Abstract
The activities of two enzymes, a 168-kDa protein and a 40-kDa protein, OmtA, purified from the filamentous fungus Aspergillus parasiticus were reported to convert the aflatoxin pathway intermediate sterigmatocystin to O-methylsterigmatocystin in vitro. Our initial goal was to determine if OmtA is necessary and sufficient to catalyze this reaction in vivo and if this reaction is necessary for aflatoxin synthesis. We generated A. parasiticus omtA-null mutant LW1432 and a maltose binding protein-OmtA fusion protein expressed in Escherichia coli. Enzyme activity analysis of OmtA fusion protein in vitro confirmed the reported catalytic function of OmtA. Feeding studies conducted with LW1432 demonstrated a critical role for OmtA, and the reaction catalyzed by this enzyme in aflatoxin synthesis in vivo. Because of a close regulatory link between aflatoxin synthesis and asexual sporulation (conidiation), we hypothesized a spatial and temporal association between OmtA expression and conidiospore development. We developed a novel time-dependent colony fractionation protocol to analyze the accumulation and distribution of OmtA in fungal colonies grown on a solid medium that supports both toxin synthesis and conidiation. OmtA-specific polyclonal antibodies were purified by affinity chromatography using an LW1432 protein extract. OmtA was not detected in 24-h-old colonies but was detected in 48-h-old colonies using Western blot analysis; the protein accumulated in all fractions of a 72-h-old colony, including cells (0 to 24 h) in which little conidiophore development was observed. OmtA in older fractions of the colony (24 to 72 h) was partly degraded. Fluorescence-based immunohistochemical analysis conducted on thin sections of paraffin-embedded fungal cells from time-fractionated fungal colonies demonstrated that OmtA is evenly distributed among different cell types and is not concentrated in conidiophores. These data suggest that OmtA is present in newly formed fungal tissue and then is proteolytically cleaved as cells in that section of the colony age.
Aflatoxins, highly toxic and carcinogenic secondary metabolites produced by the filamentous fungi Aspergillus parasiticus, A. flavus, A. nomius, and A. tamarii (11, 26), frequently contaminate food and feed crops such as corn, cotton, peanuts, and tree nuts, resulting in health risks to animals and humans (11). Most genes involved in aflatoxin biosynthesis are clustered in a 75-kb genomic DNA region that carries a positive regulator, AflR, and structural genes, including omtA (34). Although much has been learned about the molecular biology and biochemistry of aflatoxin biosynthesis, little is known about the location of aflatoxin enzymes within a fungal colony or within a fungal cell.
Early studies identified two enzymes, a 168-kDa protein and a 40-kDa protein (OmtA), in A. parasiticus capable of converting sterigmatocystin to O-methylsterigmatocystin (6, 10) in vitro; about 60% of the enzyme activity in cell extracts was associated with the 168-kDa protein, and 40% of the activity was associated with OmtA (7). Both enzymes required the cofactor S-adenosylmethionine. The 168-kDa protein was purified to near homogeneity (6) and consisted of two subunits (58 and 110 kDa). OmtA (40 kDa) was also purified to homogeneity (17, 20). The gene encoding OmtA (omtA) was cloned from a cDNA expression library using OmtA-specific polyclonal antibodies (PAb) raised against OmtA protein expressed in Escherichia coli (20, 33); omtA was later shown to reside in the aflatoxin gene cluster (34), supporting its proposed role in aflatoxin synthesis. Because sterigmatocystin and O-methylsterigmatocystin could be converted to aflatoxin B1 in vivo (5), they were generally accepted as aflatoxin pathway intermediates. Two important issues remained unresolved. (i) Although the 168-kDa protein and OmtA converted sterigmatocystin to O-methylsterigmatocystin in vitro, it was not clear which of these activities was necessary and sufficient to catalyze this reaction in vivo (12, 23, 26, 30). (ii) Because the methyl group on O-methylsterigmatocystin is removed during the subsequent oxidoreduction reaction and because structurally related molecules are incorporated into AFB1 in feeding experiments with efficiency similar to or higher than that of sterigmatocystin (12), it was not clear if this reaction was necessary for aflatoxin synthesis. To address these issues, we generated an omtA gene disruption mutant (LW1432) and a plasmid construct to express a maltose binding protein (MBP)-OmtA fusion protein in E. coli. Analysis of the enzyme activity of the fusion protein confirmed the reported catalytic function of OmtA. Feeding studies conducted with LW1432 demonstrated a critical role for OmtA and the reaction catalyzed by this enzyme in aflatoxin synthesis in vivo.
Previous studies of accumulation of aflatoxin enzymes and expression of aflatoxin genes, including omtA, were conducted primarily in liquid medium using submerged shake culture (batch fermentation) (10, 29, 33); these conditions induce aflatoxin synthesis but do not support asexual sporulation (conidiation). Because a close regulatory association has been demonstrated between aflatoxin synthesis and conidiation (9, 13, 15), we hypothesized a spatial and temporal association between omtA expression and conidiospore development. Our goal was to develop a growth model that would closely mimic regulation of toxin synthesis in soil and on the host plant. We developed a novel time-dependent colony fractionation protocol to study OmtA accumulation in fungal colonies grown on solid medium; these conditions support toxin synthesis and conidiation. This protocol also allowed analysis of OmtA distribution to different cell types in fungal colonies. OmtA-specific PAb were generated against an OmtA fusion protein (MBP-OmtA) and purified by affinity chromatography using an LW1432 protein extract. OmtA was not detected in 24-h-old colonies but was detected in 48-h-old colonies by using Western blot analysis; the protein accumulated in all regions of a 72-h-old colony, including cells (0 to 24 h old, near the colony margin) in which little conidiophore development was observed. OmtA in older parts of the colony (24 to 72 h) was partly degraded. Fluorescence-based immunohistochemical analysis conducted on thin sections of paraffin-embedded fungal cells from time-fractionated fungal colonies demonstrated that OmtA is evenly distributed among different cell types and is not concentrated in conidiophores. These data suggest that OmtA accumulates in newly formed fungal tissue and then is proteolytically cleaved as cells in that section of the colony age. The data also suggest that OmtA is localized to specific areas within a fungal cell, but it is not yet clear if these areas correspond to specific subcellular organelles. The pattern of labeling using anti-OmtA was not consistent with localization of OmtA only to nuclei, peroxisomes, or Woronin bodies.
MATERIALS AND METHODS
Fungal strains.
A. parasiticus SU1(NRRL5862, ATCC 56775) is a wild-type, aflatoxin-producing strain. A. parasiticus CS10 (ver-1 wh-1 pyrG) was derived from A. parasiticus ATCC 36537 (ver-1 wh-1) (28). LW1418 and LW1432 (ver-1 wh-1 omtA) were generated in this study by disrupting the omtA gene in CS10. AFS10 is a non-aflatoxin-producing aflR knockout strain derived from A. parasiticus NR1 (niaD) derived from SU1 (provided by Jeff Cary, U.S. Department of Agriculture, New Orleans, La.).
Construction of omtA disruption vector pLW14 and OmtA expression vector pLW12.
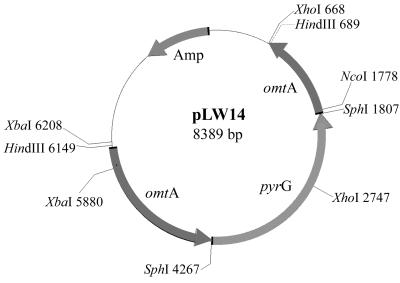
Plasmid pLW14 was constructed by inserting pyrG, which encodes orotidine monophosphate decarboxylase (28), into the coding region of omtA at the SphI site (Fig. 1). A plasmid carrying omtA genomic DNA was kindly provided by Fun Sun Chu (University of Wisconsin—Madison). The 2.5-kb pyrG fragment was generated by PCR using plasmid pPG3J (27) as the template. The primers used to amplify pyrG carried an SphI (GCATGC) restriction site (underlined) to facilitate cloning (5′-GTAGAAGTTCAGCATGCTGATGG-3′ and 5′-GAGTATCACAGTCAGGCATGCACGTC-3′). The reaction conditions for thermal cycling were 95°C for 5 min followed by 35 cycles at 95°C for 1 min, 62°C for 1 min, and 72°C for 3 min. The reaction was completed by incubation at 72°C for 10 min.
