Abstract
Using a flow cytometry-based approach, we assessed the viability of Bifidobacterium lactis DSM 10140 and Bifidobacterium adolescentis DSM 20083 during exposure to bile salt stress. Carboxyfluorescein diacetate (cFDA), propidium iodide (PI), and oxonol [DiBAC4(3)] were used to monitor esterase activity, membrane integrity, and membrane potential, respectively, as indicators of bacterial viability. Single staining with these probes rapidly and noticeably reflected the behavior of the two strains during stress exposure. However, the flow cytometry results tended to overestimate the viability of the two strains compared to plate counts, which appeared to be related to the nonculturability of a fraction of the population as a result of sublethal injury caused by bile salts. When the cells were simultaneously stained with cFDA and PI, flow cytometry and cell sorting revealed a striking physiological heterogeneity within the stressed bifidobacterium population. Three subpopulations could be identified based on their differential uptake of the probes: cF-stained, cF and PI double-stained, and PI-stained subpopulations, representing viable, injured, and dead cells, respectively. Following sorting and recovery, a significant fraction of the double-stained subpopulation (40%) could resume growth on agar plates. Our results show that in situ assessment of the physiological activity of stressed bifidobacteria using multiparameter flow cytometry and cell sorting may provide a powerful and sensitive tool for assessment of the viability and stability of probiotics.
Bifidobacteria represent one of the most important bacterial groups of the human gastrointestinal tract. They are believed to play a beneficial role in maintaining the balance of the intestinal microbiota and have been implicated in a number of health-promoting effects (26, 36). Despite their increasing use as probiotics, scientific evidence is still lacking with regard to the mechanisms by which these bacteria can contribute to promotion of health in the host. Thus, knowledge of their metabolic activities and ecology represents an important step in understanding the beneficial effects of these microorganisms and consequently may form a basis for rational selection of probiotic strains (17). This is partly due to the fact that some of the current methodologies lack the resolving power to analyze the composition and the metabolic activity of these microorganisms in situ. Assessment of active or viable microorganisms is often difficult, since no single analytical method identifies all physiological characteristics of a bacterium under a certain condition (1). Although the plate count approach is employed as the standard method for measuring bacterial viability, it only indicates how many of the cells can replicate under the conditions provided for growth. In principle, it allows one to determine viability in a retrospective manner, and there is no explicit evidence that the failure of a bacterial cell to reproduce is an indication that the cell was dead at the time of sampling (6, 14). The ability to reproduce might be repressed or blocked in a certain cell type, or reproduction might be limited to a certain set of conditions. In addition, cell populations that have been exposed to stress can enter a nonculturable state while still maintaining metabolic activity. Moreover, there is increasing evidence that a significant proportion of metabolically active bacteria in the environment cannot be cultured and are known as viable but not culturable or, according to the preference of others, active but not culturable (1, 16, 38).
Fluorescent techniques in combination with flow cytometry (FCM) have been extensively used for assessment of the viability of microorganisms from different environmental samples (3, 7, 28). FCM offers a powerful tool for analyzing a cell population at the single-cell level, since it can be used both to identify and enumerate bacterial populations from environmental samples and to characterize functional properties of the individual cell (14, 31, 34). Moreover, it allows simultaneous measurement of different physical and biochemical parameters and hence offers substantial information on the dynamics and physiological heterogeneity of a bacterial population (10, 11, 23). In addition, FCM offers the ability to physically separate selected cells by cell sorting for further molecular and physiological analysis (15, 37).
The most widely used dyes developed for assessment of cell viability include carboxyfluorescein diacetate (cFDA), a nonfluorescent precursor that readily diffuses across the cell membranes. Once inside the cell, it is converted by nonspecific esterases to a membrane-impermeant fluorescent compound. Retention of the dye by the cell indicates membrane integrity and functional cytoplasmic enzymes, while dead cells do not stain because they lack enzyme activity and the carboxyfluorescein (cF) diffuses freely through the damaged membranes (5, 8). In addition, membrane potential-sensitive probes such as bis-(1,3-dibutylbarbituric acid) trimethine oxonol [DiBAC4(3)] have been used extensively to assess bacterial susceptibility to antibiotics (13, 21, 35) and cell viability (18, 19, 20) by FCM. Oxonol is a negatively charged molecule which enters depolarized and dead cells and binds to lipid-rich compounds, resulting in bright green fluorescence. Another group of probes for viability studies consists of nucleic acid dyes, such as propidium iodide (PI), which are excluded by viable cells with intact membranes but can enter into cells with compromised membranes and bind to the DNA and RNA. The fluorescence conferred by these probes indicates the degree of cell damage, cell permeability, and ultimately cell death (9, 12, 29, 32).
The aim of this study was to assess the viability of the probiotic bacteria Bifidobacterium lactis DSM 10140 and Bifidobacterium adolescentis DSM 20083 during stress due to deconjugated bile salts (dBS) by using a rapid method based on fluorescent probes and FCM. In a first step, we evaluated the possibility of using DiBAC4(3), PI, and cFDA in single-staining assays to monitor the changes in membrane potential, membrane permeability, and enzyme activity, respectively, of the two strains under stress conditions, and we compared values for these parameters with those obtained by the plate count method. In a second step, a multiparameter FCM assay was used in combination with cell sorting to determine the contribution of each single cell to the overall physiological status of the bacterial population.
MATERIALS AND METHODS
Bacterial strains and media.
B. lactis DSM 10140 and B. adolescentis DSM 20083 were purchased from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany). The strains were grown in MRS (Oxoid) supplemented with 0.05% l-cysteine-HCl (wt/vol) and adjusted to pH 6.8 in an anaerobic chamber with an atmosphere of 10% CO2, 10% H2, and 80% N2. Stock cultures of the two strains were maintained at −80°C in MRS containing 0.05% l-cysteine-HCl (wt/vol) and 30% glycerol (vol/vol).
Stress conditions.
