Abstract
Host recognition of bacterial pathogens is a critical component of the immune response. Intracellular bacterial pathogens are able to evade the humoral immune system by residing within the host cell. Here we show the existence of an innate host surveillance mechanism in macrophages that specifically distinguishes bacteria in the cytosol from bacteria in the vacuole. Recognition of Gram-positive and Gram-negative bacterial products by this surveillance system results in transcription of the ifnb gene. The activation of cytosol-specific signaling is associated with translocation of NF-κB into the nucleus and phosphorylation of the p38 mitogen-activated protein (MAP) kinase. Activation of the p38 kinase is required for the induction of gene expression by the cytosolic surveillance pathway. Our studies suggest that infection by intracellular bacterial pathogens results in an immune response distinct from that of infection by extracellular bacterial pathogens.
Recognition of microbial patterns by an infected host is a critical determinant of an effective innate and adaptive immune response. Detection of bacterial pathogens is mediated by cell surface receptors that include the Toll-like receptors (TLRs), an evolutionarily conserved family of proteins that bind a diverse array of microbial ligands (1, 2). Recognition of multiple ligands from the same class of pathogen, like lipopolysaccharide (LPS), peptidoglycan, and flagellin from bacteria, may provide an important alternative route of activation should one of the ligands fail to act as an agonist. However, the current model of innate immune detection of bacterial pathogens focuses on extracellular bacteria and does not provide for specific recognition within a host cell.
Intracellular pathogens occupy a niche in the infected host that protects them from some immune effectors such as antibodies. The innate immune response to viruses, which are obligate intracellular pathogens, has been extensively characterized. One of the best studied transcriptional responses to viral infection is that of the gene ifnb, which encodes β-interferon (β-IFN) (3). β-IFN has pleiotropic autocrine and paracrine functions that impact the immune response, including up-regulation of the major histocompatibility complex (MHC) genes (4). In contrast, little is known about how the host may specifically recognize intracellular bacteria or what response they induce. Recent studies in epithelial cells have shown that an invasive strain of Shigella flexneri, a Gram-negative intracellular pathogen, activates nuclear factor κB (NF-κB) DNA-binding activity and subsequently, il8 gene expression, whereas a strain that remains extracellular does not (5). LPS microinjected into epithelial cells also resulted in the translocation of NF-κB. These data suggest the existence of an intracellular surveillance mechanism for LPS.
Listeria monocytogenes, a Gram-positive pathogen, is an ideal organism for the study of innate and adaptive immunity to intracellular pathogens, as there are well-characterized strains that reside in different cellular compartments. L. monocytogenes can enter host cells through passive uptake by phagocytic cells such as macrophages or by active invasion of nonphagocytic cells (6, 7). After internalization, L. monocytogenes is initially found in a membrane-bound vacuole. Escape from this vacuole into the host cytosol requires one or more bacterial virulence factors, primarily the pore-forming hemolysin, listeriolysin O (LLO; encoded by the hly gene) (8). Two bacterial phospholipases C (PLCs) also mediate escape depending on the nature of the infected cell (9, 10). Infected cells can express a variety of cytokines, including β-IFN, which alert the host immune system to the presence of an infectious agent. Macrophages play an especially important role in promoting the differentiation of immune effector cells; in general, the response of the first infected cells in a host organism shapes subsequent innate and adaptive immunity to L. monocytogenes (11).
Here we show that macrophages and epithelial cells recognize bacterial products in the host cytosol, leading to the activation of ifnb gene expression. This surveillance pathway can distinguish between nonpathogenic bacteria within a vacuole and bacteria in the cytosol, even in the presence of external bacterial-pattern-recognition receptors. Both Gram-positive and Gram-negative bacteria are sensed, resulting in the nuclear translocation of NF-κB and the phosphorylation of p38 mitogen-activated protein kinase (MAP) kinase. Inhibition of p38 phosphorylation suppresses the induction of target gene expression by the cytosolic surveillance pathway.
Materials and Methods
Bacterial Strains and Extracts.
The L. monocytogenes strains used were a wild-type strain, 10403S, or strains containing in-frame deletions of the hly gene (LLO−, DP-L2161) or the hly, plcA, and plcB genes (LLO−PLC−; DP-L2319). Single colonies were inoculated into 2 ml of BHI (brain–heart infusion) and incubated overnight at 30°C without shaking. Bacillus subtilis strains DP-B1066 (B.s.−LLO) and DP-B980 (B.s.+LLO) were generated and cultured as described (12). Escherichia coli K-12 strains DP-E3616 and DP-E3617 were generated and cultured as described (13).
Cell Culture, Infections, and Liposome Delivery.
