Abstract
Two cyclooxygenase isozymes, COX-1 and -2, are known to catalyze the rate-limiting step of prostaglandin synthesis and are the targets of nonsteroidal antiinflammatory drugs. Here we describe a third distinct COX isozyme, COX-3, as well as two smaller COX-1-derived proteins (partial COX-1 or PCOX-1 proteins). COX-3 and one of the PCOX-1 proteins (PCOX-1a) are made from the COX-1 gene but retain intron 1 in their mRNAs. PCOX-1 proteins additionally contain an in-frame deletion of exons 5–8 of the COX-1 mRNA. COX-3 and PCOX mRNAs are expressed in canine cerebral cortex and in lesser amounts in other tissues analyzed. In human, COX-3 mRNA is expressed as an ≈5.2-kb transcript and is most abundant in cerebral cortex and heart. Intron 1 is conserved in length and in sequence in mammalian COX-1 genes. This intron contains an ORF that introduces an insertion of 30–34 aa, depending on the mammalian species, into the hydrophobic signal peptide that directs COX-1 into the lumen of the endoplasmic reticulum and nuclear envelope. COX-3 and PCOX-1a are expressed efficiently in insect cells as membrane-bound proteins. The signal peptide is not cleaved from either protein and both proteins are glycosylated. COX-3, but not PCOX-1a, possesses glycosylation-dependent cyclooxygenase activity. Comparison of canine COX-3 activity with murine COX-1 and -2 demonstrates that this enzyme is selectively inhibited by analgesic/antipyretic drugs such as acetaminophen, phenacetin, antipyrine, and dipyrone, and is potently inhibited by some nonsteroidal antiinflammatory drugs. Thus, inhibition of COX-3 could represent a primary central mechanism by which these drugs decrease pain and possibly fever.
Acetaminophen is often categorized as a nonsteroidal antiinflammatory drug (NSAID), even though in clinical practice and in animal models it possesses little antiinflammatory activity (1). Like NSAIDs, however, acetaminophen inhibits pain and fever and is one of the world's most popular analgesic/antipyretic drugs. Despite acetaminophen's long use and popularity it lacks a clear mechanism of action. Flower and Vane showed that acetaminophen inhibited cyclooxygenase (COX) activity in dog brain homogenates more than in homogenates from spleen (2). This gave rise to the concept that variants of COX enzymes exist that are differentially sensitive to this drug and that acetaminophen acts centrally. Yet, even though two isozymes of COX are known, neither isozyme is sensitive to acetaminophen at therapeutic concentrations of the drug in whole cells or homogenates. Instead, COX-1 and -2 in homogenates frequently exhibit the paradoxical property of being stimulated by submillimolar concentrations of acetaminophen and inhibited by supermillimolar levels of the drug (1). This finding suggests that neither isozyme is a good candidate for the site of action of acetaminophen.
In analyzing COX-1 and -2 RNA expression in dog tissues, our laboratory observed that the cerebral cortex of dog brain contains two distinct RNAs that hybridized to a canine COX-1 cDNA. One RNA was ≈2.6 kb in size and the other was ≈1.9 kb in size, and analyses of these RNAs suggest that they encode previously uncharacterized COX-1-related proteins.
Materials and Methods
Unless otherwise stated all basic protocols used were from the manual on molecular cloning by Sambrook and Russell (3).
Isolation of RNA and Construction of a cDNA Library.
Isolation of RNA and library construction methods have been described (4). Human Multiple Tissue Northern blots (MTN) were purchased from CLONTECH.
Antisense oligonucleotides to the first intron of human and canine COX-1 genes were synthesized and end-labeled using [γ-32P]dATP.
A canine cerebral cortex cDNA library was screened using an ≈1.0-kb canine COX-1 fragment previously cloned in this laboratory by reverse transcription-coupled (RT)-PCR. The library was also screened with a 32P-labeled canine COX-1 intron 1 antisense oligonucleotide. Two full-length clones were isolated, completely sequenced, and designated COX-3 and partial COX-1a (PCOX-1a). Both were derived from the canine COX-1 gene but retain intron 1. PCOX-1a also has a 657-bp in-frame deletion spanning exons 5–8.
RT-PCR of Canine and Human Cerebral Cortex mRNA.
Canine cerebral cortex cDNA was synthesized, and primers were designed for PCR amplification. The sense primer (5′-CGGATCCGCCGCCCAGAGCTATGAG-3′) corresponded to nucleotides 15–32 of canine COX-3 sequence (submitted to GenBank under accession no. AF535138), with the 3′ end of the primer being two nucleotides downstream of the initiating methionine. The antisense primer (5′-cgccatcctggtgggggtcaggcacacgga-3′) corresponded to nucleotides 1865–1894, located 32 nucleotides upstream of the stop codon.
Northern blot analysis of human tissues with an intron 1 probe detected an ≈5.2-kb mRNA similar in size to one previously reported (5). Marathon-ready human cerebral cortex cDNA (CLONTECH) was amplified by PCR (CLONTECH, Advantage 2 PCR enzyme system), using 5′ and 3′ primers, and an ≈4.2-kb amplified fragment was recovered and found to contain the entire coding region of human COX-1 with intron 1 retained.
Expression of COX-3 and PCOX-1a in Baculovirus.
Both COX-3 and PCOX-1a were cloned into the baculovirus expression vector pBlueBac 4.5/V5-His (Invitrogen). Sf9 cells (≈1 × 106) were infected with viral stocks at a multiplicity of infection (moi) of 3 for expression of COX-3, PCOX-1a, mouse COX-1, and mouse COX-2 (6).
