Abstract
The 24-h expression of seven clock genes (Bmal1, Clock, Per1, Per2, Cry1, Cry2, and CK1ɛ) was assayed by in situ hybridization in the suprachiasmatic nucleus (SCN) and the pars tuberalis (PT) of the pituitary gland, collected every 4 h throughout 24 h, from female Soay sheep kept under long (16-h light/8-h dark) or short (8-h light/16-h dark) photoperiods. Locomotor activity was diurnal, inversely related to melatonin secretion, and prolactin levels were increased under long days. All clock genes were expressed in the ovine SCN and PT. In the SCN, there was a 24-h rhythm in Clock expression, in parallel with Bmal1, in antiphase with cycles in Per1 and Per2; there was low-amplitude oscillation of Cry1 and Cry2. The waveform of only Per1 and Per2 expression was affected by photoperiod, with extended elevated expression under long days. In the PT, the high-amplitude 24-h cycles in the expression of Bmal1, Clock, Per1, Per2, Cry1, and Cry2, but not CK1ɛ, were influenced by photoperiod. Per1 and Per2 peaked during the day, whereas Cry1 and Cry2 peaked early in the night. Hence, photoperiod via melatonin had a marked effect on the phase relationship between Per/Cry genes in the PT. This supports the conclusion that an ”external coincidence model“ best explains the way photoperiod affects the waveform of clock gene expression in the SCN, the central pacemaker, whereas an ”internal coincidence model“ best explains the way melatonin affects the phasing of clock gene expression in the PT to mediate the photoperiodic control of a summer or winter physiology.
In mammals, seasonal changes in day length (photoperiod) modulate daily and annual cycles in behavior and physiology. These effects are mediated through a central circadian pacemaker residing in the hypothalamic suprachiasmatic nuclei (SCN). This tissue controls multiple circadian outputs including rhythms in locomotor activity and the nightly production of melatonin by the pineal gland. The duration of the nocturnal melatonin signal changes quantitatively in response to changing photoperiod and is used by melatonin-responsive tissues to allow gating of seasonal changes in physiology over the course of the year (1).
The pars tuberalis (PT) of the pituitary stalk is believed to mediate the effects of photoperiod on prolactin secretion in mammals (1). Prolactin levels increase in spring in response to the reduced duration of nocturnal melatonin production, and levels of prolactin decline in autumn when the opposite applies. The central role of the PT is implied by the persistence of seasonal cycles of prolactin secretion in hypothalamic-pituitary-disconnected sheep (1) and by the localization of high concentrations of mt1 melatonin receptors to this region of the pituitary (2). The PT provides a model system for understanding the molecular basis for decoding the melatonin signal (3).
The self-sustaining rhythm generating capacity of the SCN is believed to derive from cell-autonomous, transcriptional feedback loops dependent on a small number of canonical clock genes (4). These are two period (Per) genes, Per1 and Per2, two clock/cycle-related genes, Clock and Bmal1, two mammalian cryptochrome (Cry) genes, Cry1 and Cry2, and casein kinase 1ɛ, CK1ɛ. Mutation or transgenic knockout of each of these genes has been shown to alter the free-running circadian period and/or entraining characteristics of the rodent SCN or to abolish free-running rhythmicity (4). The expression of the Period genes is robustly rhythmic and shows phase-dependent responses to photic stimuli, suggesting that light entrainment of the SCN pacemaker occurs through effects on Period gene expression (4, 5). Moreover, photoperiod-dependent changes in the timing of overt circadian rhythms are associated with effects on the duration of the elevated expression of the Period genes in the SCN (6, 7), suggesting that temporal changes in output are caused by corresponding changes in circadian gene expression in this tissue. Per1 also is expressed rhythmically in the PT, with a maximum early in the light phase. This expression is melatonin-dependent, because pinealectomy blocks the PT Per1 rhythm, without having any impact on expression in the SCN (8), and repeated daily injections of melatonin in pinealectomized animals can reinstate the cyclical pattern in the PT (9). Furthermore, in strains of mice unable to synthesize melatonin, or in transgenic mice bearing a knockout of the mt1 melatonin receptor, there is no daily rhythm in Per1 expression in the PT (9, 10).