HeLa cells were infected at a multiplicity of infection (moi) of approximately 150:1 for 60 min, resulting in an infection of 30–40%. After 60 min the monolayer was washed three times with PBS and fresh medium was added. Gentamicin was added to 50 μg/ml at 1.5 h post infection (h.p.i.) to limit the growth of extracellular bacteria. Where indicated, cycloheximide was added to infected cells at 1.5 h.p.i. to a concentration of 22.5 μg/ml to prevent postinduction transcriptional repression of the ifnb locus in HeLa cells (14). Where indicated, 10 μM SB202190 (Calbiochem), a p38 MAP kinase inhibitor, or 10 μM U0126 (Cell Signaling), a MEK-1/2 inhibitor, was used to treat cells 30 min before and for the duration of infection. The inhibitors were dissolved in DMSO. Poly(I)⋅poly(C) (100 μg/ml; Sigma) and anisomycin (30 μg/ml; Sigma) were used as positive controls for ifnb induction and p38 MAP kinase activation, respectively.
Primary bone marrow-derived macrophages (BMDM) were isolated from female mice of the strain/genotype indicated (The Jackson Laboratory) and cultured as described (8). Macrophages were infected at a moi of ≈4:1 for 30 min, resulting in an ≈99% infection rate with 1–5 bacteria per macrophage. After 30 min, the monolayer was washed three times with PBS and fresh medium was added. At 1 h.p.i., gentamicin was added to 50 μg/ml.
Liposomes were generated as described by using recombinant LLO produced in E. coli (15). The LPS used was derived from E. coli strain O111:B4 (Sigma) and the lipoteichoic acid (LTA) derived from B. subtilis (Sigma). Liposome preparations were added to macrophages at a 1:100 dilution for 1 h. After 1 h, the monolayer was washed 3 times with PBS and fresh medium was added. Liposome-treated macrophages were harvested at 6 h.p.i. for RT-PCR analysis or used to assay antigen presentation (13, 16).
Semiquantitative RT-PCR Analysis.
RNA was harvested from infected or treated cells at 6 h.p.i. with the RNeasy Mini kit (Qiagen, Valencia, CA). The RNA recovered was quantitated and used in a reverse transcriptase (RT) reaction with Moloney murine leukemia virus (MMLV) RT (Invitrogen). cDNA from the RT reaction was used for PCR amplification. Samples were removed from PCRs at varying cycle number, e.g., 25, 27, or 30 cycles, to ensure amplification in a linear range. Sequences for primers used were: β-actin F (5′-TGGCATTGTTACCAACTGGGACG), β-actin R (5′-GCTTCTCTTTGATGTCACGCACG), murine ifnb F (5′-GCACTGGGTGGAATGAGACTATTG), murine ifnb R (5′-TTCTGAGGCATCAACTGACAGGTC), human ifnb F (5′-GCTCTCCTGTTGTGCTTCTCCAC), human ifnb R (5′-CAATAGTCTCATTCCAGCCAGTGC).
Western Blotting.
Whole cell lysates were generated by adding SDS/PAGE sample buffer directly to cell monolayers. Lysates were boiled for 5 min and clarified by centrifugation at 10,000 × g for 5 min before SDS/PAGE. Proteins were transferred from the polyacrylamide gel to nitrocellulose and incubated with antibodies specific for p38 MAP kinase or phospho-p38 MAP kinase (1:1,000; Cell Signaling), followed by a horseradish peroxidase-conjugated goat anti-rabbit antibody (1:2,000; Amersham Pharmacia).
Immunofluorescence.
Cells were grown and treated on 18-mm2 glass coverslips, then fixed in 3.7% paraformaldehyde. Fixed coverslips were incubated with primary antibodies specific for NF-κB p65 (1:200; Santa Cruz Biotechnology) or L. monocytogenes (1:200; Difco), then incubated with a FITC-conjugated secondary antibody (1:100; Jackson ImmunoResearch) and rhodamine-phalloidin (1:1,000; Molecular Probes).
Results
Presence of L. monocytogenes in the Cytosol, but Not in the Vacuole, Activates Transcription of ifnb in Primary Murine BMDM.
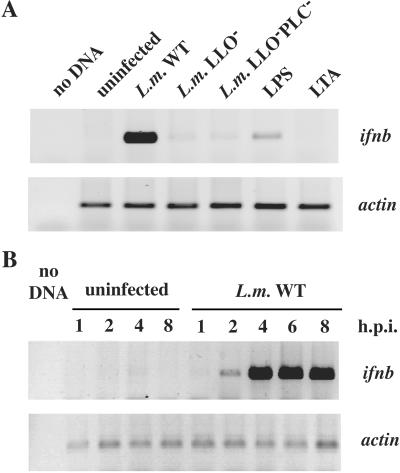
To investigate whether host cells could specifically detect the presence of L. monocytogenes in the cytosol, we infected primary murine C57BL/6 BMDM with wild-type L. monocytogenes, which are able to access the cytosol, or a LLO-deficient (LLO−) L. monocytogenes strain, that remains trapped in the vacuole. The moi was such that the majority of macrophages were infected with 1 or more bacteria. Localization of L. monocytogenes in the cytosol was determined by rhodamine-phalloidin staining of L. monocytogenes-associated F-actin clouds and tails, which are present only when bacteria are in the cytosol (17). RNA was harvested at 6 h.p.i. and analyzed by RT-PCR. The ifnb gene was induced in macrophages infected by the wild-type L. monocytogenes strain in the cytosol, but not by the LLO− strain (Fig. 1A). Differential induction of β-IFN was also observed at the protein level (E. Havell and D.A.P., unpublished observations). Because the wild-type L. monocytogenes strain grows rapidly in the cytosol, whereas the LLO− strain is unable to grow in the restrictive environment of the vacuole, the differential induction of ifnb at 6 h.p.i. could be because of larger numbers of wild-type bacteria present in the infected host cells. To address this possibility, we analyzed ifnb expression at 1, 2, 4, 6, and 8 h.p.i. (Fig. 1B). ifnb cDNA was first observed at 2 h.p.i. when the numbers of wild-type and LLO− mutant bacteria are very similar, suggesting that differential ifnb expression is not because of a larger bacterial load in cells infected by the wild-type strain. Taken together, these data show that the presence of L. monocytogenes in the cytosol triggers a specific host response in an infected cell.