In some cases, tunicamycin was added to a final concentration of 10 μg/ml to insect cells 1 h after infection, and cells were cultured and harvested after 48 h. Activity of intact cells was determined by RIA (7).
Detection of 60-, 53-, and 50-kDa COX-1-Related Proteins in Human Aorta Tissue.
Total protein (20 μg) from human aorta was analyzed by Western blotting, using COX-1 mAb (Cayman Chemical, Ann Arbor, MI) and COX-3 antipeptide polyclonal antibodies (pAb). Primary antibodies were either preincubated with a mixture of human and mouse COX-1 intron 1 peptide (described below) for 1 h at 4°C, or left unblocked. Blots were processed with appropriate rabbit-anti-mouse secondary antibody (1:2,000) or goat-anti-rabbit secondary antibody (1:10,000) from Sigma. Densitometry of the autoradiographic image was performed using the AlphaImage 2000 Documentation and Analysis System (Alpha Innotech, San Leandro, CA).
Drug Inhibition Assays.
Sf9 cells were infected with high titer viral stocks (moi = 3) and cultured for 48 h. Cells were preincubated with drug for 30 min at 25°C, arachidonic acid (100 μl, final concentration 5 or 30 μM) was then added for an additional 10-min incubation at 37°C. Supernatant was assayed for COX activity by RIA for PGE2. Assays were performed multiple times in triplicate. Inhibition curves were constructed and IC50 values were determined using PRISM 3.0 (GraphPad, San Diego).
Production of Polyclonal Anti-COX-3 Antibodies.
Peptides corresponding to the first 13 aa of human (MSRECDPGARWGC) and mouse (MSREFDPEAPRNC) COX-3, as predicted by genomic clone sequences, were synthesized and coupled to keyhole limpet hemocyanin. Peptides (a 50:50 mixture) were injected into New Zealand White rabbits. The resulting polyclonal antibodies were then affinity purified using the above peptides immobilized on a Sulfolink coupling gel (Pierce) according to the manufacturer's instructions.
Results
Two distinct mRNA species (≈2.6 and ≈1.9 kb) were detected on a Northern blot with a canine COX-1 coding region cDNA probe using RNA isolated from canine cerebral cortex (Fig. 1A, lane 1). To further investigate these transcripts, a canine cerebral cortex cDNA library was constructed and the nonamplified library was screened as described above. Eleven clones were isolated and subsequently characterized by automated DNA sequencing. All of the eleven clones were found to contain canine COX-1 cDNA sequence. However, three clones harbored an insertion of 90 nucleotides at, or near, the 5′ end of their respective cDNAs, which showed 75% sequence identity to intron 1 of either human or mouse COX-1 genes (data not shown). This extra sequence also contained 5′ and 3′ consensus splice sites indicative of a retained intron. In addition to the retention of intron 1, one of the three clones had a 657-bp in-frame deletion corresponding to exons 5–8 of the COX-1 message (sequence submitted to GenBank under accession no. AF535139).
Figure 1.
Northern blot analysis and RT-PCR. (A) Northern blot of canine cerebral cortex poly(A) RNA (1, 5.0 μg; 2, 2.5 μg) probed with (1) 32P-labeled canine COX-1 cDNA fragment and (2) 32P-labeled canine antisense oligonucleotide to intron 1. (B) PCR amplification of PCOX-1 in canine cerebral cortex. Lane 1, ethidium bromide-stained gel of amplified products corresponding to PCOX-1a containing intron 1 (upper band) and PCOX-1b (lower band) lacking intron 1; lane 2, Southern blot of the amplified products probed with antisense oligonucleotide to intron 1; lane 3, Southern blot using COX-3 cDNA as probe. (C) Human Multiple Tissue Northern blots (MTN) probed with a 32P-labeled human antisense oligonucleotide to intron 1 (HCI). The ≈5.2-kb mRNA was detected in blots 1–3 (adult tissues), and 4 (fetal tissues). Am, amygdala; B, brain; C, cerebellum: Cc, cerebral cortex; Fl, frontal lobe; Hi, hippocampus; Ht, heart; K, kidney; Lu, lung; Li, liver; M, skeletal muscle; Md, medulla; Cn, caudate nucleus; Op, occipital pole; P, placenta; Pn, pancreas; Pu, putamen; Sc, spinal cord; Th, thalamus; Tl, temporal lobe; Co, corpus callosum.
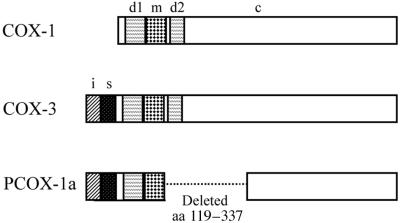
To determine whether the two detected COX mRNA transcripts (i.e., ≈2.6 and ≈1.9 kb) harbored intron 1, the Northern blot experiment was repeated using a radiolabeled antisense canine COX-1 intron 1-specific oligonucleotide probe (Fig. 1A, lane 2). Both the ≈1.9-kb and the ≈2.6-kb transcripts were detected, suggesting that multiple intron-1-containing splice variants were indeed expressed in canine cerebral cortex. The COX encoded by the ≈2.6-kb cDNA clone with nonspliced intron 1 has been designated as COX-3. We have named this newly discovered enzyme COX-3, even though it derives from the same gene as COX-1, because experience has shown that the important differences between COX-1 and -2 are pharmacological rather than genetic. Furthermore a numerical nomenclature is simpler, especially if further variants are discovered. The COX cDNA clone that harbored intron 1, lacked exons 5–8, and corresponded to the ≈1.9-kb mRNA transcript has been designated partial COX-1a or PCOX-1a (Fig. 2).
