Abstract
Hepatitis B virus (HBV) is a prototype for liver-specific pathogens in which the failure of the immune system to mount an effective response leads to chronic infection. Our understanding of the immune response to HBV is incomplete, largely due to the narrow host restriction of this pathogen and the limitations of existing experimental models. We have developed a murine model for studying human HBV replication, immunogenicity, and control. After transfection of hepatocytes in vivo with a replication-competent, over-length, linear HBV genome, viral antigens and replicative intermediates were synthesized and virus was secreted into the blood. Viral antigens disappeared from the blood as early as 7 days after transfection, coincident with the appearance of antiviral antibodies. HBV transcripts and replicative intermediates disappeared from the liver by day 15, after the appearance of antiviral CD8 + T cells. In contrast, the virus persisted for at least 81 days after transfection of NOD/Scid mice, which lack functional T cells, B cells, and natural killer (NK) cells. Thus, the outcome of hydrodynamic transfection of HBV depends on the host immune response, as it is during a natural infection. The methods we describe will allow the examination of viral dynamics in a tightly controlled in vivo system, the application of mutagenesis methods to the study of the HBV life cycle in vivo, and the dissection of the immune response to HBV using genetically modified mice whose immunoregulatory and immune effector functions have been deleted or overexpressed. In addition, this methodology represents a prototype for the study of other known and to-be-discovered liver-specific pathogens.
Hepatitis B virus (HBV) is a human hepadnavirus that causes acute and chronic hepatitis and hepatocellular carcinoma (1). Although an effective vaccine has been available for two decades, an estimated 350 million people worldwide are chronically infected. A significant proportion of chronic infections terminate in hepatocellular carcinoma, leading to more than one million deaths annually (2).
A reproducible tissue culture model of HBV infection does not exist, nor is HBV infectious for immunologically well-defined laboratory animals. Much of our current understanding of the viral life cycle after HBV infection is derived from studies of duck HBV (DHBV) (3) and woodchuck HBV (WHV) (4) infection in their natural hosts, HBV-infected chimpanzees (5–7), and HBV transgenic mice (8–10). For various reasons, however, none of these models is ideal. DHBV (11) and WHV (12, 13) are genetically divergent from HBV, and immunological studies in genetically outbred and immunologically uncharacterized ducks and woodchucks are difficult. Chimpanzee experiments are limited by cost, availability, and ethical considerations, whereas transgenic mice are immunologically tolerant to the virus, thereby compromising the greatest potential strength of a mouse model of HBV infection. We present here work describing a recently developed mouse model that alleviates many of these experimental constraints.
Materials and Methods
Animal Studies.
Breeding pairs of strains B10.D2 and NOD/LtSz-Prkdcscid/J were obtained from The Jackson Laboratory. Animals were treated according to the National Institutes of Health Guidelines for Animal Care and the Guidelines of The Scripps Research Institute. Mice were used at 6–9 weeks of age.
Quantitation of Viremia.
Viremia was measured by purification and quantitation of encapsidated viral DNA by real-time PCR as described in supporting Materials and Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Serology for Transaminase Activity and Antigenemia.
Serum alanine aminotransaminase activity (sALT) was measured at 30°C on a SpectraMax Plus spectrophotometer (Molecular Devices, Sunnyvale, CA) using the ALT Infinity Reagent (Sigma) following the manufacturer's instructions. Transaminase values are the average of n mice on days 1, 4, 7, 10, 15, and 20 (n > 5). Serum concentrations of HBsAg were quantitated by sandwich ELISA using HBsAg standards and antibodies provided in a commercial ELISA kit (Abbott) to generate a calibration curve. Serum HBeAg was detected by RIA (DiaSorin, Stillwater, MN) and quantitated by using recombinant HBeAg (Biogen) to generate a calibration curve.
Antibody ELISA.
IgG and IgM antibodies specific for HBsAg, HBeAg, and HBV core protein were detected by endpoint titration ELISA as described in supporting Materials and Methods.
In Vitro Cultivation of HBV-Specific Cytotoxic T Lymphocytes (CTLs).
Splenocytes were isolated from groups of mice (≥4 mice per group) killed on days 7, 10, 15, 20 after hydrodynamic injection and cultured as described in supporting Materials and Methods.
51Cr-Release Assay for CTL Activity.
Splenocyte cultures were harvested and quantitated after 5 days of in vitro stimulation and then used as effectors in a killing assay as described in supporting Materials and Methods.
Immmunohistochemical Staining for HbcAg.
HBV core protein was visualized by immunohistochemical staining of tissues fixed in zinc-buffered formalin by using anti-core polyclonal rabbit antibody (DAKO) exactly as described (15). The average percentage of core positive cells at each time point (days 1, 4, 7, 10, 15, and 20) was determined from ≥4 livers, counting ≥100 independent fields per liver at ×20 magnification, with an average of 75 ± 3 hepatocytes counted per field.
HBV RNA and Replicative DNA Analysis.
Viral RNA and replicative DNA intermediates were detected by Northern and Southern blot analysis of total genomic liver RNA and DNA, respectively, as described in supporting Materials and Methods. Nuclear covalently closed circular DNA (cccDNA) was analyzed following previously described methods (16, 17), which are elaborated in supporting Materials and Methods.
Ribonuclease Protection Assay.