Figure 1.
Infection of BMDM by wild-type L. monocytogenes, but not an LLO− strain, results in up-regulation of ifnb gene expression. (A) C57BL/6 macrophages were infected at a moi resulting in 1–5 bacteria per cell with L. monocytogenes or treated with E. coli LPS (1 ng/ml), or B. subtilis LTA (1 μg/ml). RNA was isolated from macrophage monolayers at 6 h.p.i. and analyzed by RT-PCR. Actin reactions shown were amplified for 15 cycles; ifnb reactions shown were amplified for 30 cycles. (B) C57BL/6 macrophages were infected as described above and RNA was isolated from macrophage monolayers. Actin reactions shown were amplified for 15 cycles; ifnb reactions shown were amplified for 30 cycles.
Induction of the Cytosol-Specific Host Response by L. monocytogenes Is Independent of LLO.
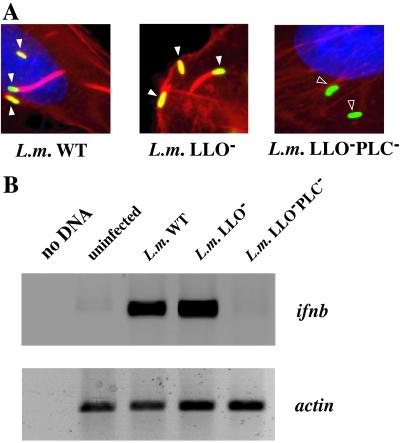
The presence of L. monocytogenes in the cytosol could result in host gene expression through the signaling activity of a bacterial virulence factor such as LLO. To test whether LLO contributes to cytosolic signaling, we examined target gene expression in infected HeLa cells. In contrast to macrophages, both wild-type and LLO− strains of L. monocytogenes can escape from the vacuole in HeLa cells, but a strain that is deficient in LLO and the two L. monocytogenes PLCs (LLO−PLC−) does not, as shown by the inability of the LLO−PLC− strain to nucleate host actin (Fig. 2A). RT-PCR analysis of HeLa cells infected with wild-type and mutant strains of L. monocytogenes revealed that ifnb transcription was induced only by those strains that escaped into the cytosol, not by the LLO−PLC− mutant strain (Fig. 2B). The il8 gene was regulated in the same manner as ifnb (data not shown), suggesting that there is a program of gene expression regulated by the presence of L. monocytogenes in the cytosol. As both the wild-type and the LLO− bacteria were able to activate ifnb and il8, we conclude that the cytosol-specific response observed in macrophages also functions in epithelial cells, and that LLO is not required.
Figure 2.
LLO is not required for ifnb gene expression in HeLa cells. (A) Indirect immunofluorescence was performed on HeLa cells at 4 h.p.i. Coverslips were incubated with a polyclonal antibody directed against L. monocytogenes and rhodamine-phalloidin. White arrowheads mark bacteria colocalizing with F-actin; open arrowheads mark bacteria that are not colocalized with F-actin. (B) HeLa cells were infected with L. monocytogenes and treated with cycloheximide at 1.5 h.p.i. RNA was isolated at 6 h.p.i. and subjected to RT-PCR analysis. Actin reactions shown were amplified for 15 cycles; ifnb reactions shown were amplified for 30 cycles.
Both Gram-Positive and Gram-Negative Nonpathogenic Bacteria Can Activate Cytosol-Specific Gene Expression.
As LLO itself did not activate the expression of ifnb, other molecules common to many bacteria, such as LTA or LPS, might be the ligand(s) detected by proteins in the host cytosol. We used two methods to address the possibility that bacterial molecular patterns, rather than specific L. monocytogenes virulence factors, might trigger a cytosol-specific response: cytosolic delivery of genetically engineered strains of nonpathogenic bacteria, or cytosolic delivery of microbial products by liposomes.