FIG. 1.
Restriction endonuclease map of plasmid pLW14.
An omtA cDNA was generated by reverse transcriptase PCR (RT-PCR). Template RNA was isolated from A. parasiticus strain SU1 cultured in YES medium (2% yeast extract, 6% sucrose, pH 5.5) for 48 to 72 h by using Trizol reagent and a procedure supplied by the manufacturer (GibcoBRL, Rockville, Md.). For first-strand cDNA synthesis, 48 μg of total RNA was incubated in the RT-PCR mix at 37°C for 2 h. All chemicals used in the RT-PCR were purchased from GibcoBRL. The 20-μl reaction mixture contained 4 μl of 5× first-strand buffer, 2 μl of 0.1 M dithiothreitol, 1 μl of 10 mM deoxynucleoside triphosphate, 2 μl of Moloney murine leukemia virus RT (200 U per μl), and 1 μl of oligo(dT) primer (0.5 μg per μl). One primer for omtA amplification contained a HindIII (AAGCTT) restriction site (underlined), and the other primer contained an XbaI (TCTAGA) restriction site (underlined) to facilitate cloning (5′-CCCTCTAGAATGGCACTACCGAGCAAAG-3′ and 5′-TGCAAGCTTCTACTTGCGCAAACGCAGT-3′). One microliter of the resulting cDNA mixture was used as a template for the PCR. The cycling conditions were as follows: 95°C for 5 min followed by 35 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 2 min. The mixture was incubated at 72°C for 10 min to complete the reaction. The omtA PCR fragment (1,260 bp) was digested with restriction enzymes XbaI and HindIII and cloned into plasmid pMAL-c2 (New England Biolabs, Beverly, Mass.) digested with the same enzymes. The resulting plasmid construct, pLW12, was transformed into E. coli DH5α. The proper construction of pLW12 in clones expressing MBP-OmtA was confirmed by restriction enzyme analysis of purified plasmid DNA isolated by the Qiagen (Valencia, Calif.) miniprep plasmid kit. The size of the fusion protein was determined by small-scale expression studies. E coli DH5α carrying pLW12 was incubated in 5 ml of Luria-Bertani broth containing ampicillin (100 μg per ml) for 16 h. One milliliter of bacterial culture was saved as noninduced control. The remaining 4 ml of culture was induced to express fusion protein by the addition of 0.3 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h.
Transformation and screening for omtA gene-disrupted strains.
Circular or linear plasmid (8 μg) digested with HindIII was used in transformation experiments using A. parasiticus CS10 as the recipient strain (28). Protoplasts were generated by digestion of mycelium (harvested 17 h after initiation of germination) with Novozyme 234, and transformation was conducted by a polyethylene glycol method as described previously (28). The selection of omtA-disrupted strains was achieved in four steps. (i) pyrG-positive clones (pyrG+; uridine prototrophs) were selected by growth on Czapek Dox agar (CZ agar) (Difco Laboratories, Detroit, Mich.), a minimal growth medium (28). (ii) Spores from 3- to 4-day-old transformant colonies grown on CZ agar plates were transferred by sterile toothpick to coconut agar medium (2) supplemented with sterigmatocystin at a concentration of 20 μg/ml and also to CZ agar medium. CS10 protoplasts regenerated and tolerated sterigmatocystin supplementation of the agar during active growth and then utilized sterigmatocystin to synthesize aflatoxin B1 as active growth slowed or ceased. (iii) Colonies that did not fluoresce blue under long-wavelength UV light (254 nm) after a 3-day incubation at 29°C in the dark on coconut agar medium were selected, and secondary metabolites were extracted and analyzed by thin-layer chromatography (TLC) by a previously published procedure (31). (iv) The genotype of suspected omtA-disrupted strains was confirmed by Southern hybridization analysis. OmtA-null mutants were further analyzed for ability to convert sterigmatocystin or O-methylsterigmatocystin to aflatoxin B1 by a feeding experiment, as described below.
Southern hybridization analysis.
Conidiospores (5 × 106) isolated from transformants were inoculated into 100 ml of YES medium and incubated with shaking at 150 rpm for 48 to 72 h at 29°C in the dark. One gram of freshly harvested mycelium was used for isolation of genomic DNA using a published procedure (28). Southern hybridization analysis was performed using a nonradioactive hybridization kit (Roche Molecular Biochemicals, Indianapolis, Ind.) and a procedure provided by the manufacturer. To identify the presence of pyrG sequences integrated within the chromosomal omtA gene, genomic DNA was digested with HindIII and EcoRI, separated by electrophoresis on a 1% agarose gel, transferred to Nytran membrane, and probed with digoxigenin-labeled 2.9-kb HindIII fragment contained in the omtA gene.
Feeding studies of omtA gene disruption strains.
One gram of mycelium from the same culture that was used in Southern hybridization analysis was inoculated into 10 ml of YES medium supplemented with either sterigmatocystin or O-methylsterigmatocystin at a concentration of 20 μg per ml and incubated at 29°C for 24 h in the dark with shaking at 150 rpm. The fungal mycelium and culture medium were extracted with 10 ml of chloroform at 4°C for 16 h. The chloroform fraction was evaporated under N2 gas and resuspended in 200 μl of acetone. Ten microliters of sample was analyzed by TLC using ether-methanol-water (96:3:1) as the solvent system.
Conversion of sterigmatocystin to O-methylsterigmatocystin by MBP-OmtA.
The pMAL protein fusion and purification system from New England Biolabs was utilized to express and purify MBP-OmtA. Large-scale preparation of MBP-OmtA was conducted for the study of enzymatic activity and antigen purification. E. coli DH5α carrying pLW12 was grown in 500 ml of rich medium (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl, 2% glucose) containing ampicillin (100 μg per ml). Fusion protein synthesis was induced by addition of IPTG (0.3 mM), cells were sonicated (Sonifier cell disrupter W-350; Fisher Scientific, Pittsburgh, Pa.), and the fusion protein was purified by amylose affinity column chromatography according to a protocol supplied by the manufacturer (New England Biolabs).
Amylose resin-purified MBP and MBP-OmtA were used for analysis of enzyme activity. In a time course experiment, three 1-ml reaction mixtures were prepared. Each reaction mixture contained 100 μg of MBP-OmtA, 20 μg of sterigmatocystin, and 600 μg of S-adenosylmethionine. Sterigmatocystin, O-methylsterigmatocystin, and S-adenosylmethionine were purchased from Sigma (St. Louis, Mo.). The reaction mixtures were incubated at room temperature for appropriate times (20-min time points for 60 min; one tube per time point). Three control reactions were also incubated for 60 min. Each contained the same reagents as above except that the MBP-OmtA, sterigmatocystin, or S-adenosylmethionine was omitted. Reactions were stopped by extraction with 4 volumes of chloroform. Chloroform extracts were dried under nitrogen gas and resuspended in 100 μl of acetone. Five microliters of extract was analyzed by TLC using chloroform-acetone (95:5) as the development system (31).
OmtA antibody production and purification.
OmtA PAb were generated in rabbits, with purified MBP-OmtA as the antigen. Each of two rabbits (New Zealand White) was injected subcutaneously with 300 μg of protein in TitreMax adjuvant (CytRx, Norcross, Ga.) at a 1:1 (vol/vol) ratio. After 35 days, the animals received one booster injection with 200 μg of protein in TitreMax adjuvant. Serum was obtained 4 weeks after the boost. The immunoglobulin G (IgG) fraction was purified from rabbit serum by precipitation with ammonium sulfate using standard procedures (3).