Per1 expression in the PT is photoperiod-responsive (11, 12), but, in contrast to the SCN, the most obvious effect of photoperiod on Per1 expression in the PT is on amplitude of expression in the early light phase—with levels 2- to 4-fold higher under long days (11, 12). This has led to the hypothesis that the duration of melatonin is decoded in the pattern of clock gene expression in melatonin-responsive tissues. Two predictions arise from this hypothesis; first, for PER1 to assume a functional role in the PT, it is necessary that other associated clock gene proteins also should be present. Second, photoperiod should strongly influence the 24-h timing of expression of Per1 and other clock genes, correlated with alteration in a seasonal output. Given the very different functions of the PT and SCN in photoperiodic time measurement, photoperiod is likely to exert differential effects on clock gene expression in these two tissues.
To test these ideas, we have taken advantage of the larger size of the brain in Soay sheep, a diurnal mammal showing robust changes in seasonal physiology, typical of wild ruminants living at temperate latitudes. This enabled us to analyze seven core clock genes (Bmal1, Clock, Per1, Per2, Cry1, Cry2, and CK1ɛ) simultaneously in the SCN and PT in individual animals. The data presented below support the predictions and also demonstrate that the temporal patterns of clock gene expression in the SCN are broadly conserved across mammalian groups, whereas the PT may function as an internal coincidence timer for seasonal physiology.
Materials and Methods
Animals.
The animal experiment was conducted in accordance with the Animals (Scientific Procedures) Act of 1986. Fifty-six young female Soay sheep were divided into two groups of 28 and housed in light-sealed rooms with free access to food and water. Spontaneous locomotor activity was recorded every 10 min by using IR sensors and a Mini Mitter VitalView system (Mini Mitter, Sunriver, OR). The animals were brought indoors in the autumn and preconditioned to short days [8-h light/16-h dark (LD 8:16)]. One group remained on short days (short-day group), and one group was switched to long days (LD 16:8) for 6 weeks (long-day group). The lighting schedule was designed to ensure that animals were not photorefractory to the lighting conditions. At the end of the photoperiod treatments, groups of animals were killed by an injection of pentobarbitone at 4-h intervals (n = 4/time point) over 28 h, starting at zeitgeber time (ZT) 19 (ZT 0 = lights on). Hence, repeat sampling at ZT 19 was achieved. Before death, blood plasma was collected from each individual for analyses of prolactin and melatonin by RIA, using the methods of McNeilly and Andrews (13) and Fraser et al. (14), respectively. The brains of animals were removed rapidly from the skull, and a block of hypothalamus was frozen by immersion in isopentane and cooled to −30°C on dry ice. Tissues were stored at −80°C until sectioning. Serial sections (20 μm) were cut on a cryostat, thaw-mounted on poly-l-lysine-coated slides, and kept at −80°C until in situ hybridization.
Riboprobe Templates.
The riboprobe template used for generating a probe for Per1 expression was prepared as described (11). Probes for Per2, Cry1, Cry2, Clock, Bmal1, and CK1ɛ were prepared by RT-PCR from RNA extracted from ovine PT, followed by cloning in pGEM-Teasy (Promega), by using the manufacturer's protocols. Primer sequences were Per2, 229–249 (f) and 762–743 (r) of GenBank accession no. NM-011066; Cry1, 673–692 and 1194–1177 of GenBank accession no. AF156986; Cry2, 771–790 and 1277–1257 of GenBank accession no. AF156987; Bmal1, 1273–1295 and 1677–1655 of GenBank accession no. AF070917; clock, 556–579 and 1009–986 of GenBank accession no. AF000998; and CK1ɛ, 1189–1212 and 1362–1348 of GenBank accession no. XM 009983. 35S-labeled antisense probes were prepared from these templates by using T7 or SP6 viral RNA-polymerase dependent on orientation of inserts, as described (11).
In Situ Hybridization.
In situ hybridization was performed as described by Messager et al. (11). All slides for a given tissue (SCN or PT) were hybridized to a single batch of labeled probe. At the end of the hybridization procedure, slides were apposed to Hyperfilm β-Max (Amersham Pharmacia). All films were exposed simultaneously to 14C-radioactive intensity standards to allow standardization of densitometric measurements across films. Films typically were exposed for intervals of between 3 days and 1 week, depending on probe and tissue (PT/SCN). The intensity of labeling on developed films was quantified by scanning densitometry by using image pro-plus 4 (Media Cybernetics, Silver Spring MD). The 14C standards allowed gray-level values in regions of interest to be converted into background subtracted radioactive intensity values (fCi/μg) according to a standard curve. Sense transcript riboprobes also were prepared and found to give only background labeling in the hypothalamus and pituitary (data not shown).
Statistics.