We first tested a Gram-positive nonpathogenic bacterium, B. subtilis, and a Gram-negative nonpathogenic bacterium, E. coli strain K-12, engineered to express LLO. The B. subtilis strain with an isopropyl β-d-thiogalactoside (IPTG)-inducible hly gene (B.s.+LLO) secretes LLO, and can escape from the vacuole into the cytosol, where limited growth occurs (12). The control B. subtilis strain (B.s.−LLO) remains trapped in the vacuole. The E. coli strain (E.c.+LLO) containing the hly gene constitutively expresses a form of LLO lacking the signal sequence so that the LLO protein remains inside the bacteria (13). Both E.c. + LLO and the control E. coli strain (E.c.−LLO) also express the T cell antigen ovalbumin (OVA) under an IPTG-inducible promoter as a reporter protein. The presence of OVA in the cytosol of the macrophage is assayed by antigen presentation of OVA on MHC class I molecules which sample peptides from the cytosol (16). The E. coli phagocytosed by macrophages are lysed in the phagosome and release OVA or OVA+LLO into the vacuole. OVA and molecules from the lysed bacteria are released into the cytosol only when LLO is present as assayed by antigen presentation of OVA (data not shown).
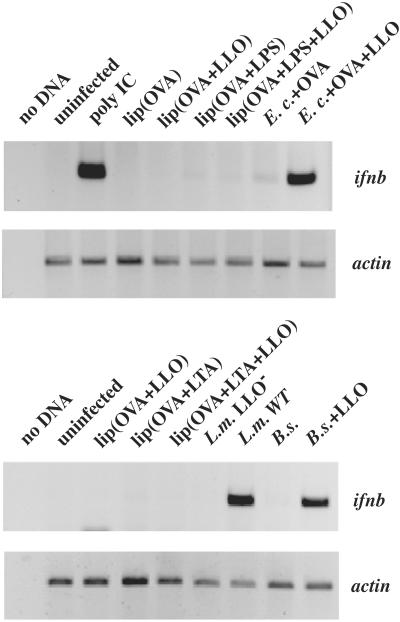
To determine whether cytosolic recognition was specific to L. monocytogenes, we exposed BMDM to B.s.+LLO or E.c.+LLO, or control strains, at a moi resulting in the majority of macrophages being infected by 1–5 bacteria. Because macrophages up-regulate ifnb in response to LPS from Gram-negative bacteria through TLR4, we used BMDM from C3H/HeJ mice that harbor a mutation in the tlr4 gene rendering these cells insensitive to extracellular LPS (18). LTA from Gram-positive bacteria added extracellularly does not up-regulate ifnb in BMDM from wild-type mice. As shown by RT-PCR analysis, macrophages that phagocytosed either B.s.+LLO or E.c.+LLO up-regulated expression of ifnb, whereas macrophages that phagocytosed the control strains, B.s.−LLO or E.c.−LLO, did not (Fig. 3).
Figure 3.
E. coli and B. subtilis in the cytosol of macrophages up-regulate ifnb expression, but LPS and LTA do not. RT-PCR analysis was performed on C3H/HeJ BMDM treated with liposomes, labeled as lip(), or infected with the E. coli, B. subtilis, or L. monocytogenes strains indicated. Macrophages treated with poly(I)⋅poly(C) (100 μg/ml) were isolated as a positive control for ifnb induction. RNA was isolated at 6 h.p.i. or 6 h post treatment and analyzed by RT-PCR. Actin reactions shown were amplified for 15 cycles; ifnb reactions shown were amplified for 30 cycles.
LTA from Gram-positive bacteria and LPS from Gram-negative bacteria are recognized by the TLR family (18, 19). To determine whether LTA and LPS are also ligands recognized by the cytosolic surveillance pathway, we delivered B. subtilis LTA or E. coli LPS to macrophages in acid-labile liposomes containing OVA in addition to the LTA or LPS. These liposomes are taken up by macrophages as particles and release their contents into the phagosome on acidification (15). The presence of OVA in the cytosol of the macrophage was determined by antigen presentation of OVA on MHC class I. Liposome contents were released into the cytosol only when LLO was contained in the liposomes (data not shown).
Liposomes were added to primary C3H/HeJ BMDM cultures for 1 h to allow phagocytosis to occur. These cultures were either assayed for antigen presentation to test for the presence of OVA in the cytosol, or harvested at 6 h.p.i. to analyze gene expression by RT-PCR. Neither LTA nor LPS in liposomes containing LLO was sufficient to induce ifnb expression (Fig. 3). The LTA- and LPS-containing liposomes were able to activate TLRs, assayed by induction of il12 expression in C57BL/6 BMDM independent of the presence of LLO, indicating that the material in the liposomes was active and in a recognizable form (data not shown). Using an LLO-independent method, scrape-loading of bacterial products into HeLa cells, yielded similar results. Scrape-loading relies on temporary disruption of host cell membranes on scraping of an established monolayer to allow access of macromolecules into the host cytosol (20). Scrape-loading LTA or LPS at concentrations that activate signaling through TLRs in macrophages did not result in cytokine induction in HeLa cells, whereas scrape-loaded E. coli or B. subtilis extracts strongly induced cytokine gene expression (data not shown). We conclude from these data that the cytosolic surveillance pathway can recognize molecules from either Gram-positive or Gram-negative bacteria, but not LTA or LPS. Furthermore, these ligands are likely to be present in all bacteria, not just pathogens, as activation of gene expression occurred when nonpathogenic bacteria or products from lysed bacteria were delivered to the cytosol.