Total liver RNA was analyzed for T cell markers by ribonuclease protection assay as described (18).
Generation and Hydrodynamic Injection of Plasmid pT-MCS-HBV1.3.
Plasmid pT-MCS-HBV1.3, which contains an HBV1.3 supergenomic DNA flanked by the inverted repeat (IR) recognition sequences of the Sleeping Beauty transposase, was generated as follows. The HBV1.3 supergenomic DNA was isolated from plasmid pHBV2 containing a head-to-tail dimer of the HBV subtype ayw (19) and then subcloned into transposon vector pT-MCS (20) by ligation of the EcoRI/BglII (2.0 kb) and NsiI/EcoRI (2.1 kb) fragments of pHBV2 to the NsiI/BglIIdigested product of pT-MCS.
A total of 13.5 μg of pT-MCS-HBV1.3 and 4.5 μg of the Sleeping Beauty transposase expression plasmid pCMV-SB (20) were injected into the tail vein of 6- to 9-week-old B10.D2 or CB17 NOD/Scid mice in a volume of saline equivalent to 8% of the body mass of the mouse (e.g., 1.6 ml for mouse of 20 g). The total volume was delivered within 5–8 seconds. Cohorts were defined by matching mice on the basis of sALT, HBsAg, and HBeAg parameters on day 1 after injection of HBV1.3 DNA. The percentage of HBc-Ag-positive hepatocytes in the liver served as a lower limit estimate of transfection efficiency and ranged on average from 5–10% within a given experiment, depending on the efficiency of the plasmid injection as well as the genetic background of the mouse. Replication levels varying by as much as 5-fold between given experiments were tolerable, as we were primarily interested in the persistence and the kinetics of disappearance of viral gene expression, replication, antigenemia, viremia, etc., following their respective maxima.
Results
Hydrodynamic Injection of pT-MCS-HBV1.3/pCMV-SB Leads to Viral Gene Expression and Replication in Vivo.
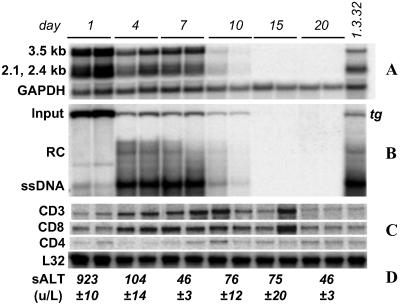
An HBV1.3 supergenomic DNA construct that has been shown to support HBV gene expression and replication in the livers of transgenic mice (15) was incorporated into a transposon delivery vector system (20) in which it was flanked by the recognition sequences (IR sequences) of the Sleeping Beauty transposase, an engineered transposase active in mammalian cells. The resulting plasmid, pT-MCS-HBV1.3, was injected intravenously into inbred B10.D2 mice under hydrodynamic conditions to transfect hepatocytes in vivo (14). A Sleeping Beauty transposase expression plasmid, pCMV-SB, was coinjected to allow transient transposase expression and somatic integration of the HBV1.3 transgene after somatic integration of the transgene, as was described (20). Consistent with the acute circulatory volume overload required for hydrodynamic transfection (14, 21), sALT activity increased sharply on day 1 (Fig. 1) and returned to baseline levels (<50 units/liter) by day 7 after transfection. Production of viral transcripts as well as RNA and DNA replicative intermediates was examined in multiple mice (n = 4) on days 1, 4, 7, 10, 15, and 20 after transfection (Fig. 1). Input DNA was observed only in episomal form in Southern blot analysis, as shown in Fig. 1B and Figs. 6 and 7, which are published as supporting information on the PNAS web site. Input DNA was most abundant on day 1, the time of peak expression of the 2.1- and 2.4-kb HBV transcripts, which encode the viral envelope proteins, and the 3.5-kb HBV transcript, which encodes the viral core and polymerase proteins and also serves as the template for viral replication. In contrast, replicative DNA intermediates were scarce on day 1 but abundant on day 4, consistent with the time required for the viral polymerase to reverse transcribe the 3.5-kb transcript to single-stranded DNA and subsequent steps in viral replication. Input DNA, HBV RNA, and replicative DNA intermediates were present at stable levels between days 4 and 7. All three species decreased by day 10 and were virtually undetectable by day 15. Disappearance of HBV RNA and DNA coincided with the influx of intrahepatic CD3 and CD8 T cell markers (Fig. 1C) and mild increases in serum transaminases (Fig. 1D).
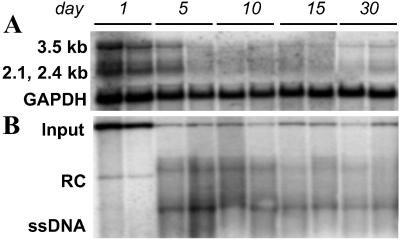
Figure 1.
Viral gene expression and replication after hydrodynamic transfection with HBV1.3 DNA persist through day 7. Liver RNA and DNA were analyzed for (A) HBV transcripts and (B) replicative intermediates by Northern and Southern blot, respectively. Liver RNA and DNA isolated on day 1 after hydrodynamic injection of saline to a 1.3.32 HBV transgenic mouse (15) were used as controls (far right lane). Disappearance of episomal input DNA, HBV transcripts, and replicative DNA intermediates coincided with (C) the influx of CD3 and CD8 T cell markers, detected by ribonuclease protection assay, and (D) mild increases in serum transaminases.