The IgG fraction of OmtA antibodies was further purified by affinity chromatography using a protein extract prepared from LW1432 grown in YES medium for 48 h. This column was prepared by conjugating 11 mg of total protein to 2 ml of Aminolink coupling gel (coupling efficiency, approximately 80%) using a procedure supplied by the manufacturer (Pierce Chemical Company, Rockford, Ill.). For antibody purification, 1 ml of IgG (8 mg) was loaded onto the gel bed in this column and an additional 2 ml of phosphate-buffered saline was loaded to cover the gel bed. The column with antibodies was incubated at room temperature for 1.5 h at room temperature. After incubation, phosphate-buffered saline was allowed to flow through, and antibody-containing fractions were identified by absorbance at 280 nm. The protein concentration was determined by a Bio-Rad (Hercules, Calif.) protein assay using dye reagent and bovine serum albumin (BSA) (fraction V; Sigma) as a standard. This highly purified antibody preparation (3 mg per ml) was used in Western blot analysis and immunofluorescence microscopy.
Western blot analysis.
To determine specificity of anti-OmtA antibodies, fungal strains SU1, CS10, and LW1418 were grown in the dark at 29°C (shaking at 150 rpm) in 100 ml of YES liquid medium for 24, 48, and 72 h, and proteins were extracted from mycelia for Western blot analysis. The mycelia were harvested and pulverized under liquid nitrogen, and the fungal proteins were extracted in TSA buffer (2 mM Tris-Cl, pH 8.0; 40 mM NaCl; 0.025% sodium azide) containing complete protease inhibitors (Roche Molecular Biochemicals). Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) using standard methods (3). For Western blot analysis, each lane on the SDS-12% polyacrylamide gel contained 30 μg of protein. The primary antibody was column-purified anti-OmtA IgG (1 μg per ml), and the secondary antibody consisted of a 10,000-fold dilution of goat anti-rabbit IgG alkaline phosphate conjugate (Schleicher & Schuell, Keene, N.H.). A BCIP (5-bromo-4-chloro-3-indolylphosphate)-nitroblue tetrazolium colorimetric detection system was utilized (Roche Molecular Biochemicals).
Time-dependent fractionation of colonies grown on solid medium.
To determine the accumulation and distribution of OmtA in fungal colonies grown on solid culture medium, conidiospores (2 × 105) of A. parasiticus SU1, AFS10, CS10, and LW1432 were inoculated onto the center of YES agar or potato dextrose agar (for SU1 only) overlaid with sterile cellophane membranes and incubated at 29°C in the dark. Some colonies were analyzed after 24 or 48 h of growth. Seventy-two-hour-old colonies were fractionated into three concentric rings based on area covered at three time points (72, 48, and 24 h). For example (see Fig. 5D), a SU1 colony with a diameter of 4.2 cm was fractionated to S1, which contained mycelia from the colony center out to a distance of 0.8 cm (ages, 48 to 72 h); S2, which contained mycelia from 0.8 to 2.5 cm (24 to 48 h); and S3, which contained mycelia from 2.5 to 4.2 cm (0 to 24 h). The harvested mycelia from appropriate sections of the colony were pulverized under liquid nitrogen, and the fungal proteins were extracted in TSA buffer containing complete protease inhibitors. Western blot analysis was performed as described above.
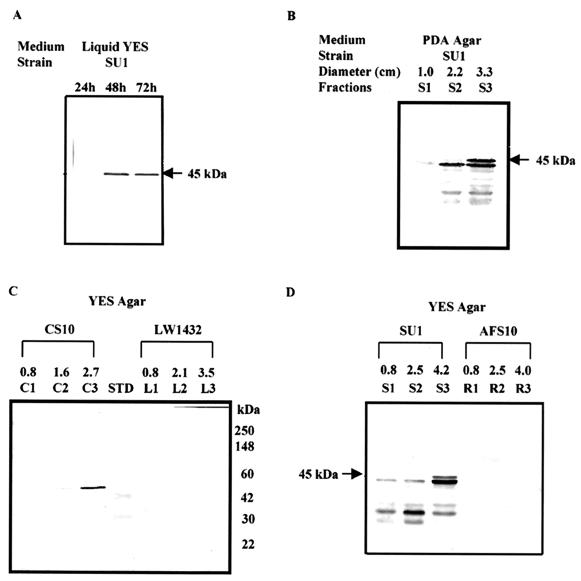
FIG. 5.
Western blot analysis of fungal protein extracts using affinity-purified OmtA PAb. (A) Analysis of SU1 crude protein extracts isolated at three time points (24, 48, and 72 h) from fungal cultures grown in YES liquid medium. The approximate mass of the primary signal (45 kDa) is shown to the right of the blots in panels A and B and to the left of the blot in panels C and D. (B) Analysis of protein extracts from time-fractionated colonies of SU1 grown on PDA medium. Seventy-two-hour-old colonies were fractionated into three concentric rings based on area covered at three time points: S1, 48 to 72 h; S2, 24 to 48 h; S3, 0 to 24 h. (C) Analysis of protein extracts from time-fractionated colonies of CS10 and LW1432 grown on YES agar medium. Lane STD contains molecular mass standards; the masses of standards are shown to the right of the panel. (D) Analysis of protein extracts from time-fractionated colonies of SU1 and AFS10 grown on YES agar medium.
Immunolocalization of OmtA protein.
Immunolocalization of OmtA protein was conducted on SU1 colony fractions. In addition, AFS10, CS10, and LW1432 colonies were fractionated following the same scheme to generate analogous fractions R1, R2, and R3; C1, C2, and C3; and L1, L2, and L3, respectively.
Preparation of paraffin-embedded fungal sections.
Samples from fungal colony fractions were embedded in Paraplast (Sigma) using a published procedure (3) with the following modifications. Fungal tissues were fixed with Streck tissue fixative (Streck Laboratory Inc., Omaha, Nebr.) at 4°C overnight and then dehydrated in a graded series of ethanol: 30% (30 min, room temperature); 50% (30 min, room temperature); 70% (overnight, 4°C); 85% (30 min, room temperature, two times); 95% (30 min, room temperature, two times); 100% (30 min, 4°C, two times), followed by incubation in 100% xylene (10 min, room temperature, three times). Fungal tissues were then incubated in a paraffin-xylene mixture (1:1; vol/vol) for 15 min, 60 min, and overnight at 60°C and finally for 8 h in 100% paraffin at 60°C (three changes of paraffin during incubation). The paraffin-embedded sample blocks were hardened in a plastic mold (VWR Scientific, Detroit, Mich.) at room temperature. Sample blocks were cut into 4-μm- thick sections using a tissue section microtome (AO Spencer 820 microtome; Fisher Scientific). The sections were attached to poly-l-lysine (Sigma)-coated coverslips (22 mm square).
Immunolabeling.
Coverslips with paraffin-embedded fungal sections were placed in a coverslip holder (EMS, Fort Washington, Pa.) for deparaffinization and antigen retrieval. Sections were deparaffinized twice in 100% xylene for 10 min and then rehydrated with a decreasing concentration of ethanol: twice in 100% ethanol for 10 min; once each in 95, 70, and 50% ethanol for 5 min each; and finally in deionized distilled H2O. Antigen retrieval was performed by heating the sections in 10 mM citrate buffer (pH 6.0) at 95°C for 5 min followed by cooling at room temperature for 20 min. Coverslips were rinsed with Tris-buffered saline (TBS) (pH 7.5) and incubated in blocking solution (1% BSA with 0.1% saponin in TBS) at 4°C overnight. The samples were immunolabeled with primary antibody (purified anti-OmtA IgG; 20 μg/ml) or anti-SKL (1:500; Zymed, South San Francisco, Calif.) at room temperature for 1.5 h, followed by secondary antibody conjugated with fluorescent probe (goat anti-rabbit IgG-Alexa 488 conjugate [5 μg/ml]; Molecular Probes, Eugene, Oreg.) at room temperature for 1 h. Coverslips were washed with TBS containing 1% BSA and 0.1% saponin after each antibody treatment followed by two washes with TBS for 10 min. Fungal nuclei were detected using SYTOX Green fluorescence dye (Molecular Probes). The samples were mounted onto microscopic slides with Prolong antifade mounting medium (Molecular Probes).