Figure 2.
Structure of COX-3 and PCOX-1a. A schematic representation of the domains of COX-3 and PCOX-1 in comparison to COX-1. s, signal peptide; d1, dimerization domain/EGF-like domain 1; d2, dimerization domain 2; m, membrane binding domain; c, catalytic domain; i, 90-bp sequence encoded by intron 1. PCOX-1b is identical to PCOX-1a except that PCOX-1b lacks intron 1. Amino acid numbering is according to residues in sheep seminal vesicle COX-1.
RT-PCR of canine cerebral cortex RNA, as well as analysis of Northern blots, indicated that COX-3 mRNA is present in this brain region at about 5% of the level of COX-1 mRNA (Fig. 1A and data not shown). Interestingly, these analyses also demonstrated that the ≈1.9-kb mRNA corresponding to PCOX-1a was actually a mixture of two mRNAs that differed in size by ≈90 nucleotides (Fig. 1B). One of these mRNAs was PCOX-1a and the other (PCOX-1b) was identical to PCOX-1a except that it lacked intron 1. PCOX-1a and -1b are expressed in equal amounts in brain cortex (Fig. 1B).
To determine whether previously uncharacterized COX-1-related mRNA transcripts were also expressed in human tissues, human Northern blot experiments were performed using a human intron-1-specific (HCI) probe. These results demonstrated the existence of previously uncharacterized ≈5.2- and ≈2.8-kb mRNA transcripts (Fig. 1C). Faint hybridization signals were also seen around 1.9 kb (data not shown). Hybridization of HCI to the ≈5.2-kb form was tissue-specific, with highest levels present in the cerebral cortex, followed by the heart. These observations differ from the characterized expression patterns of COX-1 mRNA (8).
COX enzymes are intralumenal residents of the endoplasmic reticulum and depend on N-linked glycosylation for proper folding and activity. Retention of intron 1 theoretically could prevent COX-3 and PCOX-1a expression by preventing export of these mRNAs from the nucleus or by targeting these proteins to another subcellular compartment, preventing glycosylation. Therefore, insect cells (Sf9) were infected with recombinant baculovirus expressing COX-3, PCOX-1a, and COX-1, and cell homogenates were assayed for protein expression by Western blotting. Antibodies specific for the conserved amino acid sequence (MSREXDPXA) predicted to be encoded by intron 1 in mammals were used to probe for COX-3 and PCOX-1a. This analysis demonstrated that both COX-3 and PCOX-1a are efficiently expressed in insect cells. No detectable products resulting from removal of intron 1 by splicing were detected immunologically or by RT-PCR analysis of RNA extracted from infected Sf9 cells. Moreover, the signal peptide, which in COX-3 and PCOX-1a contains an additional intron 1 encoded sequence, was not removed by signal peptidase as it is in COX-1 and -2.
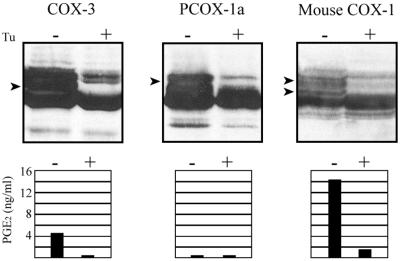
Posttranslational N-linked glycosylation of COX-3 and PCOX-1a was compared with that of COX-1 by using tunicamycin to inhibit core glycosylation. Immunoblot analysis demonstrated a decrease in or disappearance of glycosylated forms of COX-3, PCOX-1a, and COX-1 (Fig. 3 Upper; Left, Center, and Right, respectively). Expression systems assayed for PGE2 production found COX-3 to be ≈20% of that of COX-1, whereas PCOX-1a completely lacked detectable COX activity (Fig. 3 Lower). COX activity in cells treated with tunicamycin was abolished, indicating that N-linked glycosylation is necessary for COX activity of COX-3.
Figure 3.
Expression in insect cells. Western blots showing the expression of COX-3, PCOX-1a, and COX-1 in insect cells treated with (+) and without (−) tunicamycin (Upper). Arrows indicate glycosylated forms of COX-1 that are not present in cells treated with tunicamycin. Polyclonal antibodies to human and mouse COX-1 intron 1 sequence were used to probe the COX-3 and PCOX-1a blots, whereas a monoclonal antibody to ovine COX-1 (Cayman Chemical) was used to probe the mouse COX-1 blot. COX activity in insect cells expressing COX-3, PCOX-1a, and COX-1 (Lower). Cells were treated with (+) and without (−) tunicamycin.
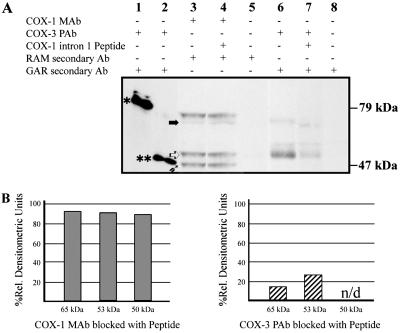
RNA studies in human tissues indicated highest levels of COX-3 message to be in the cerebral cortex and heart. Western blot analysis of human aorta (Fig. 4) by using either COX-1 monoclonal antibody or COX-3 antipeptide polyclonal antibody detected the presence of distinct 65- and 53-kDa COX-1 related proteins. Additionally, the COX-1 but not COX-3 antibody, detected a 69-kDa protein, corresponding to glycosylated COX-1, as well as a 50-kDa protein, which may represent a proteolytic fragment of COX-1 or PCOX-1b. Detection of both of the 65- and 53-kDa proteins was selectively reduced by preincubation of the antipeptide sera with its cognate peptide, whereas detection of the same proteins by the COX-1 monoclonal antibody was unaffected by this treatment.