One milliliter of the stock culture was diluted in 10 ml of MRS broth supplemented with 0.05% l-cysteine-HCl (wt/vol) and incubated at 37°C in the anaerobic chamber for 15 to 16 h. The overnight culture was then diluted 10-fold in a fresh MRS broth containing 0.05% l-cysteine HCl at 37°C, and the subcultured cells were allowed to grow anaerobically to reach the mid-exponential phase, corresponding to a concentration of approximately 108 cells/ml and an optical density at 620 nm (OD620) of 0.6 to 0.7. The bacterial culture was then centrifuged in a Mistral 3000 centrifuge (3,000 × g for 10 min at 4°C), and the pellet was washed twice with anaerobic potassium phosphate buffer (50 mM; pH 7) containing 1 mM dithiothreitol (DTT). The cells were resuspended in the same buffer to obtain the desired bacterial density. Cell suspensions of approximately 108 cells/ml were exposed to dBS, consisting of 50% sodium cholate and 50% sodium deoxycholate (Sigma-Aldrich, Steinheim, Germany), to a final concentration of 0.05, 0.1, 0.2, 0.25, or 0.3% (wt/vol) for 10 min at 37°C in anaerobic potassium phosphate buffer (50 mM; pH 7; containing 1 mM DTT). Untreated and heat-treated cells (70°C for 30 min) served as control samples.
Probes.
cFDA, PI, and DiBAC4(3) were obtained from Molecular Probes Europe BV, Leiden, The Netherlands.
DiBAC4(3) staining.
A stock solution (1 mM) of the membrane potential probe DiBAC4(3) was made up in dimethyl sulfoxide and kept at −20°C. A working solution of 250 μM was prepared in ethanol and stored at 4°C. The staining buffer contained 0.06 M Na2HPO4 and 0.06 M NaH2PO4, mixed to produce a solution with a pH of 7 and supplemented with 5 mM KCl, 130 mM NaCl, 1.3 mM CaCl2, 0.6 mM MgCl2, and 10 mM glucose (24). Samples were diluted in this buffer to approximately 106 to 107 cells/ml and were incubated anaerobically for 4 min at 37°C in the presence of 1 μM DiBAC4(3). When appropriate, 15 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP) was added as a depolarizing agent. Heat-treated cells (70°C for 30 min) were used as a positive control for DiBAC4(3) staining.
PI staining.
PI was supplied by the manufacturer as a 1-mg/ml solution in distilled water and was used as the working solution and stored in the refrigerator in the dark. Ten microliters of each sample was added to 985 μl of anaerobic potassium phosphate buffer (50 mM; pH 7; containing 1 mM DTT) in the presence of 5 μl of PI. The mixture was incubated for 15 min at 37°C in a water bath to allow staining of the cells. Samples were kept in the dark on ice and used within 1 h for FCM analysis.
cFDA staining.
A stock solution (10 mM) of cFDA was prepared by dissolving 4.6 mg of cFDA/ml in acetone and was stored at −20°C in the dark. The stock solution was further diluted in acetone to 1 mM and served as the working solution. Samples containing approximately 106 to 107 cells/ml were incubated in anaerobic potassium phosphate buffer (50 mM; pH 7; containing 1 mM DTT) in the presence of 10 μM cFDA for 30 min at 37°C in a water bath. Stained samples were kept on ice in the dark no longer than 1 h, until FCM analysis was performed.
Double staining.
When dual labeling was performed, the same dye concentrations and incubation times, described above, were used. Mixtures of heat-killed (70°C for 30 min) and freshly harvested cells were stained with cFDA and PI both in single-staining and in multistaining assays. The mixed cultures along with the unstained cultures served as controls by which to set the flow cytometer detectors and compensation. Cells electroporated in the presence of PI and subsequently stained with cFDA were used as a control for the double-stained cells. Overnight cultures of B. lactis and B. adolescentis were used to inoculate fresh MRS supplemented with 0.5% cysteine-HCl, and the suspensions were incubated anaerobically at 37°C for 3 to 4 h, until an OD620 of approximately 0.6 to 0.7 was reached. The cells were then harvested by centrifugation and washed twice with 1 mM HEPES buffer supplemented with 0.5 M sucrose, and the pellets were resuspended in the same buffer. PI (5 μM) was added to 200-μl bacterial suspensions (OD620 = 10) in a precooled Gene Pulser disposable cuvette (interelectrode distance, 0.2 cm; Bio-Rad). An electrical pulse of 1, 1.2, 1.4, 1.8, or 2.0 kV was delivered with a Gene Pulser apparatus (Bio-Rad) by using the 25-μF capacitor and setting the pulse collector at 200 Ω parallel resistance. Subsequently, the bacteria were diluted with 800 μl of MRS containing 0.05% l-cysteine-HCl and incubated anaerobically for an additional 2 h for recovery. Finally, the PI-labeled cells were centrifuged, washed twice with 50 mM anaerobic potassium phosphate buffer (50 mM; pH 7; containing 1 mM DTT), and stained with cFDA as described above. These cells were used to adjust the FCM detectors and check for PI toxicity after the sample was plated onto MRS agar plates.
FCM analysis.
Samples were analyzed with a FACSCalibur flow cytometer (Becton Dickinson Immunocytometry Systems, San Jose, Calif.) equipped with an air-cooled argon ion laser emitting 15 mW of blue light at 488 nm and with the standard filter setup. The side scatter signal was used as a trigger signal. The green fluorescence from cF- and DiBAC4(3)-stained cells was detected through a 530-nm, 30-nm-bandwidth band-pass filter (FL1 channel), and the red fluorescence of the PI signal was collected in the FL3 channel (>600-nm long-pass filter). FACSFlow solution (Becton Dickinson) was used as the sheath fluid. All bacterial analyses were performed at the low rate settings (12 μl/min), and the sample concentration was adjusted to keep the count lower than 1,000 events/s. Data were collected in list mode as pulse height signals (4 decades each on a logarithmic scale) and analyzed by using the Windows Multiple Document Interface computer program (WinMDI; Joseph Totter, Salk Institute for Biological Studies, La Jolla, Calif.; available at http://facs.Scripps.edu/software.html). The machine was checked weekly for alignment by using 0.7-μm-diameter green-yellow fluorescent beads (Polyscience, Eppelheim, Germany).
