Abstract
Differential scanning calorimetry (DSC) is used to evaluate the thermal stability and reversibility after heat treatment of transitions associated with various cellular components of Escherichia coli and Lactobacillus plantarum. The reversibility and the change in the thermal stability of individual transitions are evaluated by a second temperature scan after preheating in the DSC to various temperatures between 40 and 130°C. The viability of bacteria after a heat treatment between 55 and 70°C in the DSC is determined by both plate count and calorimetric data. The fractional viability values based on calorimetric and plate count data show a linear relationship. Viability loss and the irreversible change in DSC thermograms of pretreated whole cells are highly correlated between 55 and 70°C. Comparison of DSC scans for isolated ribosomes shows that the thermal stability of E. coli ribosomes is greater than that of L. plantarum ribosomes, consistent with the greater thermal tolerance of E. coli observed from viability loss and DSC scans of whole cells.
Thermal processing is the main technology applied in the food industry for the preservation of food materials. The main goal of thermal processing is to inactivate the spoilage and pathogenic microorganisms and produce a safe product with enhanced shelf life. An understanding of the mechanism of microbial inactivation by heat is potentially useful for optimizing heat treatments to eliminate foodborne disease and spoilage risk associated with common and emerging strains while avoiding overprocessing of the food material. Thermal inactivation of microorganisms is associated with irreversible denaturation of membranes, ribosomes, and nucleic acids. However, the patterns of macromolecular changes that induce the cell death of microorganisms during heat treatment are still not clearly known.
Differential scanning calorimetry (DSC) is a thermal analysis technique that detects, monitors, and characterizes thermally induced conformational transitions and phase transitions as a function of temperature. A number of overlapping transitions with a net endothermic effect are observed when microorganisms are heated (2, 8, 12, 14, 15). The observed transition peaks correspond to the denaturation of cellular components. Mackey et al. (12) investigated the origins of apparent individual transitions on the thermogram of Escherichia coli. Individual peaks observed in thermograms of whole cells of E. coli were assigned to cell components by comparing the transition temperatures of isolated cell components with corresponding transition temperatures in whole cells. It is thought that a strong relationship exists between thermal death of bacteria and the first major peak in DSC thermograms (temperature range of 60 to 80°C), which is attributed to ribosomal melting (13, 22). Several investigators have shown correlations between the stability of ribosomes and cell viability for Staphylococcus aureus (1), Listeria monocytogenes (21), and Salmonella enterica serovar Typhimurium (23). Furthermore, a recent DSC investigation of pressure-treated E. coli NCTC 8164 demonstrated that cell death and ribosome damage are closely related (17). Irreversible denaturation of cellular DNA requires temperatures well above the temperature of cell inactivation (12). At temperatures that cause ribosome denaturation, the DNA transition is reversible (15).
Previous DSC investigations of microorganisms employed scans to high temperatures (at or above 100°C), resulting in inactivation of the microorganisms. Most rescans did not display any peaks except for an endothermic transition attributable to DNA (2, 12, 14, 15). Although DSC thermograms were compared to the results of viability studies, no studies examined the relationship between thermal stability differences in whole cells and in isolated ribosomes or correlations between viability measures based on plate count and calorimetric data. The objectives of this study include (i) comparison using calorimetry of the thermal stability of two selected microorganisms, E. coli and Lactobacillus plantarum, in relation to the thermal stabilities of their ribosomes; (ii) investigation of the reversibility of individual transitions associated with various components of whole cells of E. coli and L. plantarum; and (iii) determination and comparison of the temperature dependence of cell viability for a linearly increasing temperature protocol from plate count and calorimetric data.
MATERIALS AND METHODS
Source and preparation of organisms.
E. coli ATCC 14948 and L. plantarum ATCC 10241 were obtained from the Culture Collection Center at the Ohio State University. A loopful of each organism was revived in 10 ml of tryptic soy broth (Difco Laboratories, Detroit, Mich.) supplemented with 0.3% (wt/wt) yeast extract (TSBY) for E. coli or MRS broth (Difco) for L. plantarum and incubated at 37°C for 18 h. Each culture was stored frozen (−80°C) in 30% (vol/vol) sterile glycerol. A loopful of each stock culture was transferred to 10 ml of tryptic soy or MRS broth and incubated for 10 h at 37°C before use.