Kinetics of Viremia.
Because viral replication in the liver is necessary but not sufficient for a good experimental model of acute HBV infection, in a separate experiment we sought to verify that the liver-specific HBV replication observed also led to physiologically relevant viremia and antigenemia in mice hydrodynamically injected with HBV1.3 DNA. Encapsidated viral DNA was purified from blood, and viral genomes were measured by a quantitative PCR assay (see supporting Materials and Methods and Table 1, which is published as supporting information on the PNAS web site). Viremia was undetectable on day 1, paralleling the low abundance of DNA replicative intermediates in the liver at this time. Viral titers on day 3 were on average 1.5 × 106 ± 0.6 × 106 copies per ml of blood, and peaked at 8 × 106 ± 4 × 106 copies per ml of blood on day 6. Viremia subsequently declined with logarithmic kinetics through day 8 (1.6 × 106 ± 0.6 × 106 copies per ml) and day 14 (6.7 × 104 ± 3.2 × 104 copies per ml), decreasing by more than three orders of magnitude by day 22 (1.8 × 103 ± 0.8 × 103 copies per ml).
Kinetics of HBcAg Expression.
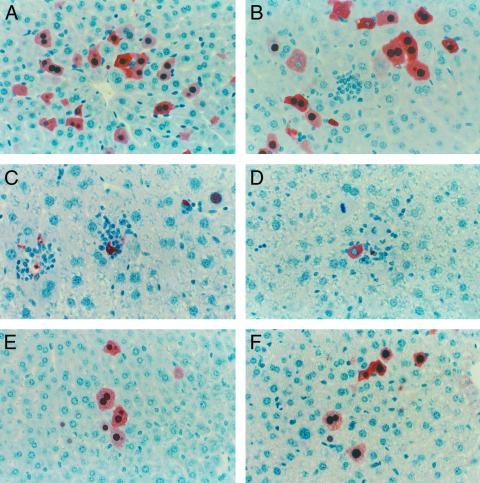
The kinetics of core protein expression was examined by immunohistochemical staining for HBV core antigen (HBcAg). HBcAg-positive hepatocytes were randomly distributed throughout the liver lobule with a tendency for localization in the centralobular area (Fig. 2A). Staining was both cytoplasmic and nuclear, with nuclear staining observed primarily in cells where cytoplasmic staining was most intense. HBcAg expression peaked on day 1 (6% ± 1% of hepatocytes HBcAg-positive; Fig. 2A) and remained at this level through day 7 when 4% ± 1% of the liver was HBcAg-positive (Fig. 2B). This modest decrease was accompanied by the appearance of small infiltrates of inflammatory cells. Between days 7 and 20, infiltration of the liver by inflammatory cells increased and the frequency of HBcAg-positive hepatocytes decreased to 1.9% ± 0.4% on day 10 (Fig. 2C), 0.09% ± 0.05% on day 15 (Fig. 2D), and 0% on day 20 and thereafter (data not shown). Notably, on day 10, HBcAg-positive hepatocytes were surrounded by inflammatory cells and appeared apoptotic (Fig. 2C). This trend continued through day 15, when inflammatory foci in the liver were most abundant and many of the associated HBcAg-positive hepatocytes appeared apoptotic. HBcAg-positive hepatocytes were completely eliminated from the liver by day 20.
Figure 2.
Transfected immunocompetent and immunocompromised mice exhibit different kinetics of HBcAg expression in the liver. (Magnification, ×40.) In B10.D2 mice, HBcAg-positive cells were maintained at approximately stable levels on (A) day 1 (5.7%), day 4 (data not shown), and (B) day 7. Infiltrates of inflammatory cells were first observed on day 7 and became more prevalent on (C) day 10 and (D) day 15 as the frequency of HBcAg-positive hepatocytes decreased and many HBcAg-positive cells appeared apoptotic. In contrast, HBcAg-positive hepatocytes persisted at stable levels and inflammatory infiltrates were absent in NOD/Scid mice, as shown here for (E) day 30 and (F) day 81.
The absence of detectable HBV RNA and DNA replicative intermediates and core protein in all other organs (brain, spleen, kidney, heart, and lung) after hydrodynamic transfection (data not shown) demonstrated that viral replication in this experimental system is successfully targeted to the liver. Other constraints on host range and tissue specificity are likely in place, however, because cccDNA, the template of viral replication in natural infections, was not detected by Southern blot analysis of the livers of hydrodynamically transfected mice (data not shown). This finding is consistent with prior reports that cccDNA is present at exceedingly low or undetectable levels in multiple, independently derived transgenic mouse lineages (16), suggesting the existence of a species restriction on the production of cccDNA.
Kinetics of Serum Antigenemia and Anti-HBV Serum Antibody Expression.