Cell sorting.
B. lactis cultures were first exposed to 0.1% dBS for 10 min at 37°C and then simultaneously stained with 10 μM cFDA and 5 μg of PI/ml, as described above. Cells were analyzed by FCM, and sort gates were defined on an FL1-versus-FL3 dot plot of cFDA- and PI-stained cells. The sorter was set to single-cell mode, and sorted cells were collected in a 50-ml sterile Greiner tube. Sorting was stopped after 2 min, which corresponded to the acquisition of 16,000 to 20,000 events for untreated samples and 4,000 to 5,000 events for stressed cells. Filter-sterilized phosphate-buffered saline, pH 7, was used as the sheath fluid. To determine the purity and the recovery rate of sorted cells from the defined gates, the samples were reanalyzed in the FCM. The sorted cells were centrifuged at 4,000 × g for 20 min, the supernatant was carefully removed, and approximately 0.5 ml was left in the bottom of the tube. To this remaining volume, which contained the sorted cells, 1 ml of MRS containing 0.05% l-cysteine-HCl was added, and the tubes were then incubated for 2 to 3 h at 37°C in the anaerobic chamber. Afterwards, the samples were plated anaerobically onto MRS agar plates containing 0.05% l-cysteine-HCl and incubated at 37°C for 72 h in anaerobic jars containing the Oxoid Gas Pack Anaerobic system.
Plate counts.
Prior to plating, samples were washed twice in anaerobic potassium phosphate buffer and then diluted with saline solution (0.8%) containing 1 g of peptone (Oxoid) and 0.05% cysteine-HCl. Portions (100 μl) of the appropriate dilutions were spread onto MRS supplemented with 0.05% HCl-cysteine and containing 1.5% agar under anaerobic conditions. Plates were incubated in anaerobic jars containing the Oxoid Gas Pack Anaerobic system for 3 days at 37°C.
RESULTS
Single staining and viability assessment of bifidobacteria. (i) Membrane potential.
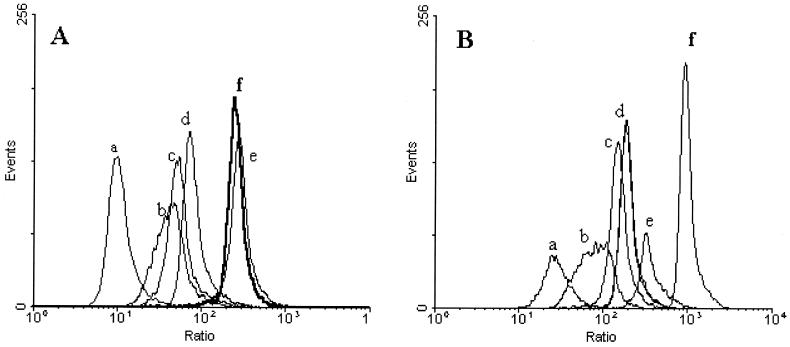
Distinct DiBAC4(3) fluorescence distributions of viable and dead bifidobacteria cells could be measured in mixed cultures of heat-killed (70°C for 30 min) and freshly harvested cells (Fig. 1A and B). Upon addition of 15 μM CCCP to freshly harvested cells, green fluorescence increased significantly over that of the untreated cells, indicating that indeed DiBAC4(3) fluorescence is due to a membrane potential-dependent dye (Fig. 1C). Furthermore, the dual-parameter dot plots show that DiBAC4(3) fluorescence and the side scatter signal are closely correlated for all cells, suggesting that DiBAC4(3) fluorescence is influenced by cell size. Thus, a new parameter proportional to the log ratio of the green fluorescence to the side scatter intensity was calculated for all cells by use of FSCPress software (Ray Hiks, Department of Medicine, University of Cambridge, Cambridge, United Kingdom; available at http://www.fcspress.com). The ratio derived from the DiBAC4(3) fluorescence allowed clear discrimination between the different ratio distributions for each treatment for both strains (Fig. 2). The median fluorescence intensity of treated cells was shifted toward higher channel numbers for both strains as the concentration of bile salts increased, revealing a gradual depolarization of the cell population related to cell death. Exposure to 0.05, 0.1, and 0.2% dBS for 10 min resulted in populations with membrane potentials intermediate between those observed for intact and heat-treated cells for both strains. When cells of B. lactis were exposed to 0.25% dBS, DiBAC4(3) staining revealed a fully depolarized (dead) population with a ratio similar to that of heat-killed cells (Fig. 2A), while for B. adolescentis the ratio was higher in heat-treated cells than in cells treated with 0.25% dBS (Fig. 2B).
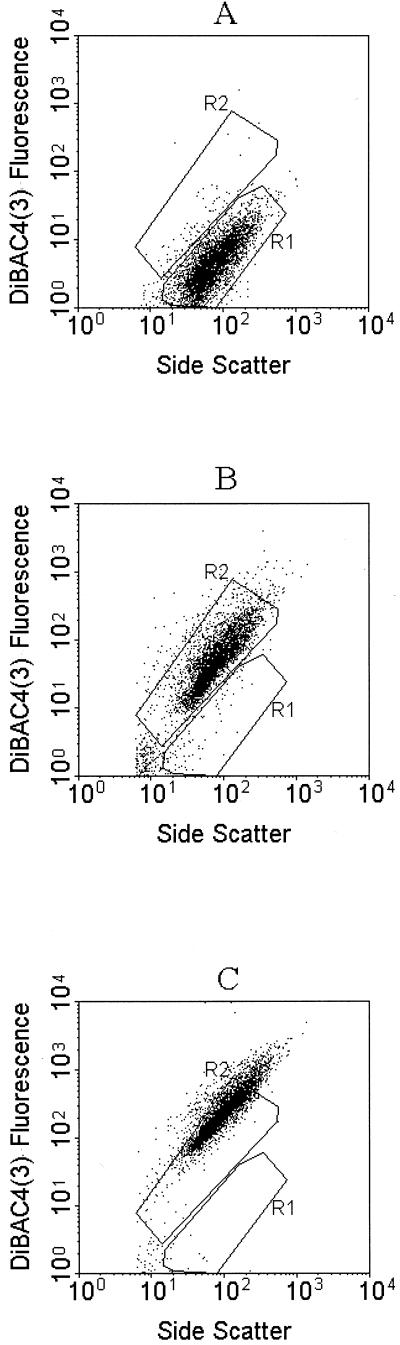
FIG. 1.