NF-κB Translocates to the Nucleus When Bacterial Products Are Present in the Host Cytosol.
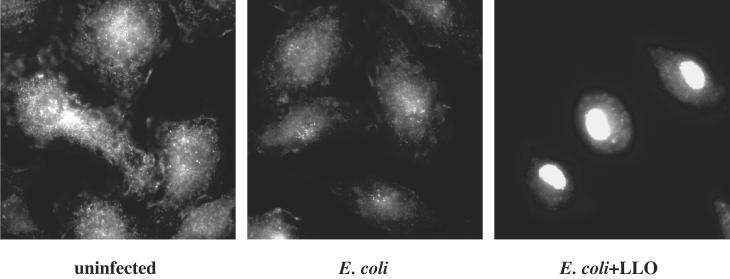
Our data show that bacterial products in the host cytosol lead to the up-regulation of target gene expression. We next investigated which host-signaling molecules might be involved in the activation of the cytosolic surveillance pathway. NF-κB is a transcription factor that regulates the expression of many cytokine genes, including ifnb and il8 (21, 22). To test whether NF-κB was activated by the presence of bacterial products in the cytosol, we looked for nuclear translocation of the p65 subunit of the NF-κB heterodimer by indirect immunofluorescence in cells that contained bacteria in the vacuole or the cytosol. C3H/HeJ macrophages were exposed to E.coli−LLO or E.coli+LLO at a moi of 5:1, resulting in the uptake of 1–5 bacteria by all macrophages observed. At 1.5 h.p.i. macrophages that had taken up the E. coli expressing LLO uniformly exhibited translocation of p65 to the nucleus (Fig 4). p65 remained in the cytosol in approximately 90% of macrophages treated with the control E. coli strain. Translocation of p65 in the remaining 10% of infected macrophages may have been because of activation of other TLRs by bacterial products from the lysed E. coli such as bacterial DNA. We conclude from these data that NF-κB translocates to the nucleus when bacterial products are released into the cytosol of macrophages.
Figure 4.
NF-κB p65 is translocated to the nucleus when bacterial products are present in the cytosol of macrophages. Indirect immunofluorescence was performed on C3H/HeJ BMDM infected with E. coli expressing OVA with or without LLO by using monoclonal antibody specific for p65 (1:200) and subsequently with a FITC-conjugated secondary goat anti-mouse antibody (1:100).
The MAP Kinase p38 Is Phosphorylated in Response to Cytosolic Bacteria.
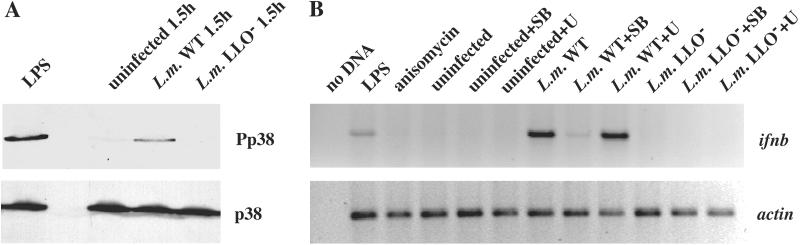
The p38 MAP kinase regulates the expression of ifnb in response to extracellular recognition of LPS by macrophages (23). To determine whether p38 MAP kinase is activated by the presence of bacteria in the cytosol, we examined the phosphorylation state of p38 MAP kinase in BMDM infected by wild-type L. monocytogenes or the LLO− strain. Western blot analysis using an antibody against the phosphorylated form of p38 indicated that at 1.5 h.p.i., p38 MAP kinase was phosphorylated in cells infected by the wild-type strain that escaped into the cytosol, but not in cells infected by the LLO− strain which remained in the vacuole (Fig. 5A). The numbers of bacteria present in cells infected by wild-type L. monocytogenes compared with the mutant strains were similar when p38 phosphorylation was observed, indicating that activation of p38 MAP kinase is not because of the differential ability of the two strains to replicate. p38 phosphorylation was also observed in HeLa cells infected with wild-type L. monocytogenes, but not the LLO−PLC− strain (data not shown). These data suggest that p38 MAP kinase is a regulator of the cytosolic surveillance pathway.
Figure 5.
The p38 MAP kinase is phosphorylated when L. monocytogenes is present in the cytosol. (A) C57BL/6 BMDM were infected with wild-type or the LLO− strain of L. monocytogenes or treated with 1 μg/ml LPS as a positive control. Cells were harvested at 1.5 h.p.i. and subjected to SDS/PAGE and Western blot analysis. A phospho-specific antibody was used to detect the phosphorylated form of p38 (Pp38). An antibody against p38 was used to detect total cellular pools of p38 by using the same whole cell lysates. (B) C57BL/6 BMDM were pretreated with p38 inhibitor, SB202190 (SB), or MEK-1 inhibitor, U0126 (U), for 30 min before infection by L. monocytogenes, or treated with LPS (1 ng/ml) or anisomycin (30 μg/ml). RNA was isolated at 6 h.p.i. or 6 h post treatment and analyzed by RT-PCR. Actin reactions shown were amplified for 15 cycles; ifnb reactions shown were amplified for 30 cycles.