Figure 4.
Western blot of human aorta lysate probed with COX-1 and -3 antibodies. (A) Western blot (lanes 3–8, 20 μg total aorta protein each lane) probed with primary, secondary, or blocked antibodies as indicated. A solid horizontal arrow indicates the 65-kDa protein, an open arrow indicates the 53-kDa proteins, and an upward diagonal solid arrow indicates the 50-kDa protein. A single asterisk denotes unglycosylated canine COX-3, and a double asterisk denotes unglycosylated canine PCOX-1a. (B) Densitometric analysis of 65-, 53-, and 50-kDa proteins. Percent relative densitometric units (% rdu) were calculated by comparison to the signal from unblocked primary antibodies. The 50-kDa protein is not detected (n/d) by unblocked or blocked COX-3 polyclonal antibody (pAb).
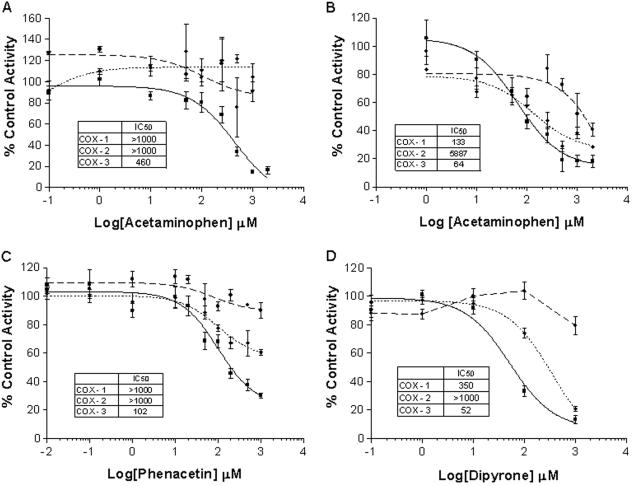
Common analgesic/antipyretic drugs and NSAIDs were tested for their ability to inhibit activity of COX-1, -2, and -3. Analyses were done in the presence of exogenously added arachidonic acid at 30- and 5-μM concentrations. At the higher concentration of substrate, only COX-3 was inhibited by acetaminophen (Fig. 5A). Moreover, COX-3 was significantly more sensitive to acetaminophen than either COX-1 or -2 at the lower substrate concentration (Fig. 5B). Acetaminophen inhibited COX-3 with an IC50 value of 64 μM when done in the presence of 5 μM arachidonic acid, whereas IC50 values for COX-1 and -2 were 2.1- and 92.4-fold higher, respectively.
Figure 5.
Drug inhibition studies. The effects of acetaminophen (A and B), phenacetin (C), and dipyrone (D) on COX-1 (♦), COX-2 (●), and COX-3 (▪) activity in insect cells. COX activity was measured by the formation of PGE2 after exposure to exogenous 5 μM (B) or 30 μM (A, C, and D) arachidonic acid for 10 min. Data are expressed as mean ± SEM (n = 6–9).
Acetaminophen is considered to be the active metabolite of phenacetin, a once popular analgesic/antipyretic drug that is no longer extensively used because of the occurrence of methemoglobinemia, renal toxicity, and suspected renal and bladder carcinogenesis (9, 10). Phenacetin is rapidly O-deethylated in the body to form acetaminophen and is further metabolized to other minor but toxic compounds. Thus, only small levels of phenacetin circulate in the blood. Interestingly, however, phenacetin was more potent at inhibiting COX-3 than was acetaminophen (Fig. 5C). Under substrate conditions of 30 μM, phenacetin inhibited COX-3 at an IC50 value of 102 μM, as opposed to 460 μM for acetaminophen tested under similar conditions. As with acetaminophen, phenacetin preferentially inhibited COX-3.
Another analgesic/antipyretic drug, dipyrone, was also significantly more potent at inhibiting COX-3 than either COX-1 or -2 (Fig. 5D). Dipyrone inhibited COX-3 with an IC50 value of 52 μM and COX-1 at a 6.6-fold higher concentration. No detectable inhibition of COX-2 by dipyrone was observed below 1 mM. Dipyrone is a pro-drug that spontaneously breaks down in aqueous solutions to structurally related pyrazolone compounds that differ in their potency as analgesic/antipyretic agents. Antipyrine and dimethylaminopyrene are related to dipyrone, and possess markedly reduced therapeutic potency and inhibition of COX-3 (Table 1). However, these compounds, like other analgesic/antipyretic agents, preferentially inhibit COX-3.
Table 1.
IC50 values of selected analgesic/antipyretic drugs and NSAIDs
| Drug | IC50, μM
|
||
|---|---|---|---|
| COX-1 | COX-2 | COX-3 | |
| Acetaminophen | >1,000 | >1,000 | 460 |
| Aminopyrine* | >1,000 | >1,000 | 688 |
| Antipyrine | >1,000 | >1,000 | 863 |
| Aspirin | 10 | >1,000 | 3.1 |
| Diclofenac | 0.035 | 0.041 | 0.008 |
| Dipyrone | 350 | >1,000 | 52 |
| Ibuprofen | 2.4 | 5.7 | 0.24 |
| Indomethacin | 0.010 | 0.66 | 0.016 |
| Phenacetin | >1,000 | >1,000 | 102 |
| Caffeine | >1,000 | >1,000 | >1,000 |
| Thalidomide | >1,000 | >1,000 | >1,000 |
All assays were carried out in the presence of 30 μM arachidonic acid.