Dual-parameter dot plot of the side scatter intensity versus DiBAC4(3) fluorescence of B. lactis. Untreated cells (A), cells incubated with 15 μM CCCP (B), and cells that had been heat killed at 70°C for 30 min (C) were stained with 1 μM DiBAC4(3) and analyzed by FCM. Data show the effects of the ionophore CCCP on the membrane potential of B. lactis and a high correlation between DiBAC4(3) fluorescence and the side scatter signal. Control cells are gated on region R1 and display little green fluorescence, while cells gated on region R2 show a marked increase in green fluorescence as a result of the addition of 15 μM CCCP or heat treatment (70°C for 30 min).
FIG. 2.
Fluorescence histogram overlays of B. lactis DSM 10140 (A) and B. adolescentis DSM 20083 (B) stained with 1 μM DiBAC4(3) for 4 min at 37°C. The x axis represents the corrected DiBAC4(3) fluorescence for the cell size, obtained by calculating the log ratio of the green fluorescence of DiBAC4(3) to the side scatter intensity. Results are shown for control untreated cells (a), for cells exposed to 0.05% (b), 0.1% (c), 0.2% (d), or 0.25% (e) dBS, and for cells that were heat treated at 70°C for 30 min (f).
(ii) Membrane integrity.
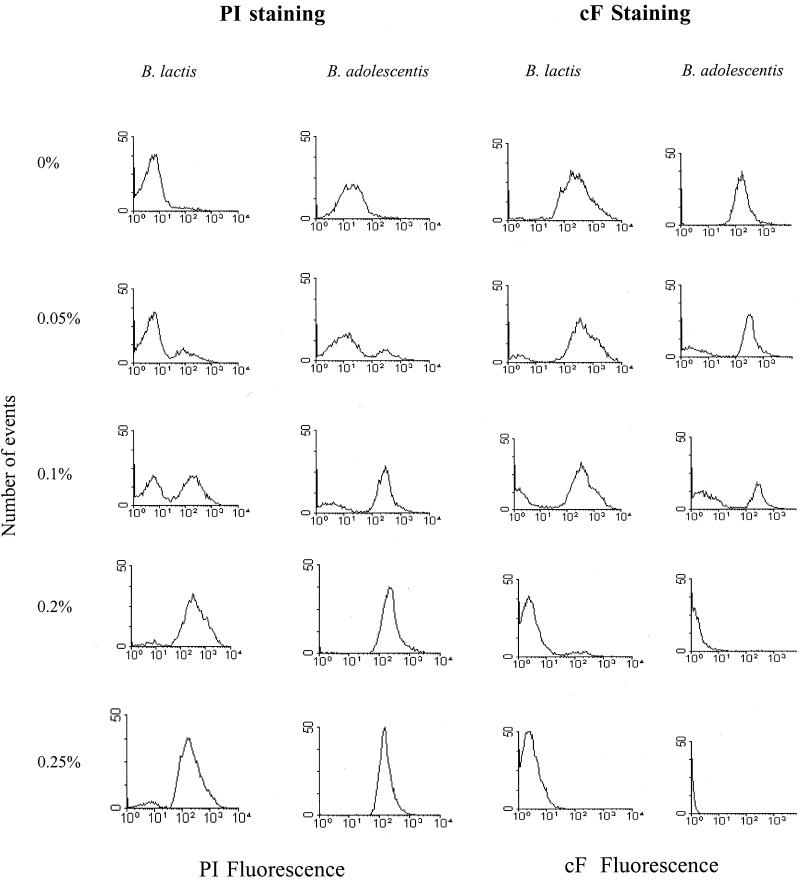
PI is commonly used as a cell death marker because it is excluded by intact plasma membranes; thus, the fluorescence conferred by the probe is generally associated with cells that have lost their membrane integrity. When bifidobacterium cells were heat treated (70°C for 30 min), the whole population was permeable to PI. The red-fluorescent events were detected above channel values of 258 and 520 of the FL3 detector for B. lactis and B. adolescentis, respectively, while a small fraction of the untreated cells (5 to 8%) was PI positive. Following bile salt treatment, a second high peak appeared above channel 255 or 520 (PI-positive subpopulation), representing the damaged, PI-permeable cells of B. lactis and B. adolescentis, respectively, as shown in Fig. 3. At dBS concentrations of 0, 0.05, 0.1, and 0.2%, the percentages of PI-permeable cells were 5, 21, 74, and 98%, respectively, for B. adolescentis and 7, 23, 56, and 91%, respectively, for B. lactis.
FIG. 3.
Fluorescence histograms of B. lactis and B. adolescentis stained with 5 μg of PI/ml or 10 μM cFDA following exposure to different concentrations of dBS.
(iii) Esterase activity.
The percentage of cF-stained cells was used to estimate the viability of dBS-treated cells. Retention of cF by the cells indicates enzyme activity as well as membrane integrity. Figure 3 shows fluorescence histograms of dBS-treated B. lactis and B. adolescentis cells stained with cFDA. A clear separation between stained and unstained subpopulations was observed with all treatments and for both strains. With increasing concentrations of dBS, the fraction with a fluorescent intensity above channel 255 of the FL1 detector decreased for B. lactis and B. adolescentis, and a cF-negative subpopulation could be observed. For B. lactis the cF-positive population decreased from 95, 73, and 40% to reach 1% after exposure to 0, 0.05, 0.1, and 0.2% dBS, respectively. At the same concentrations of dBS, B. adolescentis showed a lower percentage of cF-stained cells than B. lactis.