The p38 MAP Kinase Is Necessary for Induction of Target Gene Expression by the Cytosolic Surveillance Pathway.
While p38 phosphorylation occurs when bacteria are in the cytosol, we wanted to test the role of p38 activation in the induction of gene expression. We infected BMDM with either wild-type L. monocytogenes or the LLO− mutant strain. Infected cells were pretreated with either DMSO as a negative control, an inhibitor of p38 MAP kinase, SB202190 (SB), or an inhibitor of MEK-1 and MEK-2, U0126 (U), which prevents the phosphorylation of the related MAP kinases, ERK-1 and ERK-2. The inhibitors were present throughout the infection and no loss in macrophage viability or decrease in bacterial replication was observed. At 6 h.p.i., RNA was harvested from the infected macrophages and analyzed by RT-PCR (Fig. 5B). Infection by the LLO− mutant strain did not activate ifnb gene expression, consistent with our previous experiments. Untreated macrophages infected by the wild-type L. monocytogenes strain did up-regulate ifnb expression, but when macrophages were treated with SB202190, ifnb induction was significantly reduced. In contrast, treatment with U0126 did not suppress activation of ifnb expression by wild-type L. monocytogenes. We therefore conclude that activation of p38 MAP kinase is required for induction of gene expression in response to bacteria in the cytosol.
Discussion
We have shown that an innate host surveillance mechanism in macrophages can distinguish between the presence of bacteria in the cytosol and bacteria trapped in a vacuole. Signaling through this intracellular bacterial recognition pathway up-regulates ifnb expression, and both Gram-positive and Gram-negative bacteria in the cytosol are recognized. On entry of bacteria into the cytosol, p38 MAP kinase is phosphorylated and NF-κB p65 translocates into the nucleus. The cytosolic surveillance pathway requires p38 MAP kinase activity for activation of gene expression. Taken together, these data suggest that infection by intracellular bacterial pathogens stimulates an overlapping but distinct innate immune response to extracellular bacteria.
Our studies focus on the host cell response to intracytosolic bacteria, and we hypothesize that cytosolic surveillance is important in promoting the appropriate immune response for the resolution of infection by intracellular pathogens. We note that many intracellular pathogens, such as Mycobacterium tuberculosis and Salmonella enterica subspecies typhimurium, reside in modified vacuoles within infected host cells (24). Interestingly, macrophages infected with bacillus Calmette–Guérin (bacille Calmette–Guérin; BCG) exhibit permeability of the macrophage phagosome, suggesting that bacillus Calmette–Guérin modifies the phagosome, perhaps to facilitate nutrient exchange (25). Other bacterial mechanisms of macromolecular transport such as the type III secretion system also provide exchange between the vacuole and the host cytosol (26). Nutrient exchange mechanisms or protein translocation machinery may allow access of bacterial ligands into the cytosol, where they would be exposed to the cytosolic surveillance pathway. This hypothesis is consistent with previous studies showing that virulent strains of Chlamydia and S. enterica subspecies typhimurium can up-regulate ifnb or il8 expression (27, 28).
The identity of the bacterial ligand(s) recognized by the cytosolic surveillance pathway remains to be determined. Recent studies have shown that high concentrations of LPS, i.e., 1 mg/ml, microinjected into the cytosol trigger NF-κB translocation, suggesting that LPS is recognized by an intracellular receptor (5, 29). Although our results show a transcriptional response to Gram-positive bacteria and Gram-negative bacteria, surprisingly we do not observe target gene induction in response to LTA or LPS. Ongoing studies in our laboratory appear to exclude proteins, DNA, and RNA as possible ligands (M.O., C.H.Y., and D.A.P., unpublished observations), leading us to hypothesize that the ligand, or ligands, may be a species of carbohydrate. There may be multiple ligands, as is the case with the TLRs, which recognize many different bacterially derived molecules.
A host cell mechanism that senses bacteria in the cytosol predicts the existence of a cytosolic receptor(s) to recognize bacterial ligands. A recently identified family of mammalian genes, the Nod genes, has been implicated in the signal transduction pathway of the intracellular LPS recognition mechanism (30, 31). The nucleotide-binding oligomerization domain (NOD) proteins are intriguing candidates for regulators of innate immunity as they bear striking similarity in domain structure to a large family of plant proteins, the R proteins, that mediate resistance to phytopathogens. Overexpression of NOD1 and NOD2 in transfected cells leads to activation of NF-κB-dependent transcription in response to LPS (31). Although there is no clear evidence showing that the NOD proteins directly bind LPS, it is possible that the NOD proteins may act as intracellular receptors for LPS or other bacterial ligands. As BMDM are difficult to transfect, we were unable to determine the function of the NOD proteins in the macrophage cytosolic surveillance pathway. Whether or not the NOD proteins act directly as intracellular receptors for bacterial products, it is likely that NODs are regulators of cytosolic surveillance. Further studies will be required to clearly establish the identity of the cytosolic receptors for intracellular bacterial recognition.
