene expression of IFN-γ, IL-6, IL-12, TNF, and IL-10 in the draining lymph node (36). A recent study with the picornavirus Coxsackie B3 (CVB3) had very strong parallels with our RRV findings. Proinflammatory cytokine responses were suppressed by virus in LPS-stimulated macrophages whereas IL-10, at the mRNA and protein level, was “strongly and persistently induced by CVB3” (37).
Because ADE has been observed for several virus families and associated with disease and adverse vaccination outcomes, our findings may have broad relevance beyond RRV pathogenesis. Our results show that antiviral gene expression was significantly and specifically suppressed, and in some cases ablated, via the disruption of transcription factor complex function. Whereas there may be other novel mechanisms associated with ADE-mediated virus infection, it appears likely that the ADE pathway may augment the innate capacity of some viral genes and proteins to perturb specific antiviral responses. The RRV gene/protein responsible for the ablation of antiviral factors post-ADE infection is still unknown. There has been traditional interest in the structural protein E2 as important to RRV virulence and antibody evasion (38), but, based on the observations of NF-κB and IRF disruption for influenza, the role of nonstructural viral genes/proteins will also need to be closely considered in future studies on antiviral evasion by RRV. Ultimately, it is desirable that roles for RRV genes/proteins in both the direct inhibition of cellular antiviral responses, as described above for influenza virus, and the indirect suppression of inflammatory-antiviral activity via the enhanced stimulation of cellular proteins like IL-10 be balanced for the best possible understanding of the mechanisms that underpin antibody-enhanced infection of macrophages. As such capacities are being identified among a range of small genome RNA viruses, the importance of ADE to the general understanding of immune evasion by viruses can only increase in significance.
Supplementary Material
Acknowledgments
We thank the University of Canberra Research Office and Research Committee for their support. We also thank Dr. Frances Shannon [John Curtin School of Medical Research (JCSMR)] for reading the manuscript before submission and Dr. Joanne Banyer (JCSMR) for useful discussions. S.M. is a recipient of the Australian National Health and Medical Research Council (NHMRC) Peter Doherty Fellowship. This work was funded by an Australian Research Council grant awarded to B.A.L.
Abbreviations
- ADE
antibody-dependent enhancement
- NF-κB
nuclear factor-κB
- STAT-1
signal transducer and activator of transcription 1
- IRF-1
IFN regulatory factor 1
- NOS2
nitric-oxide synthase 2
- RRV
Ross River virus
- NMS
normal mouse serum
- IP-10
IFN-inducible protein 10
- AAF
IFN-α-activated factor
- ISGF3
IFN-stimulated gene factor 3
- EMSA
electrophoretic mobility-shift assay
- LPS
lipopolysaccharide
- TNF
tumor necrosis factor
- FACS
fluorescence-activated cell sorter
- EPA
epidemic polyarthritis
- IFA
immunofluorescent antibody
- p.i.
postinfection
- Fc-γR
Fc-γ-receptor
References
- 1.Mackenzie J S, Broom A K, Hall R A, Johansen C A, Lindsay M D, Phillips D A, Ritchie S A, Russell R C, Smith D W. Commun Dis Intell. 1998;22:93–100. doi: 10.33321/cdi.1998.22.17. [DOI] [PubMed] [Google Scholar]
- 2.Lidbury B A, Simeonovic C, Maxwell G E, Marshall I D, Hapel A J. J Infect Dis. 2000;181:27–34. doi: 10.1086/315164. [DOI] [PubMed] [Google Scholar]
- 3.Hawkes R A. Aust J Exp Biol Med Sci. 1964;42:465–482. doi: 10.1038/icb.1964.44. [DOI] [PubMed] [Google Scholar]
- 4.Morens D M, Halstead S B. J Gen Virol. 1990;71:2909–2914. doi: 10.1099/0022-1317-71-12-2909. [DOI] [PubMed] [Google Scholar]
- 5.Ochiai H, Kurokawa M, Matsui S, Yamamoto T, Kuroki Y, Kishimoto C, Shiraki K. J Med Virol. 1992;36:217–221. doi: 10.1002/jmv.1890360312. [DOI] [PubMed] [Google Scholar]
- 6.Porterfield J S. Adv Virus Res. 1986;31:335–355. doi: 10.1016/s0065-3527(08)60268-7. [DOI] [PubMed] [Google Scholar]
- 7.Robinson W E, Mitchell W M. AIDS. 1990;4:S151–S162. [PubMed] [Google Scholar]
- 8.Robinson W E, Montefiori D C, Mitchell W M. Lancet. 1988;8589:790–794. doi: 10.1016/s0140-6736(88)91657-1. [DOI] [PubMed] [Google Scholar]
- 9.Kliks S C, Nisalak A, Brandt W E, Wahl L, Burke D S. Am J Trop Med Hyg. 1989;40:444–451. doi: 10.4269/ajtmh.1989.40.444. [DOI] [PubMed] [Google Scholar]