Locomotor activity patterns were measured on a group basis only. The LD ratio of activity was calculated by using the Mini Mitter VitalView system, and the peak of activity was obtained by cosine analysis. The changes in clock gene mRNA expression and plasma concentrations of melatonin and prolactin, including the repeated ZT 19 sampling, were analyzed for effects of photoperiod and time (ZT) by two-way ANOVA and Bonferroni's posttest, using prism 3.0a for Macintosh (GraphPad, San Diego).
Results
Adaptation to Long and Short Days.
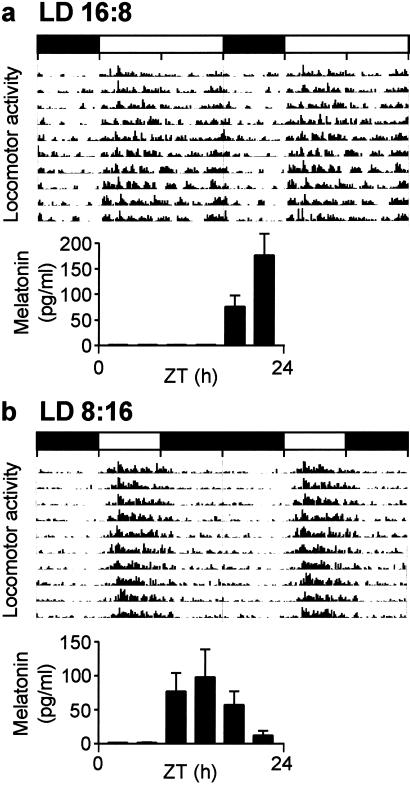
The photoperiod treatment resulted in the predicted differences in behavior and endocrine function. The animals' spontaneous locomotor activity pattern was clearly diurnal, in both the long- and short-day groups, and was characterized by bouts of synchronous activity within the group followed by rest, when the animals tended to ruminate (Fig. 1). In the long-day group, activity was increased during the first half of the light phase, with a second bout of activity before lights out; there was minimal activity during the dark phase. In the short-day group, locomotor activity was condensed into the shorter light phase so that separate morning and evening bouts were no longer discernible. The activity ratio between the light and dark phase was 2.1:1 and 4.8:1, and cosine peak activity occurred at ZT 7 and ZT 5 under long and short days, respectively.
Figure 1.
Double-plotted actogram of group-based locomotor activity in Soay sheep. (a) Long-day group (LD 16:8). (b) Short-day group (LD 8:16). The histograms illustrate blood plasma melatonin concentrations collected every 4 h throughout 24 h (ZT 0 = lights on). Horizontal bars indicate time of lights-on (open) and lights-off (filled).
Blood plasma melatonin concentrations were increased during the dark phase, inversely related to the locomotor activity patterns (Fig. 1). There was a significant difference in the melatonin profiles between groups, with a later and more truncated increase in melatonin secretion in the long-day group (P < 0.001). During the last 6-week segment of the lighting schedule, blood plasma concentrations of prolactin diverged between the long- and short-day animals, with higher values in the long-day group (data not shown). At the time of death, mean prolactin concentrations were 3.5-fold higher in the long-day compared with the short-day group (long day, LD 16:8, animals = 41.7 ± 4.1; short day, LD 8:16, animals = 12.6 ± 1.7, P < 0.001), demonstrating the predicted physiological response to photoperiod (1).
Clock Gene Expression in the SCN.