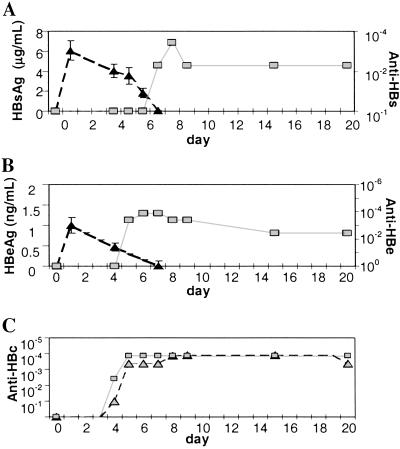
Secretion of viral antigens into the blood was also monitored over time (Fig. 3). HBV surface antigen (HBsAg) accumulated to an average concentration of 6 ± 1 μg/ml on day 1 (Fig. 3A). After this peak, serum HBsAg dropped slowly at a rate of 1.4 μg/ml per day, reaching a concentration of 37 ± 11 ng/ml by day 7 and becoming undetectable by day 8 after transfection. Interestingly, as serum levels of HBsAg declined, antibodies specific for HBsAg became detectable in the blood. Anti-HBsAg IgM antibodies were first detected on day 7 (1.2 × 10−2) and reached a maximum titer of 1.4 × 10−3 on day 8, at which time anti-HBsAg IgG antibodies also appeared (1.2 × 10−2) and serum HBsAg became undetectable (Fig. 3A).
Figure 3.
Disappearance of HBV serum antigens coincides with the induction of an HBV-specific antibody response. Antibody titers are reported as the reciprocal of the highest dilution at which samples were scored positive. (A) HBsAg (filled triangles) peaked within 24 h after transfection; the decrease between days 1 and 4 was presumably caused by concurrent decreases in HBV RNA. HBsAg antigenemia declined rapidly after day 5, becoming undetectable by day 7. This phase of antigen clearance coincided with the development of anti-HBs IgM (gray squares) and IgG (data not shown) antibodies. (B) HBeAg (filled triangles) peaked on day 1 after injection of HBV1.3 DNA, and its subsequent disappearance was accompanied by the development of anti-HBeAg IgM (gray squares) and IgG (data not shown) antibodies. (C) IgM (gray squares) and IgG (gray triangles) antibodies to HBcAg were first detectable on day 4 and rapidly reached a peak titer by day 5 that was maintained through day 20.
HBeAg was detected in the serum at 1.0 ± 0.2 ng/ml at the peak of expression on day 1 (Fig. 3B) and fell to 0.5 ± 0.1 ng/ml and 0.3 ± 0.1 ng/ml on days 4 and 7, respectively. The drop in antigenemia was again accompanied by the appearance of anti-HBeAg IgM and IgG antibodies on day 5. The anti-HBeAg IgM and IgG antibody responses peaked at titers of 1.4 × 10−4 on days 6 and 8, respectively. Decreases in HBs and HBe antigenemia between days 1 and 4 can most likely be attributed to the concurrent decline in HBV RNA; however, the decrease in both HBs and HBe antigenemia between days 4 and 7 occurred in the context of unchanging HBV RNA and HBV DNA content in the liver (Fig. 1), and is likely attributable to other causes, including an effective host immune response. The kinetic association between the induction of virus-specific antibody responses and the disappearance of HBsAg and HBeAg in the blood supports this notion. Although we did not measure serum levels of HBcAg, we did observe a vigorous antibody response to HBV core protein that appeared first as IgM antibodies to HBcAg (3.7 × 10−3 on day 4) and peaked rapidly by day 5 at a titer of 1.4 × 10−4 (Fig. 3C). IgG antibodies to HBcAg followed rapidly first appearing on day 5 and peaking by day 8 at a titer of 1.4 × 10−4 (Fig. 3C). The kinetic hierarchy of the HBV-specific humoral response in hydrodynamically transfected mice thus appears to be anti-HBcAg first, followed by anti-HBeAg, and lastly anti-HBsAg. The same sequence is characteristic of the antibody response to these viral antigens during acute HBV infection in man, though the time course of the individual responses are measured in weeks or months rather than days (22–28)
HBV-Specific CTL Response.
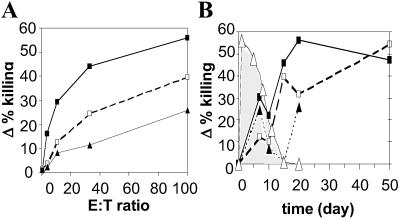
Because the CTL response plays a critical role in the resolution of acute HBV infection (29), the CTL response to HBV was analyzed on days 7, 10, 15, 20, and 50 after hydrodynamic transfection. As shown in the 51Cr-release assay illustrated in Fig. 4A, CTLs specific for at least three previously defined epitopes in the viral envelope (HBs28–39, HBs364–372) and polymerase (Pol140–148) proteins were detectable in the spleens of mice on day 7 after hydrodynamic transfection. It is interesting to note that the CTL response may have started earlier than day 7 because we observed an increase in CD3 and CD8 mRNA in the liver as early as day 4 after transfection (Fig. 1C). It is also important to point out that viral RNA (Fig. 1A), viral DNA replicative intermediates (Fig. 1B), and viral antigen expression in the liver (Fig. 2) were stable between days 4 and 7, and that small inflammatory infiltrates were first detected in the liver histologically on day 7 when the CTL response was first detected. All of these viral parameters changed rapidly thereafter, and these changes coincided with the continued evolution of the CTL response to HBV. As shown in Fig. 4B, which illustrates the cytolytic activity of the CTLs at a constant effector/target cell ratio at various times after hydrodynamic transfection, the CTL response progressively increased through day 20 and beyond except for a transient dip on day 10 for the HBs-specific CTLs and days 10 and 15 for the polymerase-specific CTLs. The induction and expression of the CTL response coincided precisely with a rapid disappearance of HBcAg-positive hepatocytes (Fig. 4B), viral RNA (Fig. 1A), and viral DNA replicative intermediates (Fig. 1B) from the liver. It also coincided with the appearance of an intrahepatic lymphocytic infiltrate that accumulated in the vicinity of HBcAg-positive hepatocytes, many of which appeared apoptotic (Fig. 2 C and D), and a slight increase in sALT activity (Fig. 1D). CTLs specific for HBs28–39, HBs364–372, and Pol140–148 continued to be observed through days 20 and 50 (Fig. 4B), when viral gene expression and replication were no longer observed (Fig. 1 A and B, and data not shown). A CTL response to HBcAg was not detected in hydrodynamically transfected mice at any time points (data not shown) consistent with the observation that the response to this protein is not robust in mice of this genetic background (30). The hierarchy of the anti-HBV CTL response in the hydrodynamic transfection model therefore appears to be HBs > pol > core. Collectively, these results strongly suggest that the HBV-specific CTLs played an important role in eliminating the HBV-positive hepatocytes from the liver.