- 10.Adams D O, Hamilton T A. Annu Rev Immunol. 1984;2:283–318. doi: 10.1146/annurev.iy.02.040184.001435. [DOI] [PubMed] [Google Scholar]
- 11.Gao J J, Filla M B, Fultz M J, Vogel S N, Russell S W, Murphy W J. J Immunol. 1998;161:4803–4810. [PubMed] [Google Scholar]
- 12.Lidbury B A, Mahalingam S. J Virol. 2000;74:8376–8381. doi: 10.1128/jvi.74.18.8376-8381.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mahalingam S, Farber J M, Karupiah G. J Virol. 1999;73:1479–1491. doi: 10.1128/jvi.73.2.1479-1491.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tone M, Powell M J, Tone Y, Thompson S A, Waldmann H. J Immunol. 2000;165:286–291. doi: 10.4049/jimmunol.165.1.286. [DOI] [PubMed] [Google Scholar]
- 15.Dignam J D, Lebovitz R M, Roeder R G. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mahalingam S, Chaudhri G, Tan C L, John A, Foster P S, Karupiah G. J Biol Chem. 2001;276:7568–7574. doi: 10.1074/jbc.M005773200. [DOI] [PubMed] [Google Scholar]
- 17.Peiris J S, Gordon S, Unkeless J C, Porterfield J S. Nature. 1981;289:189–191. doi: 10.1038/289189a0. [DOI] [PubMed] [Google Scholar]
- 18.Moore K W, de Waal Malefyt R, Coffman R L, O'Garra A. Annu Rev Immunol. 2001;19:683–765. doi: 10.1146/annurev.immunol.19.1.683. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki T, Kono H, Hirose N, Okada M, Yamamoto T, Yamamoto K, Honda Z. J Immunol. 2000;165:473–482. doi: 10.4049/jimmunol.165.1.473. [DOI] [PubMed] [Google Scholar]
- 20.Chen R, Yeh W, Yang M, Yang K D. FEMS Immunol Med Microbiol. 2001;30:1–7. doi: 10.1111/j.1574-695X.2001.tb01542.x. [DOI] [PubMed] [Google Scholar]
- 21.Yang K D, Yeh W T, Yang M Y, Chen R F, Shaio M F. J Med Virol. 2001;63:150–157. [PubMed] [Google Scholar]
- 22.King C A, Marshall J S, Alshurafa H, Anderson R. J Virol. 2000;74:7146–7150. doi: 10.1128/jvi.74.15.7146-7150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yadav M, Kamath K R, Iyngkaran N, Sinniah M. FEMS Microbiol Immunol. 1991;89:45–50. doi: 10.1111/j.1574-6968.1991.tb04969.x. [DOI] [PubMed] [Google Scholar]
- 24.Davis-Poynter N J, Farrell H E. Immunol Cell Biol. 1996;74:513–522. doi: 10.1038/icb.1996.84. [DOI] [PubMed] [Google Scholar]
- 25.Miller D M, Rahill B M, Boss J M, Lairmore M D, Durbin J E, Waldman W J, Sedmak D D. J Exp Med. 1998;187:675–683. doi: 10.1084/jem.187.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith G L. Immunol Lett. 1999;65:55–62. doi: 10.1016/s0165-2478(98)00125-4. [DOI] [PubMed] [Google Scholar]
- 27.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy D E, Durbin J E, Palese P, Muster T. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 28.Bergmann M, Garcia-Sastre A, Carnero E, Pehamberger H, Wolff K, Palese P, Muster T. J Virol. 2000;74:6203–6206. doi: 10.1128/jvi.74.13.6203-6206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talon J, Horvath C M, Polley R, Basler C F, Muster T, Palese P, Garcia-Sastre A. J Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, Li M, Zheng H, Muster T, Palese P, Beg A A, Garcia-Sastre A. J Virol. 2000;74:11566–11573. doi: 10.1128/jvi.74.24.11566-11573.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Basler C F, Wang X, Muhlberger E, Volchkov V, Paragas J, Klenk H D, Garcia-Sastre A, Palese P. Proc Natl Acad Sci USA. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Linn M L, Aaskov J, Suhrbier A. J Gen Virol. 1996;77:407–411. doi: 10.1099/0022-1317-77-3-407. [DOI] [PubMed] [Google Scholar]
- 33.Wang P, Wu P, Siegel M I, Egan R W, Billah M M. J Biol Chem. 1995;270:9558–9563. doi: 10.1074/jbc.270.16.9558. [DOI] [PubMed] [Google Scholar]
- 34.Ito S, Ansari P, Sakatsume M, Dickensheets H, Vazquez N, Donnelly R P, Larner A C, Finbloom D S. Blood. 1999;93:1456–1463. [PubMed] [Google Scholar]
- 35.Rowell J F, Griffin D E. J Immunol. 1999;162:1624–1632. [PubMed] [Google Scholar]
- 36.Grieder F B, Davis B K, Zhou X D, Chen S J, Finkelman F D, Gause W C. Virology. 1997;233:302–312. doi: 10.1006/viro.1997.8617. [DOI] [PubMed] [Google Scholar]
- 37.Hofmann P, Schmidtke M, Stelzner A, Gemsa D. J Med Virol. 2001;64:487–498. doi: 10.1002/jmv.1076. [DOI] [PubMed] [Google Scholar]
- 38.Vrati S, Faragher S G, Weir R C, Dalgarno L. Virology. 1986;151:222–232. doi: 10.1016/0042-6822(86)90044-9. [DOI] [PubMed] [Google Scholar]
- 39. Way, S. R., Lidbury, B. A. & Banyer, J. (2002) Virology, in press. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.