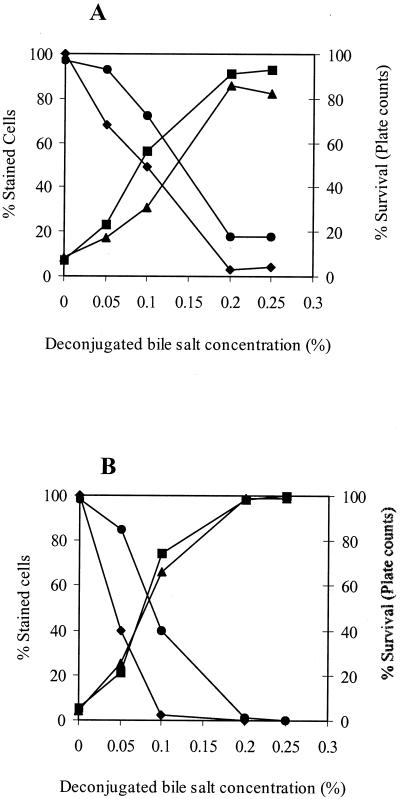
Figure 4 shows the numbers of permeable (damaged) cells (PI positive), depolarized cells [DiBAC4(3) positive], and intact (active) cells (cF positive) and the number of CFU per milliliter for both strains. The number of PI-stained bacteria correlated highly with the number of DiBAC4(3)-stained bacteria and displayed an inverse relation with the percentage of survival of the cells as determined by plate counts. The percentage of cF-stained cells showed the same trend as the results obtained by plate counts; however, it gave a higher estimation of the viability of B. lactis and B. adolescentis, by 20 to 30%, respectively, for all treatments.
FIG. 4.
Viability assessment of bile salt-stressed cells of B. lactis DSM 10140 (A) and B. adolescentis DSM 20083 (B) by FCM and plate counts. Cells were harvested at the mid-exponential phase and then exposed to 0, 0.05, 0.1, or 0.2% dBS for 10 min at 37°C in anaerobic potassium phosphate buffer (50 mM; pH 7; containing 1 mM DTT). Results are expressed as percentages of cells stained with either cF (•), PI (▴), or DiBAC4(3) (▪) and as the percentage of survival as determined by plate counts (⧫).
Multiparameter FCM.
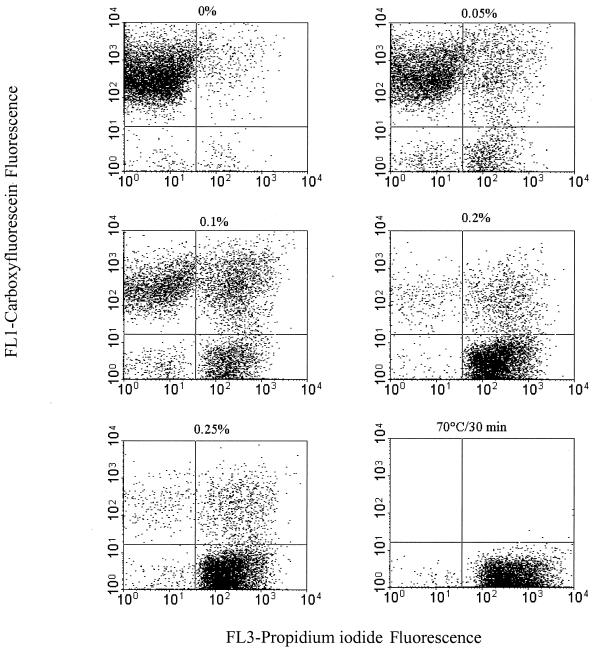
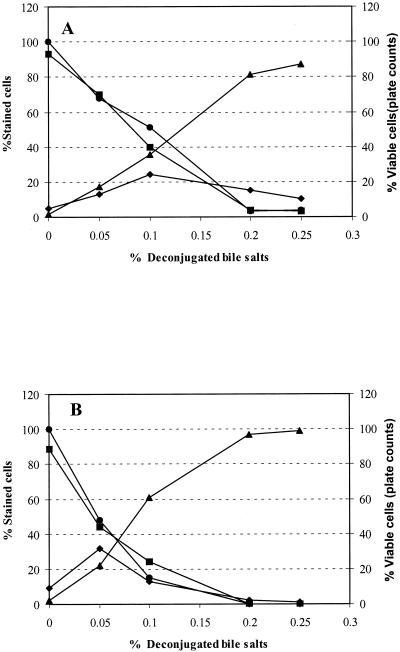
In order to validate the multiparameter assay, it was necessary to establish adequate controls. Unstained, cF-stained, PI-stained, and cF and PI double-stained cells served as controls by which to set the FCM detectors and the compensation settings. The quadrants of the dot plots were set so that the unstained cells appeared in the lower left quadrant. Cultures of both strains were exposed to 0, 0.05, 0.1, 0.2, or 0.25% dBS and then simultaneously stained with cFDA and PI. The dual-parameter dot plots of Fig. 5 indicate the existence of three main subpopulations of bile salt-treated B. lactis and show dynamic changes with increases in bile salt concentration. These subpopulations are identified based on their differential staining characteristics with PI and cF. These subpopulations consisted of cells that stained only with cF (upper left quadrant; cF+ PI−), cells that stained only with PI (lower right quadrant; cF− PI+), and a double-stained population (upper right quadrant; cF+ PI+). A fourth population that did not exceed 3% appeared in the lower left quadrant (cF− PI−); it most likely represented debris or lysed cells. The cF-stained subpopulation (cF+ PI−) decreased with increasing bile salt concentrations and accounted for 92, 67, 38, 4, and 3% of the total population after exposure to 0, 0.05, 0.1, 0.2, and 0.25% dBS, respectively. The cF+ PI− cells corresponded to the number of CFU and thus was scored as the viable subpopulation (Fig. 6A). The fraction that stained only with PI (cF− PI+) increased from 2, 13, 31, and 84% to reach 90% after exposure to 0, 0.05, 0.1, 0.2, and 0.25% dBS, respectively, and was scored as the dead population. The percent double-stained B. lactis cells (cF+ PI+) showed a subtle fluctuation during the course of the stress exposure. This fraction constituted 5, 16, 28, 11, and 9% of the total population after treatment with 0, 0.05, 0.1, 0.2, and 0.25% dBS, respectively. Similar trends were obtained with B. adolescentis (Fig. 6B), showing the succession of the three physiological states within the stressed population of B. adolescentis.