Figure 4.
HBV-specific CTLs elicited by hydrodynamic injection HBV1.3 DNA are associated with the disappearance of HBV-positive hepatocytes. (A) Representative plot illustrating Δ% killing (% release specific targets − % release nonspecific targets) versus effector/target cell ratio (E:T) for splenocytes isolated on day 7 after hydrodynamic transfection. CTLs specific for envelope-derived HBs28–39 (□) and HBs364–372 (■), as well as the polymerase-derived Pol140–148 (▴) are present on day 7, as well as on days 10, 15, 20, and 30 (data not shown). (B) Δ% killing and % HBcAg-positive hepatocytes plotted versus time for constant E:T 100. The percentage of HBcAg-positive hepatocytes (▵) was stable from day 1 through day 7 but decreased thereafter after the induction and progressive increase of CTLs against the HBs28–39 (□), HBs364–372 (■), and Pol140–148 (▴) epitopes. HBc-Ag-positive hepatocytes disappeared by day 15 after hydrodynamic injection of HBV1.3 DNA.
Persistence of Viral Gene Expression and Replication in Immunodeficient Mice.
The impact of cellular and humoral immune responses on the kinetics of HBV expression was probed by hydrodynamic transfection of CB17 NOD/Scid (severe combined immunodeficiency) mice, which lack functional B and T cells, as well as most natural killer (NK) cell activity (31). Viral transcripts and replicative DNA intermediates were detected in the livers of these mice from day 4 through at least day 30 (Fig. 5 A and B), and encapsidated viral DNA was similarly long-lived (Table 1). Examination of the kinetics of HBcAg expression by immunohistochemical staining revealed the presence of HBcAg in a significant percentage of hepatocytes between days 1 and 81 (Fig. 2 E and F and Fig. 8, which is published as supporting information on the PNAS web site) in the absence of an inflammatory reaction. In marked contrast, the disappearance of HBcAg-positive hepatocytes from the liver of immunocompetent mice between days 7 and 20 was coincident with the development of an HBV-specific T cell response (Fig. 4B) and an inflammatory infiltrate in the liver (Fig. 2 B–D). The persistence of viral gene expression for extended periods of time in the absence of an immune response strongly suggests that the immune response plays a central role in the elimination of HBV-positive hepatocytes in the immunocompetent mice.
Figure 5.
HBV gene expression and replication persist in immunocompromised mice. (A) The HBV 3.5-, 2.4-, and 2.1-kb transcripts peaked on day 1, when input DNA was most abundant, and by day 5 fell to stable levels that persisted through at least day 30. (B) HBV DNA replicative intermediates were not apparent until day 5 and were maintained at stable steady-state levels through at least day 30.
Discussion
Because of the shortcomings of currently existing models of HBV infection, we sought to develop a model that would allow us to study the immune response to HBV in an immunologically naïve small animal host. Because previous studies of HBV transgenic mice demonstrated that the murine host is capable of supporting high-level HBV replication templated by a 1.3-length supergenomic transgene, we took advantage of the hydrodynamic transfection method as a means of introducing the HBV1.3 transgene to the livers of immunocompetent mice. The results suggest that the following events were triggered by the hydrodynamic injection of HBV DNA. Within 24 h, the input DNA apparently reached the nucleus of an unknown number of hepatocytes and possibly other cells in the liver (Fig. 1B), where it was transcribed to produce an abundance of the viral transcripts (Fig. 1A). The viral RNA was rapidly translated to produce high serum titers of HBsAg (Fig. 3A) and HBeAg (Fig. 3B), and viral nucleocapsids that were detected in up to 5.7% of the hepatocytes at the 24 h time point (Fig. 2A). Viral replication was not robust at this time point, though the presence of small amounts of single-stranded DNA, the earliest replicative DNA intermediate formed by reverse transcription of the 3.5-kb pregenomic viral RNA, indicated that viral replication had begun. By day 4, the input DNA and viral RNA levels decreased substantially (Fig. 1 A and B) for unknown reasons, perhaps reflecting the activity of cellular nucleases, toxic effects of high level viral gene expression, damage secondary to hydrodynamic pressure effects, or attrition of episomal input DNA. In any event, the input DNA and viral RNA levels were stable for the next several days, during which viral replication was robust (Fig. 1B), as reflected by the presence of cytoplasmic capsids (Fig. 2) within which the enzymatic events required for viral replication are known to occur (32). Between days 1 and 4, there was also a decrease in circulating HBsAg and HBeAg (Fig. 3 A and B), which probably reflects the decreased level of viral RNA in the liver. Importantly, however, there was a sharp decrease in circulating HBsAg and HBeAg by day 7, coinciding with and presumably caused by the appearance of the corresponding antibodies heralding the onset of an immune response to the virus in immunocompetent animals. Interestingly, the sequence of appearance of the various antibody specificities (anti-core, then anti-HBe, then anti-HBs) mirrors precisely the sequence observed during a natural infection, presumably reflecting the similar intrinsic immunogenic hierarchy of these antigens in humans and mice.