Each culture was inoculated (1%, vol/vol) into TSBY or MRS broth. The cultures were incubated at 37°C. The growth phase was determined by measuring absorbance at 640 nm, using a Beckman Du-50 spectrophotometer, and matching appropriate viable counts from a standard growth curve. The cells were grown to the late exponential growth phase, as determined from the growth curve. The final concentration of cells in the medium was 1.3 × 109 ± 0.1 × 109 CFU ml−1 for E. coli and 9.0 × 108 ± 0.1 × 108 CFU ml−1 for L. plantarum. Cells in the broth were harvested by centrifugation (Beckman J2-21 centrifuge) at 10,000 × g for 10 min at 4°C. The supernatant was discarded, and the pellets were washed with sterile distilled water and centrifuged for a second time before being transferred into DSC crucibles.
Calorimetry of whole cells.
Pellets of whole cells were transferred into the empty sample crucible and were weighed (56 ± 0.3 mg [wet weight]). The dry-material content of the pellets was determined by freeze-drying (Freezone 4.5 freeze-dry system, model 77510; Labconco, Missouri) as 19% ± 0.3% for E. coli and 20% ± 0.5% for L. plantarum on a wet basis. The standard deviations were calculated based on 12 freeze dried pellets for each bacterium.
A differential scanning calorimeter (DSC 111; Setaram, Lyon, France) was used to record thermograms of microorganisms heated at 3°C min−1. All DSC measurements were conducted using fluid-tight, stainless steel crucibles. For each DSC run, the reference crucible was filled with ∼45 μl (∼80% of sample weight) of distilled water. A DSC run was performed with unsealed empty sample and reference crucibles to record an empty-crucible baseline. Crucibles were sealed using aluminum O-rings and were refrigerated at 4°C prior to DSC runs. The sample and reference crucibles were placed in the calorimeter and equilibrated at 1°C using a liquid-nitrogen cooling system. After being heated in the calorimeter, the samples were cooled rapidly with liquid nitrogen and rescanned to ascertain the reversibility of thermograms. The samples were reweighed after DSC measurements to check for loss of mass during heating. Thermograms of samples showing signs of leakage were discarded.
Heat pretreatment in the DSC of whole-cell pellets.
Heat pretreatment was performed in the DSC. Throughout the text, an unheated sample is referred to as an untreated sample. The pellet was sealed in the sample crucible, heated to the pretreatment temperature, maintained at the pretreatment temperature for 60 s, and then rapidly cooled to 1°C. The sample was rescanned from 1 to 130°C at 3°C min−1 to assess the reversibility of thermally induced transitions in bacterial cells. The reversibility of the transitions was evaluated by performing partial scans between 40 and 130°C at 5°C intervals. Additional pretreatment runs were conducted at 57.5°C for L. plantarum and at 57.5, 62.5, and 64°C for E. coli due to sharp decreases in viability observed over the temperature range of 50 to 70°C.
Measurement of cell viability after heat pretreatment.
Heat pretreatment prior to viability measurements was conducted in the DSC as described in the previous section. The crucible containing a pellet was capped (not sealed) using an aluminum ring and screw cap. The reference crucible was filled with distilled water (∼80% of the sample weight). The crucibles were refrigerated (4°C) until use. Pellets in the crucibles were heated to pretreatment temperatures between 50 and 70°C as specified in the previous section at a 3°C min−1 heating rate in the calorimeter. After rapid cooling, a portion (40 or 50 mg) of heat-treated pellet from the sample crucible was transferred using a sterile loop to a (1.5-ml) sterile polyethylene tube. Sterile peptone water was added to make a final volume of 1 ml. After careful suspension in the tube, the cells were serially diluted and plated into tryptic soy agar or MRS agar to determine viable counts. After 36 h of incubation at 37°C, viable counts of each sample were obtained by calculation of the dilution ratio. An untreated sample was used as a control.
Preparation and calorimetry of intact ribosomes.