Confocal laser scanning microscopy (CLSM).
Fluorescence image detection was performed on a Zeiss 210 laser scanning microscope with a 488-nm laser line. The 40× oil objective lens (numerical aperture = 1.3; Zeiss Plan-NeoFlura) was used to acquire all images. The Alexa 488 fluorescence probe (absorbance wavelength, 495 nm; emission wavelength, 519 nm) was detected using LP 520 or BP 520-560 barrier filters. Fluorescence image analyses of SU1, AFS10, CS10, and LW1432 were conducted under the same instrument parameter settings.
RESULTS
Generation of omtA disruption mutants.
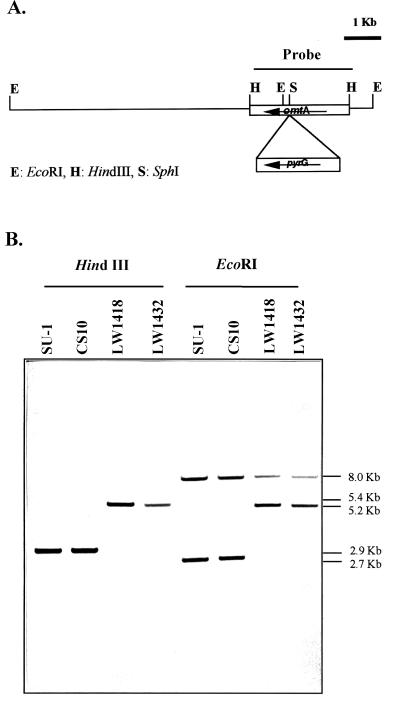
TLC analysis of cell extracts from pyrG+ transformants tentatively identified five omtA gene disruption isolates. Two isolates (LW1418 and LW1432) were identified from among 41 pyrG+ colonies transformed with circular plasmid LW14. Southern hybridization analysis of genomic DNAs isolated from LW1418 and LW1432 detected a 2.5-kb pyrG DNA fragment within the omtA gene, confirming the gene disruption event in these isolates (Fig. 2).
FIG. 2.
Southern hybridization analysis of omtA gene disruption strains. (A) Restriction endonuclease map of omtA locus in omtA disruption strains LW1418 and LW1432. (B) Southern hybridization analysis. Genomic DNAs isolated from LW1418 and LW1432, CS10 (parent strain) and SU1 (wild-type), were digested with restriction enzymes HindIII and EcoRI. DNAs were resolved and hybridized to a digoxigenin-labeled 2.9-kb HindIII omtA DNA fragment using standard methods. Strains with omtA disruption were predicted to contain a 5.4-kb HindIII fragment consisting of the 2.5-kb pyrG selectable marker inserted into the omtA locus. A 5.2-kb fragment in the omtA-disrupted strains was also predicted to replace the wild-type EcoRI fragment (2.7 kb). Numbers to the right of the blot represent the approximate size of the detected fragments in kilobase pairs.
In vivo feeding experiments.
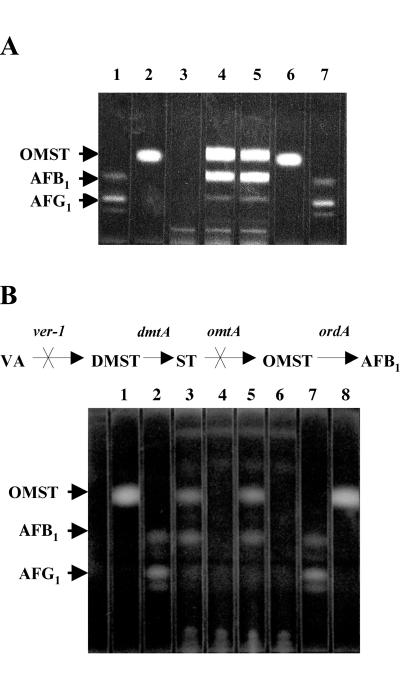
No O-methylsterigmatocystin or aflatoxin B1 could be detected in LW1418 or LW1432 supplied with (fed) exogenous sterigmatocystin; in contrast, each isolate converted exogenously supplied O-methylsterigmatocystin to aflatoxin B1 (Fig. 3B). The parental strain CS10 converted either sterigmatocystin or O-methylsterigmatocystin to aflatoxin B1 (Fig. 3A). These data suggest that LW1418 and LW1432 carry mutant copies of omtA but the genes encoding later pathway enzymes and the regulatory genes involved in aflatoxin biosynthesis are still functional.
FIG. 3.
TLC of extracts of CS10 and omtA-disrupted strains LW1418 and LW1432. (A) TLC of extracts of CS10 supplied with (fed) exogenous O-methylsterigmatocystin (OMST). Lanes 1 and 7, aflatoxin B1 (AFB1), AFB2, AFG1, and AFG2 standard mixture; lanes 2 and 6, OMST standard; lane 3, CS10 fed no pathway intermediate; lane 4, CS10 fed sterigmatocystin; lane 5, CS10 fed O-methylsterigmatocystin. Ten-microliter samples were analyzed using chloroform-acetone (95:5) as the solvent system. (B) TLC of extracts of omtA-disrupted strains LW1418 and LW1432 supplied with (fed) exogenous sterigmatocystin (ST) or OMST. Metabolic scheme for aflatoxin biosynthesis in strain LW1418 and LW1432 is shown at the top of the thin-layer chromatograph. The ver-1 and omtA genes involved in aflatoxin biosynthesis are nonfunctional in LW1418 and 1432. Lanes 1 and 8, OMST standard; lanes 2 and 7, AFB1, AFB2, AFG1, and AFG2 standard mixture; lane 3, LW1418 fed OMST; lane 4, LW1418 fed ST; lane 5, LW1432 fed OMST; lane 6, LW1432 fed ST. Ten-microliter samples were analyzed using ether-methanol-water (96:3:1) as the solvent system. Abbreviations: VA, versicolorin A; DMST, dimethylsterigmatocystin.
Purification of MBP-OmtA.
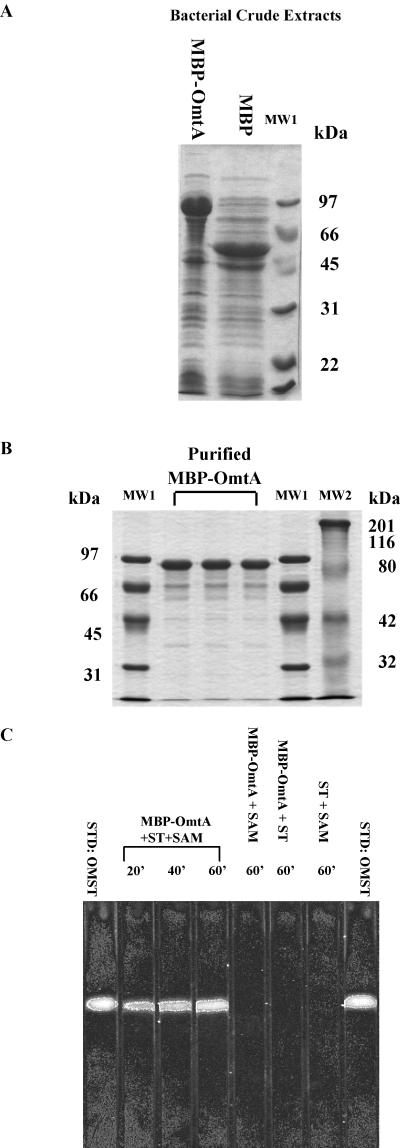
MBP-OmtA in DH5α cell extracts (Fig. 4A) was purified by amylose column chromatography. Most of the purified MBP-OmtA fusion protein was approximately 88 kDa in mass (Fig. 4B); this is consistent with a mass calculated from the fusion of 42-kDa MBP and 45-kDa OmtA predicted using nucleotide sequence data. Very little proteolytic cleavage of the protein was detected by this analysis. The fusion protein remained soluble in the bacterial cytoplasm, allowing purification of 39 mg of MBP-OmtA per liter of bacterial culture.