At the same time that antibodies were appearing, we were also able to detect HBV-specific CTLs in the spleens of the mice (Fig. 4) and the beginning of an intrahepatic inflammatory response (Fig. 2). CD3/8 markers were enriched in the liver as early as day 4 after transfection (Fig. 1C). Between days 7 and 10, the number of HBcAg-positive hepatocytes fell significantly (Fig. 4B), and this decrease was associated with the appearance of a pronounced intrahepatic inflammatory infiltrate surrounding HBcAg-positive hepatocytes that exhibited the cytological features of apoptosis (Fig. 2 C and D), as well as with a major decrease in the viral RNA (Fig. 1A) and replicative DNA (Fig. 1B) content of the liver and lastly with a slight surge in sALT activity (Fig. 1D). By day 20 after hydrodynamic transfection, all traces of virus were undetectable in the liver. These results strongly suggest that the CTL response played a large role in the elimination of virus-positive hepatocytes in the liver. This hypothesis is strongly supported by the stability of viral gene expression and replication for at least 81 days after hydrodynamic transfection of NOD/Scid mice (Fig. 5). Collectively, these results demonstrate that almost all aspects of the host–virus relationship during HBV infection can be reproduced in the context of an immunologically well-defined, inbred small animal model.
We originally used the Sleeping Beauty transposon system to deliver the HBV1.3 transgene because previous studies had indicated that transgene expression could not be sustained at significant levels for more than 24 h in the absence of transposase-mediated somatic integration (14, 20, 21). As illustrated in Figs. 1B and 6, however, episomal HBV1.3 persisted in the liver for at least 1 week after hydrodynamic transfection with no evidence of appreciable transgene integration. In subsequent experiments, we have found that hydrodynamic injection of pT-MCS-HBV1.3 with pCMV-mSB, a plasmid encoding an enzymatically dead transposase (20), leads to viral gene expression and replication with identical kinetic behavior (Fig. 7). The HBV1.3 transgene therefore appears to differ from those previously studied by the hydrodynamic transfection method (14, 20, 21).
This model affords new opportunities for the study of HBV. In particular, precise questions about HBV virology and immunology can now be addressed because the HBV genome can be precisely mutated in vitro and then examined in vivo in immunologically naïve and normal mice as well as in any of the large collection of transgenic hosts that currently exist. For example, although attention has been focused on the importance of a strong CTL response in clearance of HBV, direct evidence that such a response causes viral clearance of a natural infection has been lacking. Furthermore, the role of the innate immune system (e.g., NK cells, NKT cells, etc.) in combatting HBV, although demonstrated for other viral systems (33–35) and suggested for HBV by manipulation of the transgenic mouse model (36–38), has not been characterized because of the lack of suitable research models. The hydrodynamic transfection model permits the impact of individual components of cellular, humoral, and innate immunity on viral pathogenesis and control to be examined by simply injecting HBV plasmids into different strains of genetically modified mice (e.g., knockout mice deficient in CD8, CD4, B cells, NK cells, β-2-microglobulin, MHC class I, MHC class II, perforin, cytokines, etc.)
Interestingly, the recruitment of CD3/CD8 T lymphocytes to the liver precedes viral clearance in hydrodynamically transfected immunocompetent mice, indicating that the rate of viral clearance does not exceed the rate of viral replication until at least 3 days after initial recruitment of CD3/CD8 T cells to the liver. A formal possibility consistent with the data is that the rate of viral replication in the liver is increasing between days 4 and 7, so that even as a virus-specific T cell response is developing in the liver, the net change in viral gene products and replicative intermediates appears to be undergoing no net change during this time. Under this scenario, the rate of T cell-mediated viral clearance surpasses the rate of viral replication by day 7, making a net decrease in viral replication observable thereafter. Specifically, this scenario could be the result of a CTL response that is incipient on day 4 after transfection and that evolves to the extent that it is effective only by day 7. Additional experiments using CD8 knockout mice to characterize the mechanism of clearance in the hydrodynamic transfection model are necessary to clarify this issue.