4-dimethylaminoantipyrine.
COX-3 was also found to differ in its sensitivity to inhibition by a selection of NSAIDs. Diclofenac was the most potent inhibitor of COX-3 tested and diclofenac, aspirin, and ibuprofen preferentially inhibited COX-3 over COX-1 and -2. Thalidomide and caffeine, both of which have been described as having analgesic properties, did not inhibit COX-3. The overall results indicate that COX-3 possesses COX activity that differs pharmacologically from both COX-1 and -2, but is more similar to COX-1.
Discussion
Both COX-3 and PCOX-1a are formed by intron retention, a poorly understood form of alternative splicing. We have previously shown that COX-2 in chicken is regulated by intron 1 retention (11). In the case of chicken COX-2, retention of intron 1 prevents translation and nuclear export of the mRNA. However, both COX-3 and PCOX-1a mRNAs expressed in insect cells retain the intron and are exported from the nucleus and translated (Fig. 3). The polypeptides produced from COX-3 and PCOX-1a include sequence encoded by intron 1 and are functionally different from fully spliced COX-1. Therefore, retention of intron 1 provides a mechanism by which a previously uncharacterized COX enzyme, COX-3, can be produced in cells and tissues. Consistent with the concept that retention of intron 1 is important in creating COX-3 and/or regulating COX-1 is the finding that the DNA sequence of intron 1 from dog, human, and mouse COX-1 genes displays a high degree of conservation. In fact intron 1 is more conserved in these species than is exon 1. COX-1 and -2 genes differ in the placement of intron 1. COX-1 has ten introns, whereas COX-2 has nine. The additional intron in the COX-1 gene is intron 1, which is retained in COX-3. Highly conserved elements of intron 1 may also regulate intron retention.
COX-3 shares all of the catalytic features and important structural features of COX-1 and -2. However, the insertion of intron 1 two amino acids downstream from the initiating methionine would result in the addition of 30 aa to the signal peptide. Despite having a signal peptide and intron-1-encoded sequence retained, COX-3 comigrates with COX-1 in SDS/PAGE gels. It also appears to enter the endoplasmic reticulum where it is glycosylated, and its glycosylation is required for activity. In insect cells, COX-3 shows approximately 20% of the activity of COX-1, which in turn exhibits about 20% of the activity of COX-2. COX-1, COX-2, COX-3, and PCOX-1a all show equivalent expression in our baculovirus system, and so a lowered ability of insect cells to express active COX-1 relative to COX-2 may be due to the inability of insect cells to posttranslationally process COX-1 correctly. Whatever the mechanism, COX-3 also exhibits this problem and to a greater extent than COX-1. Subcellular localization studies done by differential centrifugation demonstrate that COX-3 and PCOX-1a are membrane-bound (data not shown).
Retention of intron 1 could alter folding and may affect dimerization and the active site. These effects could be through structural changes or altered protein targeting. COX-1 site-directed mutagenesis of either Cys-313 or -540, both of which are more than 25 Å from the heme iron, was observed to reduce the activity of the enzyme by 80–90% (12). Therefore, although COX-3 contains all of the COX-1 sequence, the retained intron sequence could significantly alter its enzymatic properties. Inhibition studies of COX-3 indicate this to be the case.
Our studies show COX-3 to be sensitive to drugs that are analgesic/antipyretic, but which have low antiinflammatory activity. Pain and fever have many etiologies that employ complex cellular and biochemical pathways. The finding that COX-3 is sensitive to analgesic/antipyretic drugs suggests that the COX-1 gene plays an integral role in pain and/or fever. Depending on the physiological context, pain pathways involve products from either the COX-1 or -2 genes. COX-2-selective drugs, for example, are clinically useful in inhibiting inflammatory pain in humans (13) and are more potent than COX-1-selective NSAIDs at inhibiting pain induced by proinflammatory agents (e.g., carrageenan) in some paw inflammation assays in rodents (14, 15). COX-1-selective drugs, in contrast, are superior to COX-2-selective agents at inhibiting visceronociception caused by a variety of chemical pain stimulators (16–18). Moreover, Ballou and colleagues (19) found that visceronociception was greatly decreased in COX-1 but not COX-2 knockout mice. Both COX-1 and -2, on the other hand, have been implicated in nociception models that measure analgesia outside the gut, such as in formalin and urate crystal tests (18–20). A role for COX-1 in pain is further supported by the fact that COX-1-selective NSAIDs [e.g., as identified by Warner and colleagues (21)]—such as aspirin, ketorolac, ketoprofen, ibuprofen, and suprofen—are clinically important analgesic agents in humans and animals. Despite their relative exclusion from the brain, these drugs may reach sufficient concentration to effect COX-3 in the brain. Furthermore, the analgesic effects of these drugs often occur at significantly lower doses than those needed to inhibit inflammation (22). Clinical and experimental association of COX-1 and pain may be functionally explained by the finding that COX-1 is a marker for subpopulations of putative nociceptor neurons in the dorsal root ganglion (23).