The activation of both the NF-κB and MAP kinase pathways appears to be important in the induction of the cytosol-specific host response. NF-κB is a known transcriptional activator of many inflammatory response genes, including ifnb, and probably performs a similar regulatory function in the response to bacteria in the cytosol. Our results show that p38 MAP kinase plays a critical role in the response to cytosolic bacterial products. These data are consistent with a recently published study identifying the p38 MAP kinase pathway in Caenorhabditis elegans as an important regulatory pathway in the innate immune response of nematodes (32). The mechanistic requirement for p38 MAP kinase in the cytosolic surveillance pathway is not clear. In the macrophage response to LPS through TLR4, p38 is thought to regulate the expression of ifnb, through phosphorylation and activation of the transcription factor IFN regulatory factor-3 (23). Recent studies also suggest that through phosphorylation of histones, p38 regulates accessibility of DNA-binding sites for NF-κB at some promoters (33). The induction of target gene expression may require p38 for the function of multiple transcription factors that participate in higher-order nucleoprotein complexes such as that described for the ifnb enhancer.
The nature of the initial host response to intracellular infection is critical in determining subsequent innate and adaptive immunity. Our studies clearly show that recognition of cytosolic bacterial ligands is distinct from signaling that occurs on binding of extracellular bacterial products. Macrophages recognize extracellular bacterial ligands and express a profile of cytokines that stimulates chemotaxis and the differentiation of other mediators of the inflammatory response. In addition, we now show that the cytosolic surveillance pathway in macrophages appears to specifically up-regulate expression of ifnb. ifnb likely represents one gene in a transcriptional program that is induced by the presence of bacteria in the cytosol. Studies of global gene expression in macrophages by cDNA microarray analysis should reveal the extent of the cytosol-specific transcription response and suggest which signaling pathways are involved in the recognition of intracellular pathogens.
In conclusion, our data demonstrate that infected host cells can distinguish between bacteria trapped in unmodified vacuoles and bacteria that escape into the cytosol. Although the mechanism of recognition remains to be determined, an intracellular surveillance system for bacterial pathogens would allow the infected host to integrate multiple signals from outside and inside the cell to identify the exact nature of the invading organism, whether it is viral, bacterial, or fungal, intracellular or extracellular. Such distinctions are likely to be important in determining both the nature of the host immune response and the resolution of infection.
Acknowledgments
We thank N. W. Thiex for technical assistance, D. Olson, A. Decatur, L. Lenz, and D. Raulet for critical review of the manuscript, and members of the Portnoy laboratory for helpful discussions. This research was supported by National Institutes of Health Grants R01 AI27655 and AI29619 (to D.A.P.), and RO1 AI47173 and R29 AI42084 (to K.-D.L.). R.G. is supported by a National Institutes of Health Minority Predoctoral Fellowship (F31 AI50250-01). M.O. is a postdoctoral fellow of the Irvington Institute for Immunological Research.
Abbreviations
- LPS
lipopolysaccharide
- LTA
lipoteichoic acid
- NF-κB
nuclear factor κB
- TLR
Toll-like receptor
- BMDM
bone marrow-derived macrophages
- LLO
listeriolysin O
- PLC
phospholipase C
- moi
multiplicity of infection
- h.p.i.
h post infection
- MAP
mitogen-activated protein
- OVA
ovalbumin
- NOD
nucleotide-binding oligomerization domain
References
- 1.Janeway C A, Jr, Medzhitov R. Annu Rev Immunol. 2002;20:197–216. doi: 10.1146/annurev.immunol.20.083001.084359. [DOI] [PubMed] [Google Scholar]
- 2.Aderem A, Underhill D M. Annu Rev Immunol. 1999;17:593–623. doi: 10.1146/annurev.immunol.17.1.593. [DOI] [PubMed] [Google Scholar]
- 3.Maniatis T, Falvo J V, Kim T H, Kim T K, Lin C H, Parekh B S, Wathelet M G. Cold Spring Harbor Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 4.Sen G C. Annu Rev Microbiol. 2001;55:255–281. doi: 10.1146/annurev.micro.55.1.255. [DOI] [PubMed] [Google Scholar]