FIG. 5.
Multiparameter dot plots of B. lactis DSM 10140 representing PI fluorescence versus cF fluorescence. Cultures were exposed for 10 min to 0, 0.05, 0.1, 0.2, or 0.25% dBS in anaerobic potassium phosphate buffer (50 mM; pH 7; containing 1 mM DTT) for 10 min at 37°C. Subsequently, all samples were stained simultaneously with 10 μM cFDA and 5 μg of PI/ml and were analyzed by FCM. Three main subpopulations, corresponding to viable cF-stained cells (upper left quadrant), injured cells double stained with PI and cF (upper right quadrant), and dead PI-stained cells (lower right quadrant), can be readily differentiated.
FIG. 6.
Viability assessment of B. lactis DSM 10140 (A) and B. adolescentis DSM 20083 (B) using multiparameter FCM and the plate count method. Results illustrate the different physiological states of bile salt-stressed cells, consisting of viable, active cells stained only with cF (▪), injured cells stained with both cF and PI (⧫), dead cells stained only with PI (▴), and culturable cells determined by the plate count method •.
Cell sorting.
To ascertain that double-positive (cF+ PI+) cells corresponded to bacteria with only partially damaged cell membranes and therefore represented the injured cells; we sorted cells from different gates. The gates were defined in the PI fluorescence-versus-cF fluorescence dot plots as follows: cells from gate 1 (G1) were stained with cF only; cells from gate 2 (G2) were double stained; cells from gate 3 (G3) were stained with PI. The cell fractions sorted from each gate were mapped by FCM reanalysis in those regions that had been defined by the respective sort gates. The FCM reanalysis demonstrated high purity (95%), while the recovery rate did not exceed 73 to 77%, as shown in Table 1. Higher sorting recovery resulted in both lower purity and reduced growth of the cells (data not shown). Sorting of bacteria from different functional stages and subsequent incubation in MRS broth revealed that all types of cells except the PI+ cF− cells are capable of resuming growth on agar plates. A significant number of the double-stained, injured subpopulation (40%) regained the capacity for cell proliferation (Table 1).
TABLE 1.
Results of sorting and recovery of B. lactis DSM 10140 cells treated with 0.1% dBS for 10 min at 37°Ca
| Gate and labelb | % Recoveryc | CFU/100 μl | % Growthd |
|---|---|---|---|
| G1 (cF+ PI−) | 77 | 149 | 47 |
| G2 (cF+ PI+) | 73 | 40 | 40 |
| G3 (cF− PI+) | 75 | 0 | 0 |
Following exposure to bile salts, cells were stained simultaneously with 10 μM cFDA and 5 μg of PI/ml and were then sorted into sterile tubes.
Sort gates of differentially stained target populations.
Calculated as (number of total sorted events/number of events reanalyzed after sorting) × 100.
Calculated as (number of CFU of the total sorted sample/number of sorted events as determined after recovery) × 100.
DISCUSSION
In this paper, we report on the use of PI, an exclusion dye, cFDA, an intracellular enzymatic stain, and DiBAC4(3), a membrane potential probe, used in single and/or multiparameter FCM analysis to assess the viability of B. lactis and B. adolescentis during bile salt stress. Clear discrimination between viable intact, permeabilized, and depolarized cells was achieved by monitoring the extents to which they retained cF and accumulated PI or DiBAC4(3) during bile salt exposure. The results of the single-staining approach provide extensive evidence that the three fluorescent probes do reflect the responses of B. lactis and B. adolescentis to the damaging effects of bile salts and thus reflect the degree of cell viability (3, 6, 12, 19).
Examining the membrane potential provided an additional means of characterizing the physiological status of bile salt-stressed cells. Correction of DiBAC4(3) fluorescence for bacterial size variations allowed for better discrimination between viable cells and cells that were depolarized (dead) as a result of bile salt stress than the initial DiBAC4(3) fluorescence distribution. We found that the ratio was higher for heat-killed B. adolescentis cells than for CCCP-treated cells or cells treated with a lethal dose of dBS, e.g., 0.25% (Fig. 2B). These results suggest that the fluorescence of DiBAC4(3) was affected differently in cells treated with dBS and CCCP than in heat-treated cells (24, 25, 31). The ratiometric method used in our study will offer an accurate approach to measuring bacterial membrane potential and assessing the viability of gram-positive bacteria.
The validity of cFDA for reflecting viability during stress has been reported for lactic acid bacteria and a number of other microbes, and high correlations between plate counts and FCM counts have been obtained (4, 8, 12, 27). In this study we showed that, for both strains and for all stress conditions, single staining with cFDA always gave a higher estimation of the number of viable cells than plate counts. This discrepancy was higher with B. adolescentis, possibly because this strain is more oxygen sensitive than B. lactis, and probably plating exerted an additional stress on this microorganism (22). It has been reported that bifidobacterium cells when exposed to oxygen could ferment carbohydrates, even though they could not increase in number by cell division due to oxygen toxicity (33). The difference observed between the FCM and plate count results suggests the presence in the stressed population of cells that could maintain cell metabolic activity, as determined by the fluorescent dyes, yet were not able to form colonies on agar plates. Indeed, stressed and starved cells can enter a nonculturable state, most likely due to “sublethal-injury” mechanisms including damage to the cell membrane, protein, and/or DNA, and can recover by repairing or replacing those damaged molecules (1, 14, 16). Temporary nonculturability has been reported for starved Micrococcus luteus (15) and Escherichia coli (27) cells by use of FCM and cell sorting.