With regard to pyresis, COX-2 but not COX-1 knockout mice demonstrate reduction in LPS- and interleukin-1-induced fevers (24, 25), and some new COX-1-selective inhibitors, such as SC-560, have proven ineffective at inhibiting LPS-induced fever in animal models (26, 27). Clinically, rofecoxib, a COX-2-selective inhibitor, inhibits naturally occurring fever (28) and also inhibits the maintenance of fever in animal models. Yet aspirin, a COX-1 preferential inhibitor, is one of the most effective antipyretic NSAIDs, and inhibits fever at doses ranging from 5–15 mg/kg (29), far below the 60–80 mg/kg used to treat inflammatory disease (30, 31). Furthermore, nimesulide, a COX-2 preferential inhibitor, was found to be antipyretic in dogs only at plasma concentrations that would also inhibit COX-1 (32). Thus, a role for COX-1 in fever may exist.
The mechanism of action of acetaminophen has been unknown and postulated to be through inhibition of a brain COX that has never been identified (1, 2). Northern blot analysis and cDNA cloning show that COX-3 is expressed in canine brain. COX-3 also appears from Northern blot studies to be expressed in specific regions of the human brain, in particular cerebral cortex (Fig. 1C). Moreover, our studies using ectopically expressed COX-3 in insect cells demonstrate that COX-3 is significantly more sensitive to acetaminophen than COX-1 or -2. The steady-state concentrations of acetaminophen after therapeutic dosage are approximately 100 μM, at which concentration only COX-3 is appreciably inhibited (Fig. 5B). Thus, inhibition of COX-3 in brain and the spinal cord may be the long sought-after mechanism of action of acetaminophen. This proposed mechanism of action also appears to extend to pyrazolone drugs such as dipyrone and related compounds aminopyrine and antipyrine. Dipyrone's active breakdown product, 4-methylaminoantipyrine, reaches concentrations of 104 μM and 86 μM in plasma and the central nervous system, respectively (33). Thus, COX-3 inhibition occurs at known physiological concentrations of pyrazolone drugs.
Analgesic/antipyretic drugs penetrate the blood–brain barrier well and accumulate in the CNS at high enough concentrations to inhibit COX-3. Carboxylate-containing NSAIDs, on the other hand, cross the blood–brain barrier poorly. Still, central analgesic mechanisms of action for carboxylate NSAIDs have been proposed in brain or spinal cord (34, 35). Because COX-3 is so sensitive to some carboxylate NSAIDs, COX-3 in the CNS may be an essential target of both analgesic/antipyretics and standard NSAIDs. Furthermore, the sensitivity of COX-3 to analgesic/antipyretic drugs and NSAIDs observed in these studies (Fig. 5, Table 1) suggests that highly selective inhibitors can be made for COX-3.
Human COX-3 is mainly expressed as an ≈5.2-kb mRNA and has a tissue-specific pattern of expression (Fig. 1C). This ≈5.2-kb mRNA is an alternatively polyadenylated human COX-1 message reported and partly characterized in its 3′ region (5). It appears, therefore, that the retention of intron 1 may influence the site at which the mRNA is polyadenylated. This finding suggests that the 3′ untranslated regions of the mRNA may play a functional role in expression of COX-3 and perhaps PCOX-1a. The functional significance and the mechanism by which intron retention and alternative polyadenylation are coordinated need to be elucidated. It is also interesting to note that the ≈5.2-kb mRNA has been shown to be regulatable (36) and hence may be regulated in response to physiological stimuli and signal transduction. Indeed, the levels of COX-3 mRNA in human and canine cerebral cortex are relatively low. This may be due to cell type-specific expression such as has been shown for COX-1 immunoreactive protein in a subpopulation of putative nociceptor neurons (23). However, COX-3 in human will require further experimentation because some of the published sequences differ by one nucleotide in intron 1 and hence are out of frame. These may constitute genuine polymorphisms or sequencing errors. Alternatively, intron 1 may be out of frame in humans, requiring other mechanisms such as ribosomal frame shifting to produce a functional COX-3 protein.
We have immunologically identified a 65-kDa protein in human aorta, which we postulate is COX-3, and ≈53 kDa proteins, which we postulate to be PCOX-1a. These proteins are detected by both COX-3 antipeptide polyclonal antibody and a COX-1 monoclonal antibody and appear to be present at about 25% of the level of COX-1. The 65-kDa protein is smaller than would be predicted if the protein is glycosylated to the same extent as COX-1, suggesting that hypoglycosylation or other differences exist between the 65-kDa protein and COX-1. The 53-kDa proteins are present as a doublet, and are of a higher molecular weight than that predicted by the PCOX-1a protein primary sequence. This suggests that, like canine PCOX-1a expressed in insect cells, the human protein may be glycosylated, and that different glycosylation states may exist, giving rise to the doublet observed. A 50-kDa protein is also detected only by the COX-1 monoclonal antibody, and is a candidate for being PCOX-1b. It appears to be present at about 15% of the level of COX-1.
PCOX-1a is identical to COX-3 except for a deletion of 219 aa in the catalytic domain of the protein, corresponding to exons 5–8. It lacks detectable cyclooxygenase activity, as shown by its inability to make prostaglandins from arachidonic acid. The deleted portion contains structural helices HE, H1, H2, H3, H5, and part of H6 defined for COX-1 and -2. Of these helices, H2 and H5 form part of the core peroxidase catalytic site. Because of the lack of H2 and H5, PCOX-1 proteins most likely lack detectable peroxidase activity. In this way they are similar to plant pathogen-induced oxygenase (PIOX) enzymes and Gaeumannomyces graminis linoleate diol synthase, which also lack peroxidase activity (37, 38).