- 5.Philpott D J, Yamaoka S, Israel A, Sansonetti P J. J Immunol. 2000;165:903–914. doi: 10.4049/jimmunol.165.2.903. [DOI] [PubMed] [Google Scholar]
- 6.Braun L, Cossart P. Microbes Infect. 2000;2:803–811. doi: 10.1016/s1286-4579(00)90365-4. [DOI] [PubMed] [Google Scholar]
- 7.Tilney L G, Portnoy D A. J Cell Biol. 1989;109:1597–1608. doi: 10.1083/jcb.109.4.1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Portnoy D A, Jacks P S, Hinrichs D J. J Exp Med. 1988;167:1459–1471. doi: 10.1084/jem.167.4.1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith G A, Marquis H, Jones S, Johnston N C, Portnoy D A, Goldfine H. Infect Immun. 1995;63:4231–4237. doi: 10.1128/iai.63.11.4231-4237.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marquis H, Doshi V, Portnoy D A. Infect Immun. 1995;63:4531–4534. doi: 10.1128/iai.63.11.4531-4534.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mocci S, Dalrymple S, Nishinakamura R, Murray R. Immunol Rev. 1997;158:107–114. doi: 10.1111/j.1600-065x.1997.tb00996.x. [DOI] [PubMed] [Google Scholar]
- 12.Bielecki J, Youngman P, Connelly P, Portnoy D A. Nature. 1990;345:175–176. doi: 10.1038/345175a0. [DOI] [PubMed] [Google Scholar]
- 13.Higgins D E, Shastri N, Portnoy D A. Mol Microbiol. 1999;31:1631–1641. doi: 10.1046/j.1365-2958.1999.01272.x. [DOI] [PubMed] [Google Scholar]
- 14.Whittemore L A, Maniatis T. Mol Cell Biol. 1990;10:1329–1337. doi: 10.1128/mcb.10.4.1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee K D, Oh Y K, Portnoy D A, Swanson J A. J Biol Chem. 1996;271:7249–7252. [PubMed] [Google Scholar]
- 16.Sanderson S, Shastri N. Int Immunol. 1994;6:369–376. doi: 10.1093/intimm/6.3.369. [DOI] [PubMed] [Google Scholar]
- 17.Dabiri G A, Sanger J M, Portnoy D A, Southwick F S. Proc Natl Acad Sci USA. 1990;87:6068–6072. doi: 10.1073/pnas.87.16.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poltorak A, He X, Smirnova I, Liu M Y, Huffel C V, Du X, Birdwell D, Alejos E, Silva M, Galanos C, et al. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- 19.Schwandner R, Dziarski R, Wesche H, Rothe M, Kirschning C J. J Biol Chem. 1999;274:17406–17409. doi: 10.1074/jbc.274.25.17406. [DOI] [PubMed] [Google Scholar]
- 20.McNeil P L, Murphy R F, Lanni F, Taylor D L. J Cell Biol. 1984;98:1556–1564. doi: 10.1083/jcb.98.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stein B, Baldwin A S., Jr Mol Cell Biol. 1993;13:7191–7198. doi: 10.1128/mcb.13.11.7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lenardo M J, Fan C M, Maniatis T, Baltimore D. Cell. 1989;57:287–294. doi: 10.1016/0092-8674(89)90966-5. [DOI] [PubMed] [Google Scholar]
- 23.Navarro L, David M. J Biol Chem. 1999;274:35535–35538. doi: 10.1074/jbc.274.50.35535. [DOI] [PubMed] [Google Scholar]
- 24.Duclos S, Desjardins M. Cell Microbiol. 2000;2:365–377. doi: 10.1046/j.1462-5822.2000.00066.x. [DOI] [PubMed] [Google Scholar]
- 25.Teitelbaum R, Cammer M, Maitland M L, Freitag N E, Condeelis J, Bloom B R. Proc Natl Acad Sci USA. 1999;96:15190–15195. doi: 10.1073/pnas.96.26.15190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hueck C J. Microbiol Mol Biol Rev. 1998;62:379–433. doi: 10.1128/mmbr.62.2.379-433.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rodel J, Assefa S, Prochnau D, Woytas M, Hartmann M, Groh A, Straube E. FEMS Immunol Med Microbiol. 2001;32:9–15. doi: 10.1111/j.1574-695X.2001.tb00527.x. [DOI] [PubMed] [Google Scholar]
- 28.Devitt A, Lund P A, Morris A G, Pearce J H. Infect Immun. 1996;64:3951–3956. doi: 10.1128/iai.64.10.3951-3956.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Girardin S E, Tournebize R, Mavris M, Page A L, Li X, Stark G R, Bertin J, DiStefano P S, Yaniv M, Sansonetti P J, Philpott D J. EMBO Rep. 2001;2:736–742. doi: 10.1093/embo-reports/kve155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Girardin S E, Sansonetti P J, Philpott D J. Trends Microbiol. 2002;10:193–199. doi: 10.1016/s0966-842x(02)02334-x. [DOI] [PubMed] [Google Scholar]
- 31.Inohara N, Ogura Y, Chen F F, Muto A, Nunez G. J Biol Chem. 2001;276:2551–2554. doi: 10.1074/jbc.M009728200. [DOI] [PubMed] [Google Scholar]
- 32.Kim D H, Feinbaum R, Alloing G, Emerson F E, Garsin D A, Inoue H, Tanaka-Hino M, Hisamoto N, Matsumoto K, Tan M W, Ausubel F M. Science. 2002;297:623–626. doi: 10.1126/science.1073759. [DOI] [PubMed] [Google Scholar]
- 33.Saccani S, Pantano S, Natoli G. Nat Immunol. 2002;3:69–75. doi: 10.1038/ni748. [DOI] [PubMed] [Google Scholar]