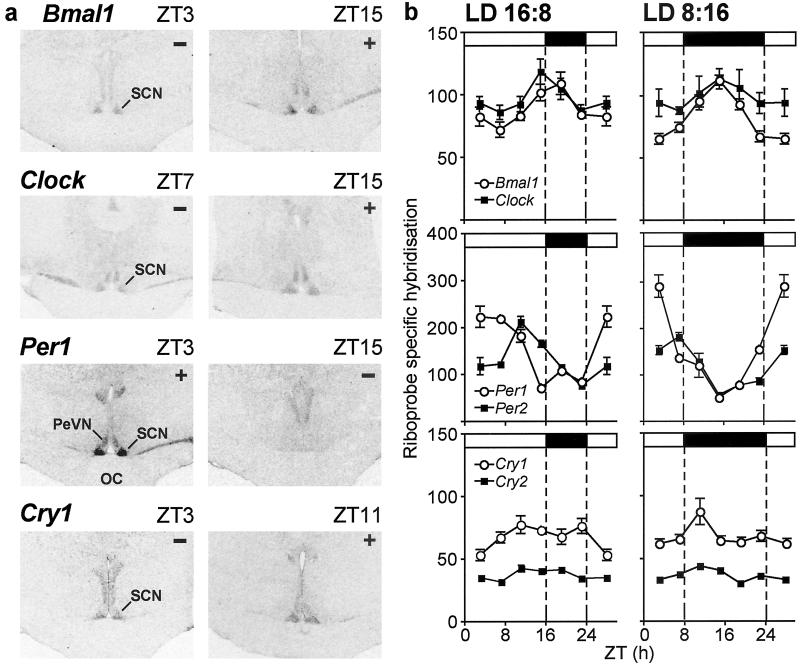
All seven measured clock genes were expressed in the ovine SCN. For Bmal1, Clock, Per1, Per2, Cry1, and Cry2, but not CK1ɛ, expression, there was significant variation during the 24-h cycle (P < 0.05). Examples of Bmal1, Clock, Per1, and Cry1 minima and maxima in the hybridization signal in the SCN are shown in Fig. 2a, and the overall 24-h profiles are shown in Fig. 2b. Bmal1 expression peaked at ZT 15–19, and Clock expression peaked at ZT 15 under both photoperiods. The pattern of expression in Per1 and Per2 was in antiphase with Bmal1/Clock, with maxima at ZT 3 for Per1, and at ZT 7 and ZT 11 for Per2 in the short- and long-day groups, respectively. The variation in Cry1 and Cry2 expression over the 24-h cycle was of low amplitude, peaking at ZT 11.
Figure 2.
(a) Representative peak (+) and nadir (−) autoradiograms of Bmal1, Clock, Per1, and Cry1 in situ hybridization in coronal sections of the SCN from Soay sheep acclimated to long days (ZT 0 = lights-on). (b) Profiles (24 h) of the expression of Bmal1 and Clock (Top), Per1 and Per2 (Middle), and Cry1 and Cry2 (Bottom) (mean ± SEM) in the SCN in Soay sheep under long days (LD 16:8, Left) and short days (LD 8:16, Right). Data from the repeated ZT 19 samplings are merged into a single time point, and the ZT 3 time point is double-plotted. Horizontal bars indicate time of lights-on (open) and lights-off (filled) (ZT 0 = lights-on). OC, optic chiasm; PeVN, periventricular nucleus.
Two-way ANOVA revealed a photoperiod × time (ZT) interaction only for Per1 and Per2 expression in the SCN (P < 0.001). There were more sustained levels of increased expression in Per1 and Per2 and a later increase of Per2 under long days compared with short days (Fig. 2b). The maximal expression in Per1 was decreased under long days (P < 0.01).
Clock Gene Expression in the PT.
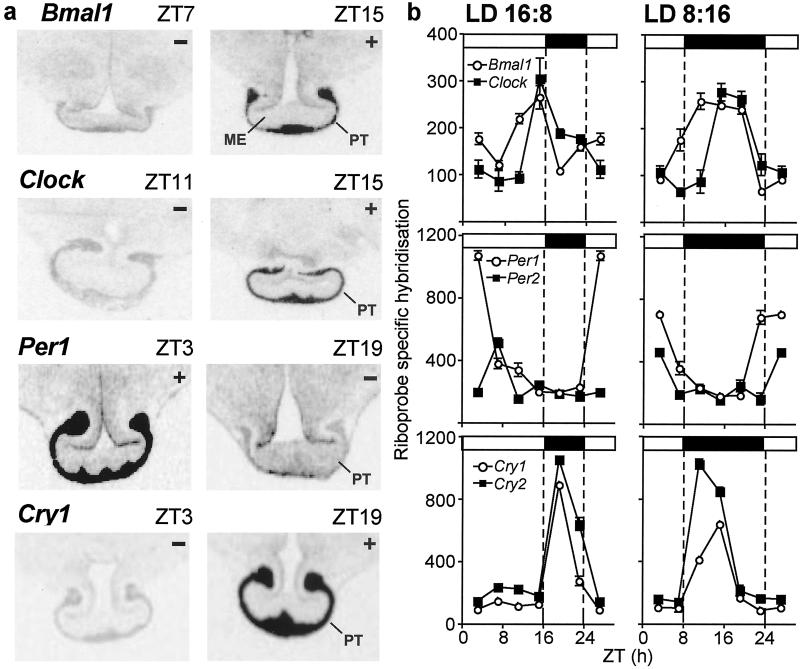
All seven measured clock genes were expressed in the PT; examples of Bmal1, Clock, Per1, and Cry1 minima and maxima in the hybridization signal in the PT are shown in Fig. 3a, and the overall 24-h profiles are shown in Fig. 3b. There was a significant photoperiod × time (ZT) interaction for Bmal1, Clock, Per1, Per2, Cry1, and Cry2, but not CK1ɛ, in the PT (P < 0.001). Bmal1 and Clock expression peaked at ZT 11–15, and Per1 and Per2 expression was at a maximum at ZT 3–7, with inverse timing similar to that of the SCN. The duration of elevated Bmal1 and Clock expression was more sustained under short days compared with long days. The peak in Per1 expression was of higher amplitude under long days, opposite to the situation in the SCN. Cry1 and Cry2 expression peaked at ZT 19 under long days and ZT 11–15 under short days, which was early in the dark phase in both photoperiods (Fig. 3b).