FIG. 4.
Conversion of sterigmatocystin (ST) to O-methylsterigmatocystin (OMST) by affinity-purified MBP-OmtA fusion protein. (A) SDS-PAGE of IPTG-induced bacterial crude extracts containing MBP-OmtA or MBP. (B) SDS-PAGE of affinity-purified MBP-OmtA. Lanes MW1 and MW2 represent molecular mass standards; molecular mass is indicated to the left and right of the gel, respectively. (C) TLC of the conversion of ST to OMST by affinity-purified MBP-OmtA and control reactions. Five microliters of reaction mixture was analyzed after appropriate time periods by TLC using chloroform-acetone (95:5) as the developing system. Control reactions included all reagents except ST, SAM, or MBP-OmtA, respectively. OMST standards are located in lanes on both ends of the TLC (STD:OMST).
Enzymatic conversion of sterigmatocystin to O-methylsterigmatocystin by MBP-OmtA.
Purified OmtA fusion protein efficiently converted sterigmatocystin to O-methylsterigmatocystin in the presence of S-adenosylmethionine (Fig. 4C) within 60 min. The cofactor S-adenosylmethionine was required for this conversion. Without exogenous S-adenosylmethionine, no O-methylsterigmatocystin could be detected in the presence of the MBP-OmtA. As expected, no O-methylsterigmatocystin was detected in the reaction that contained S-adenosylmethionine without substrate (sterigmatocystin), in a reaction without added MBP-OmtA, or in a reaction with MBP but without the MBP-OmtA (data not shown).
Production of anti-MBP-OmtA PAb.
To obtain highly specific anti-OmtA PAb, amylose-purified MBP-OmtA was used as the antigen to produce PAb in two rabbits. The IgG fraction of the rabbit serum was further purified by affinity chromatography using a protein extract from LW1432 (omtA gene disruption strain). The specificity of anti-OmtA PAb was tested on SU1 (wild type) cultured in YES liquid medium for 24, 48, or 72 h. No OmtA could be detected in Western blot analysis of protein extracts prepared from SU1 grown on YES liquid medium for 24 h while one primary band (approximately 45 kDa) could be detected in SU1 grown for 48 or 72 h in the same medium (Fig. 5A).
Western blot analysis of OmtA in colonies grown on solid medium.
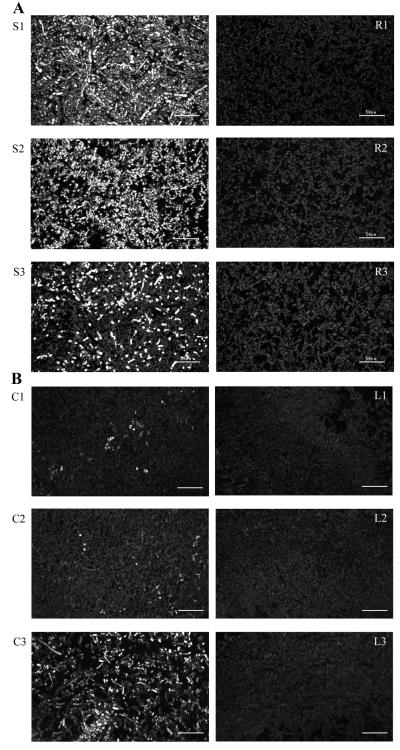
The accumulation and distribution of OmtA was analyzed in time-fractionated colonies grown on solid YES or PDA medium for 72 h (Fig. 5B to D). OmtA protein was detected in fractions S1, S2, and S3 of a 72-h-old SU1 colony but not in corresponding colony fractions isolated from AFS10 (aflR knockout) (Fig. 5D) or LW1432 (omtA knockout) (Fig. 5C). OmtA in colony fractions S1 (48 to 72 h) and S2 (24 to 48 h) showed increased levels of smaller peptides and correspondingly less full-length protein. In contrast, in fraction S3 (0 to 24 h) of a 72-h-old colony, full-length OmtA was found at higher levels. A similar result was observed using SU1 grown on PDA medium and CS10 (parent strain of LW1432) grown on YES agar medium; very little OmtA protein could be detected in fractions S1 and C1 (48 to 72 h), while more full-length protein was detected in the youngest colony fraction (S3 and C3; 0 to 24 h) (Fig. 5C). No OmtA protein could be detected in any colony fraction of either LW1432 or AFS10.
CLSM.
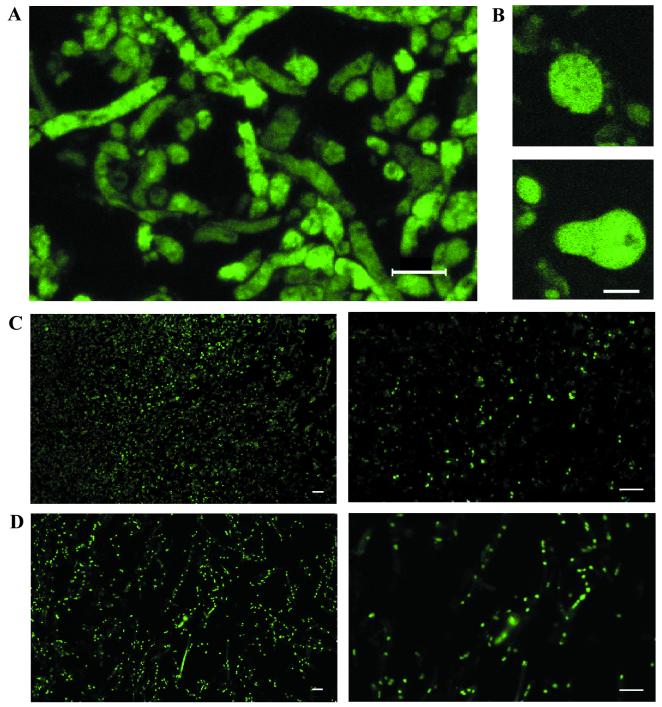
Immunofluorescence microscopy was conducted on samples prepared from fractionated colonies grown on YES solid medium. To compare the fluorescence labeling intensities of SU1 and CS10 and their non-aflatoxin-producing counterparts, AFS10 and LW1432, respectively, the samples were viewed under the lowest zoom level during the CLSM (zoom = 20) in order to acquire maximum cell number within a field. Under the same contrast and brightness settings, the samples prepared from the control strain AFS10 (fractions R1, R2, and R3) and LW1432 (fractions L1, L2, and L3) did not show significant fluorescent signals compared to samples prepared from the same colony fractions from wild-type SU1 (fractions S1, S2, and S3) or CS10 (fractions C1, C2, and C3) (Fig. 6). These data are consistent with Western blot analysis of protein extracts isolated from the same colony fractions (Fig. 5C and D) (i.e., OmtA was only detected in SU1 and CS10 but not in AFS10 or LW1432). OmtA was observed in the substrate-level mycelium throughout a 72-h-old colony grown on YES medium; the protein was also detected in the conidiospore-bearing structures (particularly in vesicles [Fig. 7B ]) located in fractions S1 and S2 at nearly equal intensity as in substrate level mycelium. The fluorescent signal was confined to discrete areas (patches) within cells (Fig. 7A). However, due to limitations in resolution of CLSM, it was not clear if these areas are associated with particular organelles. Similar samples were also probed with anti-SKL antibodies that are expected to detect proteins targeted to peroxisomes and Woronin bodies (Fig. 7C) (16, 32). The observed fluorescent pattern was consistent with localization to these organelles. In addition, we used SYTOX Green to stain nuclei in fungal fractions (Fig. 7D); again the fluorescent pattern was consistent with the expected results. Neither anti-SKL nor SYTOX Green generated the same patchy pattern as anti-OmtA. In summary, the data suggest that OmtA protein is evenly distributed in all cell types in a fungal colony and does not accumulate to the highest levels in conidiophores. The fluorescent signal is not consistent with localization of OmtA to only peroxisomes, Woronin bodies, or nuclei. However, the data do not rule out the possibility that OmtA is localized to other cell locations as well as these specific organelles.