Multicolor FCM has been successfully used to assess physiological heterogeneity within bacterial populations during bacterial fermentations (10, 11) and to gain insight into the mechanism of action of antibiotics on Staphylococcus aureus and M. luteus (25). Our multiparameter FCM results clearly illustrated the succession of cell changes that occurred in a bile salt-stressed bifidobacterium population and revealed physiological heterogeneity within the cell population (23). Cell sorting confirmed that the bile salt-treated cell populations contained a mixture of viable cells, dead cells, and an injured (stressed) subpopulation stained with PI and cF. Regrowth of the injured cells following sorting confirmed that a fraction of the stressed cells adopted a latent state in which they could not reproduce but could be induced to a physiologically active state after recovery (1, 6, 15, 16). The percent recovery of injured cells (40%) and the percent viable cells (47%) did not reach higher values presumably because of the additional stress caused by sorting and plating (11, 23). The electroporated PI- and cF-stained bifidobacterium cells were able to grow on agar plates (data not shown), showing that the concentration of PI (5 μg/ml) used in our experiment was not toxic for bifidobacteria. Multiparametric results show that cell permeability as monitored by PI is a sensitive marker of cell damage, yet it is a poor indicator of cell death of stressed bacteria (14, 25, 31). Therefore, we assume that bile salts induced sublethal injury within the bifidobaterium population, possibly through a reversible and transient membrane permeabilization which resulted in a loss of viability, as defined by plate counts, but these cells could regain growth after being sorted and resuscitated. The precise mechanism of action of bile salts is unknown, but these compounds act as detergents for the digestion of fats in the intestinal tract and are reported to have inhibitory effects on a number of bacteria. Recently, it was shown that bile acids induce expression of specific stress response genes in E. coli, possibly in response to membrane perturbation, oxidative stress, or DNA damage (2). Bile salt-stressed B. adolescentis NCC481 showed a remarkably increased resistance to lethal concentrations of bile salts, most likely through induction of a mechanism allowing the cells to build up a protection against the solubilization of their membrane proteins (30).
In this study we showed that multiparameter FCM combined with cell sorting provides a way not only to distinguish between live and dead cells but also to discriminate between different physiological states of a stressed bifidobacterium population. Moreover, it clearly illustrates the dynamics in the physiology of microbial populations during dBS treatment. In conclusion, this method may provide a novel tool for assessing the viability and stability of bacteria during the processing and storage of probiotic products. Furthermore, we aim to use this approach, along with molecular techniques such as fluorescent in situ hybridization, to analyze the activity and stability of these microorganisms within the complex ecosystem of the gastrointestinal tract.
REFERENCES
- 1.Barer, M. R., and C. R. Harwood. 1999. Bacterial viability and culturability. Adv. Microb. Physiol. 41:93-137. [DOI] [PubMed] [Google Scholar]
- 2.Bernstein, C., H. Bernstein, C. M. Payen, S. E. Beard, and J. Schneider. 1999. Bile salt activation of stress response promoters in Escherichia coli. Curr. Microbiol. 39:68-72. [DOI] [PubMed] [Google Scholar]
- 3.Breeuwer, P., and T. Abee. 2000. Assessment of viability of microorganisms employing fluorescence techniques. Int. J. Food Microbiol. 55:193-200. [DOI] [PubMed] [Google Scholar]
- 4.Bunthof, C. J., S. van Schalkwijk, W. Meijer, T. Abee, and J. Hugenholtz. 2001. Fluorescent method for monitoring cheese starter permeabilization and lysis. Appl. Environ. Microbiol. 67:4264-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bunthof, C. J., K. Bloemen, P. Breeuwer, F. M. Rombouts, and T. Abee. 2001. Flow cytometry assessment of the viability of lactic acid bacteria. Appl. Environ. Microbiol. 67:2326-2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davey, H. M., D. H. Welchart, D. B. Douglas, and A. S. Kaprelyants. 1999. Estimation of microbial viability using flow cytometry, p. 11.3.11-11.3.20. In P. Robinson, Z. Darzynkiewics, P. Dean, A. Orfao, P. Rabinovitch, H. Tank, and L. Wheeless (ed.), Current protocols in cytometry, Wiley, New York, N.Y.
- 7.Davey, H. M., and D. B. Kell. 1996. Flow cytometry and cell sorting of heterogeneous microbial populations: the importance of single-cell analyses. Microbiol. Rev. 60:641-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Diaper, J. P., and C. Edwards. 1994. The use of fluorogenic esters to detect viable bacteria by flow cytometry. J. Appl. Bacteriol. 77:221-228. [Google Scholar]
- 9.Ericsson, M., D. Hanstorp. P. Hagberg, J. Enger, and T. Nyström. 2000. Sorting out bacterial viability with optical tweezers. J. Bacteriol. 182:5551-5555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hewitt, C. J., and G. Nebe-von-Caron. 2001. An industrial application of multiparameter flow cytometry: assessment of cell physiology state and its application to the study of microbial fermentations. Cytometry 44:179-187. [DOI] [PubMed] [Google Scholar]
- 11.Hewitt, C. J., G. Nebe-von-Caron, A. W. Niennow, and C. M. McFarlane. 1999. The use of multi-parameter flow cytometry to compare the physiological response of Escherichia coli W3110 to glucose limitation during batch, fed-batch and continuous culture cultivations. J. Biotechnol. 75:251-264. [DOI] [PubMed] [Google Scholar]
- 12.Humphreys, M. J., R. Allman, and D. Lloyd. 1994. Determination of the viability of Trichomonas vaginalis using flow cytometry. Cytometry 15:343-348. [DOI] [PubMed] [Google Scholar]