Figure 3.
(a) Representative peak (+) and nadir (−) autoradiograms of Bmal1, Clock, Per1, and Cry1 in situ hybridization in coronal sections of the mediobasal hypothalamus, with the rostral PT attached from Soay sheep acclimated to long days (ZT 0 = lights-on). (b) Profiles (24 h) of the expression of Bmal1 and Clock (Top), Per1 and Per2 (Middle), and Cry1 and Cry2 (Bottom) (mean ± SEM; when error bars are small, they may be contained within the symbol) in the PT in Soay sheep under long days (LD 16:8, Left) and short days (LD 8:16; Right). Data from the repeated ZT 19 samplings are merged into a single time point, and the ZT 3 time point is double-plotted. Horizontal bars indicate time of lights-on (open) and lights-off (filled) (ZT 0 = lights-on). ME, median eminence.
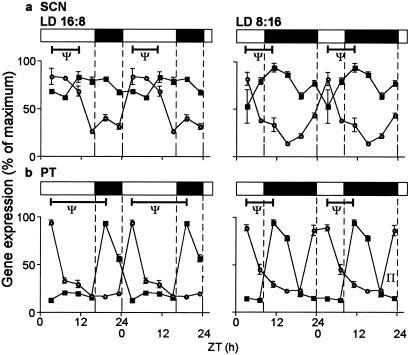
Photoperiod influenced the relative timing of period and cryptochrome gene expression in the PT, because Per1/Per2 expression increased in the early light phase and Cry1/Cry2 expression increased in the early dark phase. This produced differences between photoperiods in the phasing of the two sets of genes in the PT, unlike the situation in the SCN, and is summarized for Per1 and Cry2 in Fig. 4. In the PT under long days, the interval (Ψ) between peaks in Per1 and Cry2 was 16 h, whereas under short days, the corresponding interval was 8 h. A difference between photoperiods also was apparent for the Per1–Cry1 (16 vs. 12 h) and Per2–Cry2 intervals (12 vs. 8 h).
Figure 4.
Double plots of 24-h profiles of expression of Per1 (○) and Cry2 (■, mean ± SEM, expressed as % of maximum) in the ovine SCN (a) and PT (b) under long days (LD 16:8) and short days (LD 8:16). Note that in the light-responsive SCN, photoperiod caused a change in waveform in Per1, but not in Cry2, whereas in the melatonin-responsive PT, photoperiod caused a marked change in the phase relationship in peak expression of Per1 and Cry2 (ψ). Horizontal bars indicate time of lights-on (open) and lights-off (filled).
Discussion
The large size of the sheep brain allowed us to undertake a comprehensive in situ hybridization analysis of clock gene expression in a diurnal mammalian species, and all seven established clock genes were found to be expressed in the ovine SCN and PT.
SCN.
The Per genes showed the highest amplitude variation in expression, with Per1 peaking earlier in the light phase than Per2 under both photoperiods. The expression of both Clock and Bmal1 also varied significantly with time, peaking during the night approximately 12–16 h later than the Per genes. This matches well with the antiphase relationship previously reported between Per and Bmal1 expression in rodents (4). Although significant variation in Clock expression has not been reported in the rodent SCN, variation in other tissues has been reported with phasing relative to Bmal1 and Per expression similar to that reported here (15). Temporal variation in Cry expression in the ovine SCN was of low amplitude, peaking later than that of the Per genes, again agreeing with the rodent data (4). Together with Per1 and Per2 data in the diurnal 13-lined ground squirrel (Spermatophilus tridecemlineatus) (16), this demonstrates that the internal phase relationships between 24-h cycles of clock gene expression in the SCN and their phasing relative to the light–dark cycle are conserved across mammals, whether they are diurnal or nocturnal.