FIG. 6.
OmtA protein localization in time-fractionated colonies of A. parasiticus SU1, AFS10 (afR knockout), CS10, and LW1432 (omtA knockout) grown on YES agar for 72 h. Paraffin-embedded fungal sections were immunolabeled with affinity-purified OmtA PAb (20 μg/ml) followed by Alexa 488-conjugated goat anti-rabbit IgG. (A) Fluorescence images of SU1 (S1, S2, and S3) and AFS10 (R1, R2, and R3). (B) Fluorescence images of CS10 (C1, C2, and C3) and LW1432 (L1, L2, and L3). All colony fractions were analyzed under the same instrument settings. Bar = 50 μm.
FIG. 7.
Protein localization using OmtA PAb and anti-SKL and detection of nuclei using SYTOX Green. (A) Z-series overlay image of SU1 (fraction S2) immunolabeled with OmtA PAb. The image consists of an overlay of 10 consecutive optical sections taken at a Z interval of 800 nm (step size). (B) Fluorescence images of vesicles of conidiophores of SU1 (fraction S1) immunolabeled with OmtA PAb. (C) Immunolabeling of SU1 (fraction S2) with anti-SKL. The magnification for the left panel is ×400 and that for the right panel is ×2,000. (D) Fluorescence image of nuclei in SU1 (fraction S2) stained with SYTOX Green. The magnification for the left panel is ×400 and that for the right panel is ×2,000. The bar in each panel represents 10 μm.
DISCUSSION
The initial goal of this study was to determine if OmtA is necessary and sufficient to convert sterigmatocystin to O-methylsterigmatocystin in vivo and if this reaction is necessary for aflatoxin synthesis. When the same concentration of either sterigmatocystin or O-methylsterigmatocystin was fed to the parent strain CS10 (wild-type OmtA and OrdA activities), nearly equal concentrations of O-methylsterigmatocystin and aflatoxin B1 were generated. This observation could be due to one of at least three potential reasons. (i) The intermediates were fed in excess of the capacity of the pathway enzyme to carry out complete conversion in the 24-h period allotted. (ii) The entry of the intermediates into the cell or into the enzyme active sites was rate-limiting. (iii) The intermediates stimulate a feedback repression system downregulating activities of one or both enzymes. Nevertheless, it was clear that CS10 could convert either sterigmatocystin or O-methylsterigmatocystin to aflatoxin B1, while LW1432 and LW1418 could convert O-methylsterigmatocystin to aflatoxin B1 but were unable to convert sterigmatocystin to either O-methylsterigmatocystin or aflatoxin B1. These data clearly demonstrate that OmtA is necessary for efficient conversion of sterigmatocystin to O-methylsterigmatocystin in vivo in A. parasiticus CS10 under the specified growth and assay conditions. The data also demonstrate that this reaction is necessary for aflatoxin biosynthesis. It is not yet clear if OmtA alone is sufficient to carry out this reaction in vivo. We cannot rule out the possibility that the 168-kDa protein contributes to this reaction, but its contribution appears negligible compared to that of OmtA. In support of this idea, LW1432 (omtA disruption) could not convert exogenously supplied sterigmatocystin to detectable levels of either O-methylsterigmatocystin or aflatoxin B1 but could convert O-methylsterigmatocystin to aflatoxin B1 in vivo. In contrast, CS10 (parent of LW1432) converted either sterigmatocystin or O-methylsterigmatocystin to aflatoxin B1. These data demonstrate that the omtA disruption is gene specific and does not affect the activity of late pathway enzymes or regulators of aflatoxin synthesis.
Because a close regulatory association between aflatoxin synthesis and conidiation has been demonstrated in several studies (13, 15), we hypothesized a close temporal and spatial association between OmtA expression and conidiospore development. The second part of this study was designed to address this hypothesis. First, it was necessary to develop highly specific OmtA antibodies and a method for analysis of protein accumulation and distribution in cells and fungal colonies grown on solid growth medium.
We initially experienced specificity problems with PAb (20) raised against native OmtA that resulted in severe cross-reactivity in Western blot analysis and artifacts in protein localization. Because aflatoxin enzymes are present at low concentration in the fungus (17), purification of OmtA from the fungus likely resulted in copurification of at least trace amounts of other proteins that, although undetectable by SDS-PAGE, still possessed strong antigenicity.
To increase specificity, we generated PAb against affinity-purified MBP-OmtA and purified them by affinity chromatography with a column carrying fungal proteins isolated from LW1432 (omtA knockout). This procedure helped eliminate cross-reactive antibodies. Specificity was demonstrated via Western blot analysis. OmtA could not be detected in either AFS10 or LW1432, strains that do not synthesize this protein. OmtA PAb did not detect OmtA after 24 h in YES liquid or solid medium but did detect a protein of appropriate size (45 kDa) at 48 and 72 h. This pattern of accumulation is similar to that observed for other aflatoxin proteins, including Nor-1 (35), Ver-1 (18, 19), and versicolorin B synthase (unpublished data), suggesting that accumulation of these proteins is coordinately regulated. In addition, analysis of fungal extracts from SU1, CS10, AFS10, and LW1432 did not show significant cross-reaction of OmtA PAb to other cellular proteins, including DmtA; sequence identity between OmtA and DmtA was reported previously (24).
Time-dependent colony fractionation provided a practical method for monitoring protein accumulation and distribution in fungal tissues of different ages; similar information could not be obtained in liquid shake culture (batch fermentation). Knowledge of OmtA distribution was essential to identify fungal tissues that were rich in the target protein and allowed successful immunohistochemistry in this study and immunoelectron microscopy in a study now under way. Since these protocols require very small quantities of fungal tissue (1 mm3 for immunoelectron microscopy, for example) one could easily miss the relevant protein if it was not uniformly distributed in a colony.
Using this colony fractionation protocol, OmtA PAb, and Western blot analysis, we observed OmtA in all fractions of a 72-h-old colony (fractions S1, S2, and S3) grown on YES agar. The youngest colony fraction (S3; 0 to 24 h) contained mostly full-length OmtA, while fraction S1 (48 to 72 h) contained less full-length OmtA and more OmtA-derived peptides. In fraction C1 (CS10; 48 to 72 h) and the oldest fraction of SU1 (48 to 72 h) grown on PDA medium, OmtA was almost undetectable. Similarly, OmtA was not observed in the oldest fraction of a 90-h-old colony grown on YES medium. No OmtA protein was detected in 24-h-old colonies grown on YES medium; OmtA was detected in 48-h-old colonies on the same medium. Together the data suggest that OmtA biosynthesis in A. parasiticus SU1 grown on solid YES medium initiates after 24 h. OmtA then accumulates to relatively high levels in the youngest fraction, for example, in C3 and S3 (0 to 24 h) of a 72-h-old colony. As cells in that fraction age (at 48 or 72 h), OmtA synthesis appears to decline, and the protein is proteolytically cleaved and disappears completely by 90 h. The rate of OmtA proteolysis appears to be age and growth medium dependent. OmtA proteolysis was not observed to the same extent in cells grown in liquid culture.