The protocol described by Mackey et al. (12) with modification of buffer solutions was applied to prepare the intact ribosomes for both bacteria. The cell pellets obtained by centrifugation of 3.5 liters of late-exponential-phase cultures were washed and resuspended in 20 mM HEPES buffer (pH 7.5) containing 6 mM MgCl2 and 50 mM NH4Cl. The cell suspension was disrupted by being passed two or three times through a previously cooled French press (AMINco SLM Instruments, Inc., Urbana, Ill.). DNase (RNase free) (Sigma) was added (0.4 mg ml−1), and the material was centrifuged (Beckman L85-55M ultracentrifuge) at 32,500 × g for 30 min. The supernatant (cell extract) was centrifuged at 150,000 × g for 3.5 h to obtain a pellet of crude intact ribosomes. The water content of the ribosome pellet was determined to be 65.7% for the E. coli ribosome and 64.9% for the L. plantarum ribosome on a wet-cell basis. Pellets of intact ribosomes were placed in the calorimeter sample crucible. The reference crucible was filled with an amount of HEPES buffer equal to the amount of buffer in the sample. The crucibles were heated from 1 to 140°C at a 4°C min−1 in the calorimeter.
Data analysis.
DSC thermograms were corrected for differences in the empty crucibles by subtracting an empty-crucible baseline. Total heat data corresponding to the envelope of endothermic peaks (enthalpy, expressed as Joules per gram) between approximately 45 and 130°C for E. coli and 45 and 110°C for L. plantarum were determined by integrating the temperature-versus-heat-flow curve using software provided by the instrument manufacturers. A curved baseline with three temperature points was utilized to calculate the apparent enthalpy of both whole cells and the intact ribosomes. Use of a curved baseline which takes into account the apparent heat capacity change before and after the transition(s) of interest is explained in detail by Lee and Kaletunç (10). Peak temperatures for the thermally induced transitions were also determined.
RESULTS
Thermograms of E. coli and L. plantarum whole cells.
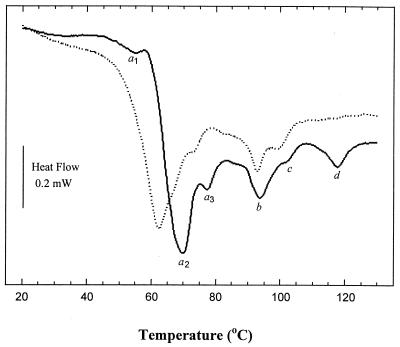
Figure 1 shows the DSC thermograms for untreated E. coli and L. plantarum pellets. The major peak in the DSC thermograms of both bacteria was observed over a temperature range of 40 to 80°C (Fig. 1). Several differences exist between the DSC thermograms of E. coli and L. plantarum. The first peak, a1 (peak temperature, 56°C), which is proposed to be the denaturation of the smallest ribosomal subunit (30S) in E. coli (12), was not observed in the thermogram of L. plantarum as a separate peak or shoulder. The major peak, peak a2, appeared at a higher temperature in the E. coli thermogram (70°C) than in the L. plantarum thermogram (63°C). A peak (peak b) similar to the peak reported by Mackey et al. (12) as the melting of DNA in E. coli existed, although at slightly different temperatures, in thermograms of both E. coli (94°C) and L. plantarum (93°C). Similarly, peak c, a peak suggested by Mackey et al. (12) to be related to denaturation of a cell wall component, appeared at 102.5°C in E. coli thermogram and at 100°C in the L. plantarum thermogram. Figure 1 also shows that peak d (peak temperature, 118°C), which appeared in the E. coli thermogram, was absent from the L. plantarum thermogram. Also apparent from Fig. 1 is a difference in the apparent heat capacity of the live and inactivated cells (difference between the baseline before and after the transition) of about 0.6 J g−1 K−1 for both organisms.
FIG. 1.
Thermograms of whole cells of E. coli (solid line) and L. plantarum (dotted line) obtained by DSC (1 to 150°C at a heating rate of 3°C min−1).
Thermograms of isolated ribosomes.