In rodents, expression of the Per genes is acutely light-inducible during the subjective night, and Per1 and Per2 have nonredundant roles in mediating photic entrainment (5, 17). Correspondingly, previous studies have demonstrated that photoperiod modulates the waveforms of expression of both Per1 and Per2 in the SCN of rodents. The present study extends this finding to sheep, and of all clock genes studied in this tissue, only the Per genes were photoperiod-responsive. The principal effect of photoperiod on Per gene expression in the SCN of both sheep and rodents appears to be a decompression of the elevated portion of the expression rhythm for both genes under long days, and in Siberian hamsters, this long-day decompression of the PER1 and PER2 protein waveforms persisted, at least in part, when animals were placed in constant darkness (DD) (7). Together, these observations are consistent with a model for SCN function in which photoperiod influences pacemaker activity as a result of coincidence between light and photoinducible phases in the Per expression rhythms (external coincidence model).
Recently, studies on Per1 and Per2 mutant mice (17) have produced tentative support for a model of SCN function, which proposes that the Per1 and Per2 genes constitute two separate clock gene oscillators, one coupled to dawn (M, morning; Per1), and one coupled to dusk (E, evening; Per2), and that photoperiod modulates the temporal relationship between these components (internal coincidence model, ref. 18). This model, based on theoretical concepts, has been criticized because of a lack of empirical data for multiple clock genes measured within the same individuals (19). Our comprehensive study of clock gene expression in the ovine SCN does not allow us to exclude the internal coincidence model, but the expression profiles suggest that any photoperiodic effects on the Per1–Per2 phase relationship or cryptochrome expression in the SCN are, at best, subtle.
PT.
Whereas the overall temporal phasing of Bmal1/Clock and Per1/Per2 expression in the ovine PT was similar to that in the SCN, clock gene expression generally underwent higher amplitude variation over the 24-h cycle in the PT, which has been demonstrated for other peripheral tissues (20). An exception to this was CK1ɛ, which showed no significant temporal variation in either tissue, and this finding is consistent with its reported permissive role in determining the period of circadian rhythms (21).
The PT expresses a high concentration of mt1 melatonin receptors, and studies have shown that the 24-h pattern of melatonin secretion dictates the pattern of Per1 gene expression in this tissue (8, 9, 12). Acute melatonin treatment inhibits Per1 expression in ovine PT cells, probably through suppression of cAMP-mediated signaling (3). In contrast, prolonged melatonin treatment and subsequent melatonin withdrawal potently stimulate Per1 expression, probably because of the chronic sensitizing effects of melatonin on adenylyl cyclase (9, 22). This finding suggests that longer melatonin signals associated with short days might result in greater induction of Per1 at the end of the night. Paradoxically, however, the opposite is apparent here and in previous studies in rodents (6, 11). An explanation for this pattern of gene expression is suggested by the different rates of decline of melatonin levels at the end of the night on the two photoperiods. Under long days, melatonin levels fall precipitately with the onset of light, and a narrow, high-amplitude peak of Per1 expression in the ovine PT is seen subsequently. Under short days, a broad, lower-amplitude peak of Per1 expression is associated with a more gradual decline in melatonin levels during the later part of the dark phase. Per2 expression in the ovine PT also peaked in the early light phase, slightly after Per1. Unlike Per1, there is no evidence that Per2 expression is responsive to cAMP signaling (23, 24), suggesting different control mechanisms for the induction of the two Per genes.
The most striking difference between the PT and the SCN was the high-amplitude temporal variation in Cry expression seen in the former tissue and its responsiveness to photoperiod. Under both long and short days, peak Cry expression occurred at the onset of darkness, associated with the increase in melatonin secretion. This temporal control resulted in different phasing between peak expression of Per1/Per2 (linked to lights-on) and Cry1/Cry2 (linked to lights-off) under the two photoperiods. Studies in the rodent SCN indicate that the BMAL1/CLOCK protein complex acts as the transactivator for both the Per and Cry genes and that the induction of the Per1/Per2 genes occurs only slightly in advance of Cry1/Cry2 (4). This was not the case in the ovine PT, where photoperiod (presumably through melatonin) dictated the interval between peaks in Per1/Per2 and Cry1/Cry2 expression. Furthermore, the Bmal1 and Clock expression rhythms showed a lengthening of increased expression under short days but no consistent temporal relationship to peak Cry1/Cry2 expression. This finding suggests that, in the PT, the transcriptional drive for Cry1/Cry2 genes occurs through different mechanisms than that in the SCN.