OmtA proteolysis may occur after accidental contact with fungal proteases released during mechanical disruption of fungal tissues. However, because the tissue is quickly frozen before disruption and because protease inhibitors are added to the extraction buffer, we hypothesize that OmtA proteolysis occurs as part of a natural process during fungal growth and development. To generate developmental structures in solid culture, fungi may require proteases to digest unneeded structures or metabolic proteins (27). For example, a mutation in one protease, subtilisin-related serine proteinase (ALP2), resulted in smaller conidiophore vesicles (50% reduction) and a lower number of conidia (80% reduction) in A. nidulans (27). We observed that SU1 fungal colonies on PDA, in which colonies grew more slowly than on YES medium (smaller diameter), also had more severe OmtA proteolysis. Conidiophore structures also could be found in fraction C3 on YES medium, whereas they were absent in S3 on YES medium.
Septal pores form channels between adjacent cells in the mycelium and appear to play an important role in cell-to-cell communication in filamentous fungi (4). Cytoplasm, mitochondria, and nuclei migrate throughout the mycelium via septal pores (4, 21). Because we do not see accumulation of OmtA in 24-h-old colonies on YES medium but do see accumulation of OmtA in cells in the youngest fraction S3 (0 to 24 h) of a 72-h old fungal colony, we hypothesize that migration of OmtA together with cytoplasm and organelles occurs from older cells (fraction S2, 24 to 48 h) to younger cells (fraction S3, 0 to 24 h) on solid medium. Alternatively, it is also possible that a specific regulatory factor(s) involved in aflatoxin biosynthesis (e.g., AflR) moves from the aflatoxin-producing cells in fraction S2 to fraction S3, inducing omtA expression in these younger tissues.
OmtA-specific PAb were also used in immunofluorescence microscopy. The data provided strong evidence that our purification strategy resulted in PAb that were sensitive and specific. Although the paraffin embedment procedure and sectioning technique have been used for microscopic analysis of a variety of organisms (8), this is the first reported use in the study of a fungus grown on solid medium and the first application in a study of A. parasiticus. We previously utilized a fungal cell preparation technique for immunolabeling that required digestion of the cell wall using Novozyme (14). However, in these early localization studies, variation in cell wall digestion resulted in significant artifacts. The paraffin embedment and sectioning procedures developed and described in this study successfully preserved fungal mycelium, developmental structures, organelle structure, and protein antigenicity; they also eliminated the need for cell wall digestion, in turn generating consistent and reproducible immunolabeling results. This technique provides a useful tool to localize proteins and possibly other compounds in fungal cells and colonies.
Despite the close regulatory relationship between sporulation and aflatoxin production, our data suggest that OmtA is not produced exclusively in conidiophores as hypothesized. On the contrary, CLSM micrographs showed that OmtA is distributed throughout the colony and in both conidiophores and vegetative hyphae. Western blot analysis indicated that an abundance of full-length OmtA was observed in fraction S3, even though conidiophores were nearly absent in this area. In fraction S2, which showed the highest intensity of OmtA detected by CLSM, OmtA accumulated to similar levels in vegetative hyphae and conidiophores.
Cells immunolabeled with OmtA PAb and analyzed by CLSM showed patches of fluorescence within fungal cells, suggesting that OmtA is confined to subcellular compartments. An alternative explanation is that OmtA is present in cytoplasm and is thus excluded from cell organelles. To gain more information about subcellular localization, we labeled paraffin- embedded sections to identify specific organelles; for example, anti-SKL antibodies (16) were used to label peroxisomes and SYTOX Green was used to detect nuclei. Both probes generated small regularly shaped signals consistent with the expected organelles, indicating that the double-membraned nuclei as well as single-membraned microbody organelles were well-preserved. These observations strongly suggest that we have minimized artifacts due to poor sample preparation. Images using both probes were different from those obtained with OmtA PAb. The anti-SKL-labeled organelles were recently demonstrated to be Woronin bodies by immunoelectron microscopy (data not shown).
There are some indirect data available that suggest that at least some aflatoxin proteins are localized in organelles. For example, in A. parasiticus, the aflatoxin enzyme OrdA was found to be membrane associated during protein purification (10). Based on amino acid sequence, another aflatoxin protein, AflJ, was predicted to contain a C-terminal microbody targeting signal, NRY, and three membrane-spanning regions (22). By comparing the protein sequence of purified OmtA with the predicted amino acid sequence derived from the OmtA cDNA, this protein was proposed to contain a leader sequence that apparently is processed to generate a 42-kDa protein (33). This leader sequence may be required for this enzyme to interact with membranes (33). Based on computer-assisted analysis using prediction of protein localization sites(PSORT, version 6.4 [http://psort.nibb.ac.jp/psort]), we also speculate that putative peroxisomal targeting signals are present in aflatoxin proteins, including Nor-1, AvnA, OmtA, and OrdA (unpublished data). In contrast, OmtA (10) and Nor-1 (35) were observed in the postmicrosomal cytoplasmic fraction in cell fractionation studies. Therefore, the distribution of aflatoxin enzymes within a fungal cell is not clearly understood.
Localization to specific organelles has been demonstrated for enzymes involved in secondary metabolism. Penicillin is a secondary metabolite produced by several filamentous fungi, including A. nidulans. In a previous study, the enzyme 6-aminopenicillanic acid acyltransferase, which catalyzes the final step of penicillin biosynthesis in Penicillium spp., was localized to a membrane-bound organelle, the microbody (25); the authors suggested that penicillin is synthesized in this organelle. In a separate study, prehelminthosporol, a phytotoxin produced by Bipolaris sorokiniana was localized in the Woronin body (1), which is thought to be derived from the peroxisome (32). Prehelminthosporol has been shown to disrupt plant plasma membranes and inhibit growth of gram-positive bacteria. Detailed subcellular localization of OmtA and several other aflatoxin enzymes is now being conducted using immunoelectron microscopy in A. parasiticus. These data should help clarify the cellular site of aflatoxin synthesis in this filamentous fungus.
Acknowledgments
L.-W.L. and C.-H.C. contributed equally to this work.
We thank Fun Sun Chu (University of Wisconsin—Madison) for kindly providing the omtA plasmid and Jeff Cary (USDA) for providing the aflR knockout strain, AFS10.
This work was supported by the Michigan Agricultural Experiment Station (MSU), the National Food Safety and Toxicology Center (MSU), the Center for Environmental Toxicology (MSU), and the National Institutes of Health (RO1 CA52003-11).
REFERENCES
- 1.Akesson, H., E. Carlemalm, E. Everitt, T. Gunnarsson, G. Odham, and H.-B. Jansson. 1996. Immunocytochemical localization of phytotoxins in Bioplaris sorokiniana. Fungal Genet. Biol. 20:205-216. [Google Scholar]
- 2.Arseculeratne, S. N., L. M. De Silva, S. Wijesundera, and C. H. S. R. Bandunatha. 1969. Coconut as a medium for the experimental production of aflatoxin. Appl. Microbiol. 18:88-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2001. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 4.Belozerskaya, T. A. 1998. Cell-to-cell communication in differentiation of mycelial fungi. Membr. Cell Biol. 11:831-840. [PubMed] [Google Scholar]
- 5.Bhatnagar, D., S. P. McCormick, L. S. Lee, and R. A. Hill. 1987. Identification of O-methylsterigmatocystin as an aflatoxin B1 and G1 precursor in Aspergillus parasiticus. Appl. Environ. Microbiol. 53:1028-1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhatnagar, D., A. H. Ullah, and T. E. Cleveland. 1988. Purification and characterization of a methyltransferase from Aspergillus parasiticus SRRC 163 involved in aflatoxin biosynthetic pathway. Prep. Biochem. 18:321-349. [DOI] [PubMed] [Google Scholar]
- 7.Bhatnagar, D., T. E. Cleveland, and E. B. Lillehoj. 1989. Enzymes in aflatoxin B1 biosynthesis: strategies for identifying pertinent genes. Mycopathologia 107:75-83. [DOI] [PubMed] [Google Scholar]
- 8.Bratthauer, G. R. 1999. Processing of tissue specimens, p. 77-84. In L. C. Javois (ed.), Methods in molecular biology. Humana Press, Totowa, N.J. [DOI] [PubMed]
- 9.Cary, J., J. Linz, and D. Bhatnagar. 2000. Aflatoxins: biological significance and regulation of biosynthesis. In J. Cary, D. Bhatnagar, and J. Linz (ed.), Microbial foodborne disease: mechanisms of pathogenesis and toxin synthesis. Technomic Publishing, Lancaster, Pa.