We have previously observed that different populations of inflammatory cells may be responsible for early noncytolytic viral clearance and late cytolytic viral pathogenesis in the chimpanzee model of HBV infection (7), a system in which infection and viral spread allows virtually 100% of hepatocytes to be infected at the peak. Under those circumstances, it was postulated that in an early innate immune response, NK cells may have mediated an IFN-γ-dependent down-regulation of viral replication and that a subsequent virus-specific T cell response led to the eventual clearance of HBV by cytolytic and/or noncytolytic mechanisms. The murine immune system is apparently capable of both modes of immune control because they have both been induced in the transgenic mouse model (37, 39). In the case of the hydrodynamic transfection model, compression of the immune response to 2 weeks may prevent detection of two kinetically distinct stages corresponding to the innate and adaptive immune responses observed in the chimpanzee. Our experiments using the hydrodynamic transfection model do demonstrate that a virus-specific CTL response occurs. The relationship between the HBV-specific CTL response and viral clearance remains to be determined, as do the existence and functional significance of any innate immune response. Fortunately, the hydrodynamic model lends itself easily to analysis in genetically manipulated mice and the importance of the CTL response as well as the presence and the importance of any innate immune response can all be tested.
The hydrodynamic injection of mutant viral genomes will also facilitate the study of HBV gene regulation and replication. Furthermore, the hydrodynamic transfection model may be useful in directed evolution experiments in which random mutagenesis is used to generate libraries of HBV variants that are then subjected to selective pressure in vivo in order to map regulatory and structural elements or to identify variants with altered function. This approach might also be useful in developing new model systems. Libraries of HBV variants mutagenized in the envelope genes might yield viral genomes that are adapted to the mouse.
Although we have reported the application of hydrodynamic transfection to the study of HBV, this methodology may be useful in the study of other liver-specific pathogens, as has been shown by Taylor and colleagues for hepatitis delta virus (40). This and other replicating viral genomes may be probed to identify the structural and functional determinants mediating replication, and viral antigen genes can be expressed to characterize the immunobiology of nonreplicating viruses and other liver-specific pathogens with this system. Finally, the tissue tropism of viruses might also be probed by using hydrodynamic transfection. This method may especially prove valuable in the characterization of newly discovered viruses as long as they have the potential to replicate in the liver.
Supplementary Material
Acknowledgments
We are grateful to Mark Kay for his generous gift of the pT-MCS and pCMV-SB plasmids and to K. Kakimi, A. Martin, C.Y. Cho, and N. S. Gray for stimulating discussions and helpful comments on the manuscript. We also thank M. Chadwell and S. Wieland for assistance with immunohistochemical staining and quantititative PCR, respectively, and C. Padilla for assistance with manuscript preparation. This is manuscript 14521-MEM of The Scripps Research Institute. This work was supported by National Institutes of Health Grants CA40489 and CA76403 (to F.V.C.). P.L.Y. was supported by National Institutes of Health Training Grant AI07354-10.
Abbreviations
- HBV
hepatitis B virus
- CTL
cytotoxic T lymphocyte
- sALT
serum alanine aminotransaminase
- HBsAg
HBV surface antigen
- HBeAg
HBV e antigen
- HBcAg
HBV core antigen
- HBs
HBV small envelope protein
- cccDNA
covalently closed circular DNA
- NK
natural killer
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Ganem D, Varmus H E. Annu Rev Biochem. 1987;56:651–693. doi: 10.1146/annurev.bi.56.070187.003251. [DOI] [PubMed] [Google Scholar]
- 2.Beasley R P, Hwang L Y, Lin C C, Chien C S. Lancet. 1981;2:1129–1133. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 3.Mason W S, Seal G, Summers J. J Virol. 1980;36:829–836. doi: 10.1128/jvi.36.3.829-836.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Summers J, Smolec J M, Snyder R. Proc Natl Acad Sci USA. 1978;75:4533–4537. doi: 10.1073/pnas.75.9.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker L F, Chisari F V, McGrath P P, Dalgard D W, Kirschstein R L, Almeida J D, Edington T S, Sharp D G, Peterson M R. J Infect Dis. 1973;127:648–662. doi: 10.1093/infdis/127.6.648. [DOI] [PubMed] [Google Scholar]
- 6.Bertoni R, Sette A, Sidney J, Guidotti L G, Shapiro M, Purcell R, Chisari F V. J Immunol. 1998;161:4447–4455. [PubMed] [Google Scholar]