The observed photoperiod-induced differences in the timing of rhythms in Per1/Per2 and Cry1/Cry2 gene expression may have important consequences for the clockwork in the PT, because the fate of newly synthesized PER and CRY proteins depends on their opportunity to form complexes that are stable in the nucleus (25, 26). In the mouse SCN, Per2 expression is required for maintaining circadian oscillation in Bmal1 expression, suggesting a stimulatory role for PER2 protein in the transcription of Bmal1 (4). Thus, the altered coincidence between the PER2 and CRY proteins may affect the Bmal1 waveform. The data for the ovine PT are consistent with this prediction, with the duration of elevated Bmal1 expression being longer under short days compared with long days.
Overall, and in contrast to the case in the SCN, our PT data strongly support an internal coincidence model for photoperiodic time measurement. Here, photoperiod-induced changes in the phasing of Cry and Per expression appear to occur as a consequence of the changing melatonin signal. Our working hypothesis is that these central changes in the PT will cause long-term alterations in the transcriptional control of downstream genes, which would produce two distinct cellular states, one associated with long days and one associated with short days. These cellular states remain uncharacterized, but because the PT is thought to regulate prolactin secretion via a paracrine mechanism (1–3), and the appropriate prolactin response to day length was seen in our animals, we speculate that differential production of a prolactin-releasing factor may be a downstream consequence of the phenomenon described here.
In conclusion, this study describes the 24-h pattern of expression of Bmal1, Clock, Per1, Per2, Cry1, Cry2, and CKIɛ in the SCN and the PT of a long-lived, diurnal mammal and its adaptation to photoperiod. The inverse relationship and timing of cycles in Bmal1/Clock and Per1/Per2 are similar to those of nocturnal rodents, indicating that the core clockwork is conserved across species. In the ovine SCN, photoperiod affected the waveforms of Per1 and Per2 expression, but with no clear effect on their phase relationship, whereas in the PT, photoperiod had a pronounced effect on the phase relationship between sharply defined peaks of Per and Cry expression. Based on this evidence, we propose that although an external coincidence model for photoperiodism may be sufficient to account for photic effects on the primary oscillator (the SCN), an internal coincidence model accounts better for the way melatonin mediates the effects of photoperiod in the melatonin-target tissue, the PT. The core clock gene system therefore is used differently in the circadian clock and the photoperiodic relay within the pituitary gland.
Acknowledgments
We gratefully acknowledge the skilled efforts of the staff at the Marshall building, who conducted these experiments, and Mrs. Kirsten Brown, who performed the in situ hybridization procedures. This work was supported by the Biotechnology and Biological Sciences Research Council (S.M. and D.H.) and the Medical Research Council (H.A. and G.L.).
Abbreviations
- CK1ɛ
casein kinase 1ɛ
- PT
pars tuberalis
- SCN
suprachiasmatic nucleus
- ZT
zeitgeber time
- LD
light/dark
Footnotes
References
- 1.Lincoln G. In: Melatonin After Four Decades. Olcese J, editor. Boston: Kluwer; 1999. pp. 137–153. [Google Scholar]
- 2.Morgan P J, Barrett P, Howell H E, Helliwell R. Neurochem Int. 1994;24:101–146. doi: 10.1016/0197-0186(94)90100-7. [DOI] [PubMed] [Google Scholar]
- 3.Hazlerigg D G, Morgan P J, Messager S M. J Biol Rhythms. 2001;16:326–335. doi: 10.1177/074873001129002042. [DOI] [PubMed] [Google Scholar]