Although the peroxidase activity of cyclooxygenase is needed to create the protein radical used in the cyclooxygenase reaction, continued peroxidase activity is not essential for continued cyclooxygenase activity (39). Because only one turnover of the peroxidase active site is required for cyclooxygenase activity in COX-1 and -2, there may be enough residual peroxidase activity in PCOX-1 proteins to prime them.
We have previously found that cyclooxygenases bind to nucleobindin (40). Nucleobindin is a candidate for binding to PCOX-1 proteins as well. Additionally, a form of COX-1 has been described that colocalizes with prostacyclin synthase in filamentous structures of cultured endothelial cells (41). This filamentous form of COX-1 has no cyclooxygenase activity, and is a candidate for being a PCOX-1 protein.
We previously identified an acetaminophen-inhibited COX enzyme activity in a murine macrophage cell line (J774.2) treated with diclofenac (42). We proposed that this activity was a variant of COX-2, because a protein immunoreactive with anti-COX-2 sera was co-induced with the activity. However, this activity was insensitive to aspirin and showed reduced sensitivity to diclofenac, indomethacin, and flurbiprofen—properties not shared with COX-3. This finding suggests that additional distinct acetaminophen-inhibitable COX forms exist that are likely derived from COX-2.
Acknowledgments
We thank and acknowledge the assistance of Marcus Rampton, John Clinger, Jenny Taylor, Joel Wilson, and Eme Ekpo for assistance with Western blots and radioimmunoassays. We thank Kenneth D. Westover for suggesting the acronym PCOX. We gratefully acknowledge the financial support of the National Institutes of Health (Grant AR46688), the Brigham Young University Cancer Research Center, the Brigham Young University Technology Transfer Office, the Office of Research and Creative Work, and the College of Physical and Mathematical Sciences.
Abbreviations
- COX
cyclooxygenase
- NSAID
nonsteroidal antiinflammatory drug
- PCOX-1
partial COX-1
- RT
reverse transcription-coupled
Footnotes
References
- 1.Botting R M. Clin Infect Dis. 2000;31:8202–8210. doi: 10.1086/317520. [DOI] [PubMed] [Google Scholar]
- 2.Flower R J, Vane J R. Nature (London) 1972;240:410–411. doi: 10.1038/240410a0. [DOI] [PubMed] [Google Scholar]
- 3.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. 3rd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 2001. [Google Scholar]
- 4.Simmons D L, Levy D B, Yannoni Y, Erikson R L. Proc Natl Acad Sci USA. 1989;86:1178–1182. doi: 10.1073/pnas.86.4.1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hla T. Prostaglandins. 1996;51:81–85. doi: 10.1016/0090-6980(95)00158-1. [DOI] [PubMed] [Google Scholar]
- 6.Simmons D L, Xie W, Chipman J G, Evett G E. In: Prostaglandins, Leukotrienes, Lipoxins and PAF. Bailey J M, editor. New York: Plenum; 1991. pp. 67–78. [Google Scholar]
- 7.Salmon J A. Prostaglandins. 1978;15:383–397. doi: 10.1016/0090-6980(78)90122-3. [DOI] [PubMed] [Google Scholar]
- 8.O'Neill G P, Ford-Hutchinson A W. FEBS. 1993;330:156–160. doi: 10.1016/0014-5793(93)80263-t. [DOI] [PubMed] [Google Scholar]
- 9.Moyad M A. Semin Urol Oncol. 2001;19:280–293. [PubMed] [Google Scholar]
- 10.Cohen S M, Shirai T, Steineck G. Scand J Urol Nephrol Suppl. 2000;205:105–115. doi: 10.1080/00365590050509869. [DOI] [PubMed] [Google Scholar]
- 11.Xie W, Chipman J G, Robertson D L, Erikson R L, Simmons D L. Proc Natl Acad Sci USA. 1991;88:2692–2696. doi: 10.1073/pnas.88.7.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kennedy T A, Smith C J, Marnett L J. J Biol Chem. 1994;269:27357–27364. [PubMed] [Google Scholar]
- 13.Matheson A J, Figgitt D P. Drugs. 2001;61:833–865. doi: 10.2165/00003495-200161060-00019. [DOI] [PubMed] [Google Scholar]
- 14.Riendeau D, Percival M D, Brideau C, Charleson S, Dube D, Ethier D, Falgueret J P, Friesen R W, Gordon R, Greig G, et al. J Pharmacol Exp Ther. 2001;296:558–566. [PubMed] [Google Scholar]
- 15.Smith C J, Zhang Y, Koboldt C M, Muhammad J, Zweifel B S, Shaffer A, Talley J J, Masferrer J L, Seibert K, Isakson P C. Proc Natl Acad Sci USA. 1998;95:13313–13318. doi: 10.1073/pnas.95.22.13313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kusuhara H, Matsuyuki H, Okumoto T. Prostaglandins Other Lipid Mediat. 1998;55:43–49. doi: 10.1016/s0090-6980(98)00004-5. [DOI] [PubMed] [Google Scholar]