- 7.Guidotti L G, Rochford R, Chung J, Shapiro M, Purcell R, Chisari F B. Science. 1999;284:825–829. doi: 10.1126/science.284.5415.825. [DOI] [PubMed] [Google Scholar]
- 8.Chisari F V. Mol Biol Med. 1989;6:143–149. [PubMed] [Google Scholar]
- 9.Chisari F V. Curr Top Microbiol Immunol. 1991;168:85–101. doi: 10.1007/978-3-642-76015-0_5. [DOI] [PubMed] [Google Scholar]
- 10.Chisari F V. Curr Top Microbiol Immunol. 1996;206:149–173. doi: 10.1007/978-3-642-85208-4_9. [DOI] [PubMed] [Google Scholar]
- 11.Mandart E, Kay A, Galibert F. J Virol. 1984;49:782–792. doi: 10.1128/jvi.49.3.782-792.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson W S. Ann NY Acad Sci. 1980;354:371–378. doi: 10.1111/j.1749-6632.1980.tb27978.x. [DOI] [PubMed] [Google Scholar]
- 13.Di Q, Summers J, Burch J B, Mason W S. Virology. 1997;229:25–35. doi: 10.1006/viro.1996.8422. [DOI] [PubMed] [Google Scholar]
- 14.Liu F, Song Y, Liu D. Gene Ther. 1999;6:1258–1266. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- 15.Guidotti L G, Matzke B, Schaller H, Chisari F V. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raney A K, Kline E F, Tang H, McLachlan A. J Virol. 2001;75:2900–2911. doi: 10.1128/JVI.75.6.2900-2911.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Y Y, Summers J. J Virol. 2000;74:5257–5265. doi: 10.1128/jvi.74.11.5257-5265.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hobbs M V, Weigle W O, Noonan D J, Torbett B E, McEvilly R J, Koch R J, Cardenas G J, Ernst D N. J Immunol. 1993;150:3602–3214. [PubMed] [Google Scholar]
- 19.Chisari F V, Filippi P, Buras J, McLachlan A, Popper H, Pinkert C A, Palmiter R D, Brinster R L. Proc Natl Acad Sci USA. 1987;84:6909–6913. doi: 10.1073/pnas.84.19.6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yant S R, Meuse L, Chiu W, Ivics Z, Izsvak Z, Kay M A. Nat Genet. 2000;25:35–41. doi: 10.1038/75568. [DOI] [PubMed] [Google Scholar]
- 21.Liu D, Knapp J E. Curr Opin Mol Ther. 2001;3:192–197. [PubMed] [Google Scholar]
- 22.Ibrahim A B, Vyas G N, Perkins H A. Infect Immun. 1975;11:137–141. doi: 10.1128/iai.11.1.137-141.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aldershvile J, Dietrichson O, Hardt F, Nielsen J O, Skinhoj P. Scand J Gastroenterol. 1977;12:917–922. doi: 10.3109/00365527709181350. [DOI] [PubMed] [Google Scholar]
- 24.Aldershvile J, Nielsen J O. J Virol Methods. 1980;2:97–105. doi: 10.1016/0166-0934(80)90043-9. [DOI] [PubMed] [Google Scholar]
- 25.McMahon B J, Bender T R, Berquist K R, Schreeder M T, Harpster A P. J Clin Microbiol. 1981;14:130–134. doi: 10.1128/jcm.14.2.130-134.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salberg M, Norder H, Weiland O, Magnius L. J Med Virol. 1989;29:296–302. doi: 10.1002/jmv.1890290414. [DOI] [PubMed] [Google Scholar]
- 27.Vento S, Rondanelli E G, Ranieri S, O'Brien C J, Williams R, Eddleston A L. Lancet. 1987;2:119–122. doi: 10.1016/s0140-6736(87)92329-4. [DOI] [PubMed] [Google Scholar]
- 28.Milich D R, Sallberg M, Maruyama T. Springer Semin Immunopathol. 1995;17:149–166. doi: 10.1007/BF00196163. [DOI] [PubMed] [Google Scholar]
- 29.Chisari F V. J Clin Invest. 1997;99:1472–1477. doi: 10.1172/JCI119308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhrober A, Wild J, Pudollek H P, Chisari F V, Reimann J. Int Immunol. 1997;9:1203–1212. doi: 10.1093/intimm/9.8.1203. [DOI] [PubMed] [Google Scholar]
- 31.Shultz L D, Schweitzer P A, Christianson S W, Gott B, Schweitzer I B, Tennent B, McKenna S, Mobraaten L, Rajan T V, Greiner D L, et al. J Immunol. 1995;154:180–191. [PubMed] [Google Scholar]
- 32.Hirsch R C, Lavine J E, Chang L J, Varmus H E, Ganem D. Nature. 1990;344:552–555. doi: 10.1038/344552a0. [DOI] [PubMed] [Google Scholar]
- 33.Salazar-Mather T P, Orange J S, Biron C A. J Exp Med. 1998;187:1–14. doi: 10.1084/jem.187.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brown M G, Dokun A O, Heusel J W, Smith H R, Beckman D L, Blattenberger E A, Dubbelde C E, Stone L R, Scalzo A A, Yokoyama W M. Science. 2001;292:934–937. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- 35.Biron C A, Brossay L. Curr Opin Immunol. 2001;13:458–464. doi: 10.1016/s0952-7915(00)00241-7. [DOI] [PubMed] [Google Scholar]
- 36.Pasquetto V, Guidotti L G, Kakimi K, Tsuji M, Chisari F V. J Exp Med. 2000;192:529–536. doi: 10.1084/jem.192.4.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kakimi K, Guidotti L G, Koezuka Y, Chisari F V. J Exp Med. 2000;192:921–930. doi: 10.1084/jem.192.7.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guidotti L G, Chisari F V. Annu Rev Immunol. 2001;19:65–91. doi: 10.1146/annurev.immunol.19.1.65. [DOI] [PubMed] [Google Scholar]
- 39.Guidotti L G, Ishikawa T, Hobbs M V, Matzke B, Schreiber R, Chisari F V. Immunity. 1996;4:25–36. doi: 10.1016/s1074-7613(00)80295-2. [DOI] [PubMed] [Google Scholar]
- 40.Chang J, Sigal L J, Lerro A, Taylor J. J Virol. 2001;75:3469–3473. doi: 10.1128/JVI.75.7.3469-3473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.