- 4.Reppert S M, Weaver D R. Annu Rev Physiol. 2001;63:647–676. doi: 10.1146/annurev.physiol.63.1.647. [DOI] [PubMed] [Google Scholar]
- 5.Albrecht U, Zheng B, Larkin D, Sun Z S, Lee C C. J Biol Rhythms. 2001;16:100–104. doi: 10.1177/074873001129001791. [DOI] [PubMed] [Google Scholar]
- 6.Messager S, Hazlerigg D G, Mercer J G, Morgan P J. Eur J Neurosci. 2000;12:2865–2870. doi: 10.1046/j.1460-9568.2000.00174.x. [DOI] [PubMed] [Google Scholar]
- 7.Nuesslein-Hildesheim B, O'Brien J A, Ebling F J, Maywood E S, Hastings M H. Eur J Neurosci. 2000;12:2856–2864. doi: 10.1046/j.1460-9568.2000.00173.x. [DOI] [PubMed] [Google Scholar]
- 8.Messager S, Garabette M L, Hastings M H, Hazlerigg D G. NeuroReport. 2001;12:579–582. doi: 10.1097/00001756-200103050-00029. [DOI] [PubMed] [Google Scholar]
- 9.von Gall C, Garabette M L, Kell C A, Frenzel S, Dehghani F, Schumm-Draeger P-M, Weaver D R, Korf H-W, Hastings M H, Stehle J H. Nat Neurosci. 2002;5:234–238. doi: 10.1038/nn806. [DOI] [PubMed] [Google Scholar]
- 10.Sun Z S, Albrecht U, Zhuchenko O, Bailey J, Eichele G, Lee C C. Cell. 1997;91:1003–1011. doi: 10.1016/s0092-8674(00)80366-9. [DOI] [PubMed] [Google Scholar]
- 11.Messager S, Ross A W, Barrett F, Morgan P J. Proc Natl Acad Sci USA. 1999;96:9938–9943. doi: 10.1073/pnas.96.17.9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan P J, Ross A W, Graham E S, Adam C, Messager S, Barrett P. J Neuroendocrinol. 1998;10:319–323. doi: 10.1046/j.1365-2826.1998.00232.x. [DOI] [PubMed] [Google Scholar]
- 13.McNeilly A S, Andrews P. J Endocrinol. 1974;60:359–367. doi: 10.1677/joe.0.0600359. [DOI] [PubMed] [Google Scholar]
- 14.Fraser S, Cowen P, Franklin M, Franey C, Arendt J. Clin Chem. 1983;29:396–397. [PubMed] [Google Scholar]
- 15.Young M E, Razeghi P, Taegtmeyer H. Circ Res. 2001;88:1142–1150. doi: 10.1161/hh1101.091190. [DOI] [PubMed] [Google Scholar]
- 16.Mrosovsky N, Edelstein K, Hastings M H, Maywood E S. J Biol Rhythms. 2001;16:471–478. doi: 10.1177/074873001129002141. [DOI] [PubMed] [Google Scholar]
- 17.Steinlechner S, Jacobmeier B, Scherbarth F, Dernbach H, Kruse F, Albrecht U. J Biol Rhythms. 2002;17:202–209. doi: 10.1177/074873040201700303. [DOI] [PubMed] [Google Scholar]
- 18.Daan S, Albrecht U, van der Horst G T J, Illnerova H, Roenneburg T, Wehr T A, Schwartz W J. J Biol Rhythms. 2001;16:105–116. doi: 10.1177/074873001129001809. [DOI] [PubMed] [Google Scholar]
- 19.Hastings M. J Biol Rhythms. 2001;16:117–123. doi: 10.1177/074873001129001818. [DOI] [PubMed] [Google Scholar]
- 20.Oishi K, Sakamoto K, Okada T, Nagase T, Ishida N. Biochem Biophys Res Commun. 1998;253:199–203. doi: 10.1006/bbrc.1998.9779. [DOI] [PubMed] [Google Scholar]
- 21.Lowrey P L, Shimomura K, Antoch M P, Yamazaki S, Zemenides P D, Ralph M R, Menaker M, Takahashi J S. Science. 2000;288:483–491. doi: 10.1126/science.288.5465.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hazlerigg D G, Gonzalez-Brito A, Lawson W, Hastings M H, Morgan P J. Endocrinology. 1993;132:285–292. doi: 10.1210/endo.132.1.7678217. [DOI] [PubMed] [Google Scholar]
- 23.Takekida S, Yan L, Maywood E S, Hastings M H, Okamura H. Eur J Neurosci. 2000;12:4557–4561. doi: 10.1046/j.0953-816x.2000.01324.x. [DOI] [PubMed] [Google Scholar]
- 24.Yagita K, Okamura H. FEBS Lett. 2000;465:79–82. doi: 10.1016/s0014-5793(99)01724-x. [DOI] [PubMed] [Google Scholar]
- 25.Shearman L P, Sriram S, Weaver D R, Maywood E S, Chaves I, Zheng B H, Kume K, Lee C C, Van Der Horst G T J, Hastings M H, Reppert S M. Science. 2000;288:1013–1019. doi: 10.1126/science.288.5468.1013. [DOI] [PubMed] [Google Scholar]
- 26.Miyazaki K, Mesaki M, Ishida N. Mol Cell Biol. 2001;21:6651–6659. doi: 10.1128/MCB.21.19.6651-6659.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]