- 13.Jepras, R. I., F. E. Paul, S. C. Pearson, and M. J. Wilkinson. 1997. Rapid assessment of antibiotic effects of Escherichia coli by bis-(1,3-dibutylbarbituric acid) trimethine oxonol and flow cytometry. Antimicrob. Agents Chemother. 41:2001-2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joux, F., and P. Lebaron. 2000. Use of fluorescent probes to assess physiological functions of bacteria at single-cell level. Microbes Infect. 2:1523-1535. [DOI] [PubMed] [Google Scholar]
- 15.Kaprelyants, A. S., G. V. Mukamolova, H. M. Davey, and D. B. Kell. 1996. Quantitative analysis of the physiological heterogeneity within starved cultures of Micrococcus luteus by flow cytometry and cell sorting. Appl. Environ. Microbiol. 62:1311-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kell, D. B., A. S. Kaprelyants, D. H. Weichart, C. R. Harwood, and M. R. Barer. 1998. Viability and activity in readily culturable bacteria: a review and discussion of practical issues. Antonie Leeuwenhoek 73:169-187. [DOI] [PubMed] [Google Scholar]
- 17.Klaenhammer, T., R., and M. J. Kullen. 1999. Selection and design of probiotics. Int. J. Food Microbiol. 50:45-57. [DOI] [PubMed] [Google Scholar]
- 18.Lopez-Amoros, R., J. Comas, and J. Vives-Rego. 1995. Flow cytometric assessment of Escherichia coli and Salmonella typhimurium starvation-survival in seawater using rhodamine 123, propidium iodide, and oxonol. Appl. Environ. Microbiol. 61:2521-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lopez-Amoros, R., S. Castel, J. Comas-Riu, and J. Vives-Rego. 1997. Assessment of E. coli and Salmonella viability and starvation by confocal laser microscopy and flow cytometry using rhodamine 123, DiBAC4(3), propidium iodide, and CTC. Cytometry 29:298-305. [DOI] [PubMed] [Google Scholar]
- 20.Mason, D. J., R. Lopez-Amoros, R. Allman, J. M. Stark, and D. Lloyd. 1995. The ability of membrane potential dyes and calcafluor white to distinguish between viable and non-viable bacteria. J. Appl. Bacteriol. 78:309-315. [DOI] [PubMed] [Google Scholar]
- 21.Mason, D. J., R. Allman, J. M. Stark, and D. Lloyd. 1994. Rapid estimation of bacterial antibiotic susceptibility with flow cytometry. J. Microsc. 176:8-16. [DOI] [PubMed] [Google Scholar]
- 22.Meile, L., W. Ludwig, U. Rueger, C. Gut, P. Kaufmann, G. Dasen, S. Wenger, and M. Teuber. 1997. Bifidobacterium lactis sp. nov., a moderately oxygen tolerant species isolated from fermented milk. Syst. Appl. Microbiol. 20:56-64. [Google Scholar]
- 23.Nebe-von-Caron, G., P. J. Stephens, C. H. Hewitt, J. R. Powell, and R. A. Badley. 2000. Analysis of bacterial function by multi-colour fluorescence flow cytometry and single cell sorting. J. Microbiol. Methods 42:97-114. [DOI] [PubMed] [Google Scholar]
- 24.Novo, D., N. G. Perlmutter, R. H. Hunt, and H. M. Shapiro. 1999. Accurate flow cytometric membrane potential measurement in bacteria using diethyloxacarbocyanine and ratiometric technique. Cytometry 35:55-63. [DOI] [PubMed] [Google Scholar]
- 25.Novo, D. J., N. G. Perlmutter, R. H. Hunt, and H. M. Shapiro. 2000. Multiparameter flow cytometric analysis of antibiotic effects on membrane potential, membrane permeability, and bacterial counts of Staphylococcus aureus and Micrococcus luteus. Antimicrob. Agents Chemother. 44:827-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ouwehand, A. C., P. V. Kirjavainen, C. Shortt, and S. Salminen. 1999. Probiotics: mechanisms and established effects. Int. Dairy J. 9:43-52. [Google Scholar]
- 27.Porter, J., C. Edwards, and R. W. Pickup. 1995. Rapid assessment of physiological status in Escherichia coli using fluorescent probes. J. Appl. Microbiol. 79:399-408. [DOI] [PubMed] [Google Scholar]
- 28.Porter, J., D. Deere, R. Pickup, and C. Edwards. 1996. Fluorescent probes and flow cytometry: new insights into environmental bacteriology. Cytometry 23:91-96. [DOI] [PubMed] [Google Scholar]
- 29.Ritz, M., J. L. Tholozan, M. Fedeeighi, and M. F. Pilet. 2001. Morphological and physiological characterization of Listeria monocytogenes subjected to high hydrostatic pressure. Appl. Environ. Microbiol. 67:2240-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schmidt, G., and R. Zink. 2000. Basic features of the stress response in three species of bifidobacteria: B. longum, B. adolescentis, and B. breve. Int. J. Food Microbiol. 55:41-45. [DOI] [PubMed] [Google Scholar]
- 31.Shapiro, H. M. 2000. Microbial analysis at the single-cell level: tasks and techniques. J. Microbiol. Methods 42:3-16. [DOI] [PubMed] [Google Scholar]
- 32.Shapiro, H. M. 1995. Practical flow cytometry, 3rd ed. Alan R. Liss, Inc. New York, N.Y.
- 33.Shimamura, S., F. Abe, N. Ishibashi, H. Miyakawa, T. Yaeshima, T. Araya, and M. Tomita. 1992. Relationship between oxygen sensitivity and oxygen metabolism of bifidobacterium species. J. Dairy Sci. 75:3296-3306. [DOI] [PubMed] [Google Scholar]
- 34.Sincock, S. A., and J. P. Robinson. 2001. Flow cytometric analysis of microorganisms. Methods Cell Biol. 64:511-537. [DOI] [PubMed] [Google Scholar]
- 35.Suller, M. T. E., and D. Lloyd. 1999. Fluorescence monitoring of antibiotic-induced bacterial damage using flow cytometry. Cytometry 35:235-241. [DOI] [PubMed] [Google Scholar]
- 36.Tannock, G. W. 1999. Microecology of Lactobacilli and Bifidobacteria inhabiting the digestive tract: essential knowledge for successful probiotic research, p 17-31. In L. A. Hanson and R. H. Yolken (ed.), Probiotics, other nutritional factors, and intestinal microflora. Nestlé Nutrition Services, Lippincott-Raven, Philadelphia, Pa.
- 37.Wallner, G., B. Fuchs, S. Spring, W. Beisker, and R. Amann. 1997. Flow sorting of microorganisms for molecular analysis. Appl. Environ. Microbiol. 63:4223-4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward, D., R. Weller, and M. M. Bateson. 1990. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]