- 10.Cleveland, T. E., A. R. Lax, L. S. Lee, and D. Bhatnagar. 1987. Appearance of enzyme activities catalyzing conversion of sterigmatocystin to aflatoxin B1 in late-growth-phase Aspergillus parasiticus cultures. Appl. Environ. Microbiol. 53:1711-1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eaton, D. L., and J. D. Groopman (ed.). 1994. The toxicology of aflatoxins: human health, veterinary, and agricultural significance. Academic Press, San Diego, Calif.
- 12.Gengan, R. M., A. A. Chuturgoon, D. A. Mulholland, and M. F. Dutton. 1999. Synthesis of sterigmatocystin derivatives and their biotransformation to aflatoxins by a blocked mutant of Aspergillus parasiticus. Mycopathologia 144:115-122. [DOI] [PubMed] [Google Scholar]
- 13.Guzman-de-Pena, D., and J. Ruiz-Herrera. 1997. Relationship between aflatoxin biosynthesis and sporulation in Aspergillus parasiticus. Fungal Genet. Biol. 21:198-205. [DOI] [PubMed] [Google Scholar]
- 14.Harris, S. D., J. L. Morrell, and J. E. Hamer. 1994. Identification and characterization of Aspergillus nidulans mutants defective in cytokinesis. Genetics 136:517-532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hicks, J. K., J.-H. Yu, N. P. Keller, and T. H. Adams. 1997. Aspergillus sporulation and mycotoxin production both require inactivation of the FadA G protein-dependent signaling pathway. EMBO J. 16:4916-4923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keller, G.-A., S. Krisans, S. J. Gould, J. M. Sommer, C. C. Wang, W. Schliebs, W. Kunau, S. Brody, and S. Subramani. 1991. Evolutionary conservation of a microbody targeting signal that targets proteins to peroxisomes, glyoxysomes, and glycosomes. J. Cell Biol. 114:893-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Keller, N. P., H. C. Dischinger, Jr., D. Bhatnagar, T. E. Cleveland, and A. H. J. Ullah. 1993. Purification of a 40-kDa methyltransferase activity in the aflatoxin biosynthetic pathway. Appl. Environ. Microbiol. 59:479-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang, S.-H., C. D. Skory, and J. E. Linz. 1996. Characterization of the function of the ver-1 and ver-1B genes, involved in aflatoxin biosynthesis in Aspergillus parasiticus. Appl. Environ. Microbiol. 62:4568-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang, S.-H., T.-S. Wu, R. Lee, F. S. Chu, and J. E. Linz. 1997. Analysis of mechanism regulating expression of the ver-1 gene, involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 63:1058-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, B. H., N. P. Keller, D. Bhatnagar, T. E. Cleveland, and F. S. Chu. 1993. Production and characterization of antibodies against sterigmatocystin O-methyltransferase. Food Agric. Immunol. 5:155-164. [Google Scholar]
- 21.Markham, P. 1995. Organelles of filamentous fungi, p. 75-98. In N. A. R. Gow and G. W. Gooday (ed.), The growing fungus, 1st ed. Chapman & Hall, London, United Kingdom.
- 22.Meyers, D. M., G. Obrian, W. L. Du, D. Bhatnagar, and G. A. Payne. 1998. Characterization of aflJ, a gene required for conversion of pathway intermediates to aflatoxin. Appl. Environ. Microbiol. 64:3713-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mito, R. E., and C. A. Townsend. 1997. Enzymology and molecular biology of aflatoxin biosynthesis. Chem. Rev. 97:2537-2555. [DOI] [PubMed] [Google Scholar]
- 24.Motomura, M., N. Chihaya, T. Shinozawa, T. Hamasaki, and K. Yabe. 1999. Cloning and characterization of the O-methyltransferase I gene (dmtA) from Aspergillus parasiticus associated with the conversion of demethylsterigmatocystin to sterigmatocystin and dihydrodemethylsterigmatocystin to dihydrosterigmatocystin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 65:4987-4994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Muller, W. H., T. P. van der Krift, A. J. J. Krouwer, H. A. B. Wosten, L. H. M. van der Voort, E. B. Smaal, and A. J. Verkleij. 1991. Localization of the pathway of the penicillin biosynthesis in Penicillium chrysogenum. EMBO J. 10:489-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Payne, G. A., and M. P. Brown. 1998. Genetics and physiology of aflatoxin biosynthesis. Annu. Rev. Phytopathol. 36:329-362. [DOI] [PubMed] [Google Scholar]
- 27.Reichard, U., G. T. Cole, T. W. Hill, R. Ruchel, and M. Monod. 2000. Molecular characterization and influence on fungal development of ALP2, a novel serine proteinase from Aspergillus fumigatus. Int. J. Med. Microbiol. 290:549-558. [DOI] [PubMed] [Google Scholar]
- 28.Skory, C. D., J. S. Horng, J. J. Pestka, and J. E. Linz. 1990. Transformation of Aspergillus parasiticus with the homologous gene (pyrG) involved in pyrimidine biosynthesis. Appl. Environ. Microbiol. 56:3315-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skory, C. D., P.-K. Chang, J. Cary, and J. E. Linz. 1992. Isolation and characterization of a gene from Aspergillus parasiticus associated with the conversion of versicolorin A to sterigmatocystin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 58:3527-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweeney, M. J., and A. D. W. Dobson. 1999. Molecular biology of mycotoxin biosynthesis. FEMS Micobiol. Lett. 175:149-163. [DOI] [PubMed] [Google Scholar]
- 31.Trail, F., P.-K. Chang, J. Cary, and J. E. Linz. 1994. Structural and functional analysis of the nor-1 gene involved in the biosynthesis of aflatoxin by Aspergillus parasiticus. Appl. Environ. Microbiol. 60:3315-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valenciano, S., J. R. De Lucas, I. Van der Klei, M. Veenhuis, and F. Laborda. 1998. Characterization of Aspergillus nidulans peroxisomes by immunoelectron microscopy. Arch. Microbiol. 170:370-376. [DOI] [PubMed] [Google Scholar]
- 33.Yu, J., J. W. Cary, D. Bhatnagar, T. E. Cleveland, N. P. Keller, and F. S. Chu. 1993. Cloning and characterization of a cDNA from Aspergillus parasiticus encoding an O-methyltransferase involved in aflatoxin biosynthesis. Appl. Environ. Microbiol. 59:3564-3571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu, J., P.-K. Chang, J. W. Cary, M. Wright, D. Bhatnagar, T. E. Cleveland, G. A. Payne, and J. E. Linz. 1995. Comparative mapping of aflatoxin pathway gene clusters in Aspergillus parasiticus and Aspergillus flavus. Appl. Environ. Microbiol. 61:2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou, R. 1997. The function, accumulation, and localization of the Nor-1 protein involved in aflatoxin biosynthesis; the function of the fluP gene associated with sporulation in Aspergillus parasiticus. Ph.D. dissertation. Michigan State University, East Lansing.