- 17.Goto K, Ochi H, Yasunaga Y, Matsuyuki H, Imayoshi T, Kusuhara H, Okumoto T. Prostaglandins Other Lipid Mediat. 1998;56:245–254. doi: 10.1016/s0090-6980(98)00054-9. [DOI] [PubMed] [Google Scholar]
- 18.Ochi T, Motoyama Y, Goto T. Eur J Pharmacol. 2000;391:49–54. doi: 10.1016/s0014-2999(00)00051-0. [DOI] [PubMed] [Google Scholar]
- 19.Ballou L R, Botting R M, Goorha S, Zhang J, Vane J R. Proc Natl Acad Sci USA. 2000;97:10272–10276. doi: 10.1073/pnas.180319297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Martinez R V, Reval M, Campos M D, Terron J A, Dominguez R, Lopez-Munoz F J. J Pharm Pharmacol. 2002;54:405–412. doi: 10.1211/0022357021778475. [DOI] [PubMed] [Google Scholar]
- 21.Warner T D, Giuliano F, Vojnovic I, Bukasa A, Mitchell J A, Vane J R. Proc Natl Acad Sci USA. 1999;96:7563–7568. doi: 10.1073/pnas.96.13.7563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buckley M M, Brogden R N. Drugs. 1990;39:86–109. doi: 10.2165/00003495-199039010-00008. [DOI] [PubMed] [Google Scholar]
- 23.Chopra B, Giblett S, Little J G, Donaldson L F, Tate S, Evans R J, Grubb B D. Eur J Neurosci. 2000;12:911–920. doi: 10.1046/j.1460-9568.2000.00979.x. [DOI] [PubMed] [Google Scholar]
- 24.Li S, Wang Y, Matsumura K, Ballou L R, Morham S G, Blatteis C M. Brain Res. 1999;825:86–94. doi: 10.1016/s0006-8993(99)01225-1. [DOI] [PubMed] [Google Scholar]
- 25.Li S, Ballou L R, Morham S G, Blatteis C M. Brain Res. 2001;910:163–173. doi: 10.1016/s0006-8993(01)02707-x. [DOI] [PubMed] [Google Scholar]
- 26.Steiner A A, Li S, Llanos Q J, Blatteis C M. Neuroimmunomodulation. 2001;9:263–275. doi: 10.1159/000054289. [DOI] [PubMed] [Google Scholar]
- 27.Dogan M D, Ataoglu H, Akarsu E S. Brain Res Bull. 2002;57:179–185. doi: 10.1016/s0361-9230(01)00739-0. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz J I, Chan C-C, Mukhopadhyay S, McBride K J, Jones T M, Adcock S, Moritz C, Hedges J, Grasing K, Dobratz D, et al. Clin Pharmacol Ther. 1999;65:653–660. doi: 10.1016/S0009-9236(99)90087-5. [DOI] [PubMed] [Google Scholar]
- 29.Vigano A, Dalla Villa A, Cecchini I, Biasini G C, Principi N. Eur J Clin Pharmacol. 1986;31:359–361. doi: 10.1007/BF00981138. [DOI] [PubMed] [Google Scholar]
- 30.Doughty R A, Giesecke L, Athreya B. Am J Dis Child. 1980;134:461–463. doi: 10.1001/archpedi.1980.02130170011005. [DOI] [PubMed] [Google Scholar]
- 31.Gunsberg M, Bochner F, Graham G, Imhoff D, Parsons G, Cham B. Clin Pharmacol Ther. 1984;35:585–593. doi: 10.1038/clpt.1984.81. [DOI] [PubMed] [Google Scholar]
- 32.Toutain P L, Cester C C, Haak T, Laroute V. J Vet Pharmacol Ther. 2001;24:43–55. doi: 10.1046/j.1365-2885.2001.00304.x. [DOI] [PubMed] [Google Scholar]
- 33.Cohen O, Zylber-Katz E, Caraco Y, Granit L, Levy M. Eur J Clin Pharmacol. 1998;54:549–553. doi: 10.1007/s002280050511. [DOI] [PubMed] [Google Scholar]
- 34.Bovill J G. Eur J Anaesthesiol Suppl. 1997;15:9–15. doi: 10.1097/00003643-199705001-00003. [DOI] [PubMed] [Google Scholar]
- 35.Jurna I, Brune K. Pain. 1990;41:71–80. doi: 10.1016/0304-3959(90)91111-U. [DOI] [PubMed] [Google Scholar]
- 36.Plant M H, Laneuville O. Biochem J. 1999;344:677–685. [PMC free article] [PubMed] [Google Scholar]
- 37.Sanz A, Moreno J I, Castresana C. Plant Cell. 1998;10:1523–1537. doi: 10.1105/tpc.10.9.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliw E H, Su C, Sahlin M. Adv Exp Med Biol. 1999;469:679–685. doi: 10.1007/978-1-4615-4793-8_98. [DOI] [PubMed] [Google Scholar]
- 39.Landino L M, Crews B C, Gierse J K, Hauser S D, Marnett L J. J Biol Chem. 1997;272:21565–21574. doi: 10.1074/jbc.272.34.21565. [DOI] [PubMed] [Google Scholar]
- 40.Ballif B A, Mincek N V, Barratt J T, Wilson M L, Simmons D L. Proc Natl Acad Sci USA. 1996;93:5544–5549. doi: 10.1073/pnas.93.11.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liou J Y, Shyue S K, Tsai M J, Chung C L, Chu K Y, Wu K K. J Biol Chem. 2000;275:15314–15320. doi: 10.1074/jbc.275.20.15314. [DOI] [PubMed] [Google Scholar]
- 42.Simmons D L, Botting R M, Robertson P M, Madsen M L, Vane J R. Proc Natl Acad Sci USA. 1999;96:3275–3280. doi: 10.1073/pnas.96.6.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]