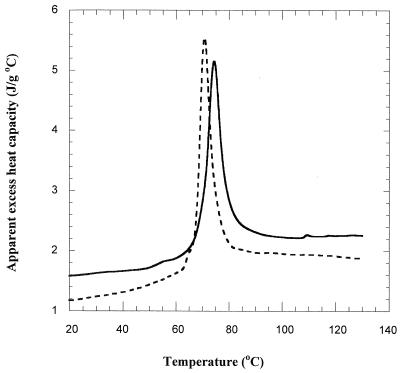
Intact ribosomes from both bacteria were isolated, and DSC thermograms of ribosomes suspended in HEPES buffer (pH 7.5) were collected and compared with those of whole cells. Two endothermic transitions were observed for E. coli ribosomes (Fig. 2). The L. plantarum ribosome thermogram displayed an endothermic peak with a shoulder on the ascending side of the peak. Comparison of denaturation peaks for ribosomes suspended in HEPES buffer (pH 7.5) showed that the peak temperature of the transition was higher for the E. coli ribosome (74.3°C) than for the L. plantarum ribosome (70.7°C), indicating higher thermal stability. The area of the peak which corresponds to the enthalpy of ribosome denaturation also was different for each ribosome. E. coli ribosomes displayed a higher enthalpy of denaturation (26.6 J g of dry ribosome−1) than did L. plantarum ribosomes (23.5 J g of dry ribosome−1). A heat capacity change as a result of ribosome denaturation (difference between the baseline before and after the transition) was observed for both E. coli (0.4 J g−1 K−1) and L. plantarum (0.33 J g−1 K−1).
FIG. 2.
Thermograms of isolated intact ribosomes of E. coli (solid line) and L. plantarum (dashed line) obtained by DSC (1 to 140°C at a heating rate of 4°C min−1).
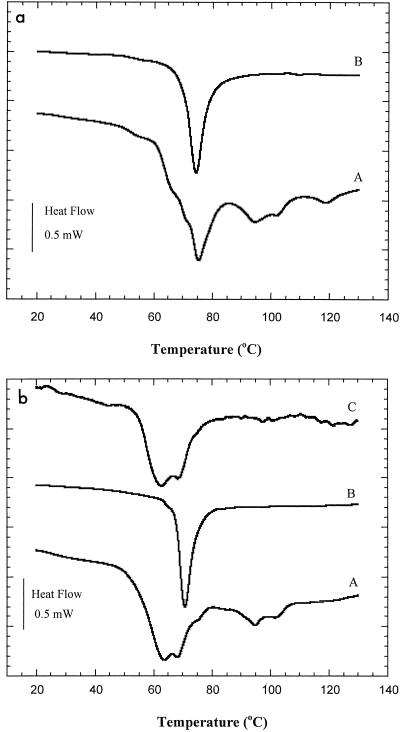
DSC thermograms of intact ribosomes were compared to those of whole cells (Fig. 3, thermograms A and B). For E. coli, the peak temperature (75°C) of the major peak in the thermogram of the whole-cell pellet washed in HEPES buffer coincided with the peak temperature (74.3°C) of the ribosome denaturation peak of the isolated intact ribosomes suspended in HEPES buffer (Fig. 3a). However, for L. plantarum, while a single peak was observed for ribosomes suspended in HEPES buffer at pH 7.5, the whole-cell thermogram showed dual peak temperatures (63.5 and 68.2°C). The DSC thermogram of L. plantarum ribosomes suspended in potassium phthalate buffer at pH 4 (Fig. 3b, thermogram C) displayed a profile (peak temperatures, 62.4 and 68.4°C) which strongly resembled the major peak in the thermogram of L. plantarum whole cells.
FIG. 3.
(a) Thermograms of whole cells (A) and isolated intact ribosomes (B) of E. coli obtained by DSC after a wash with HEPES buffer (pH 7.5). (b) (A and B) Thermograms of whole cells (A) and isolated intact ribosomes (B) of L. plantarum obtained by DSC after a wash with HEPES buffer (pH 7.5). (C) Thermogram of ribosomes obtained by DSC in potassium phthalate buffer (pH 4).
Effect of heat pretreatment on the DSC profiles of E. coli and L. plantarum.
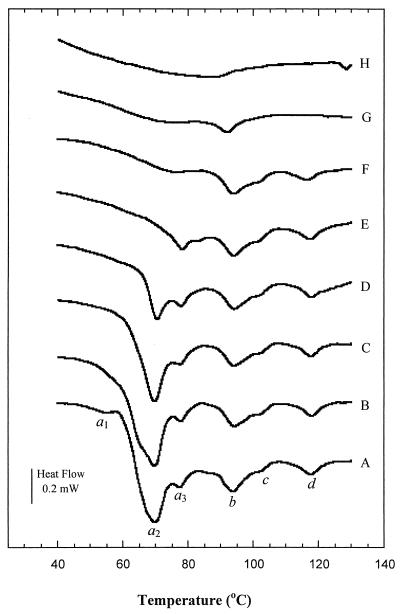
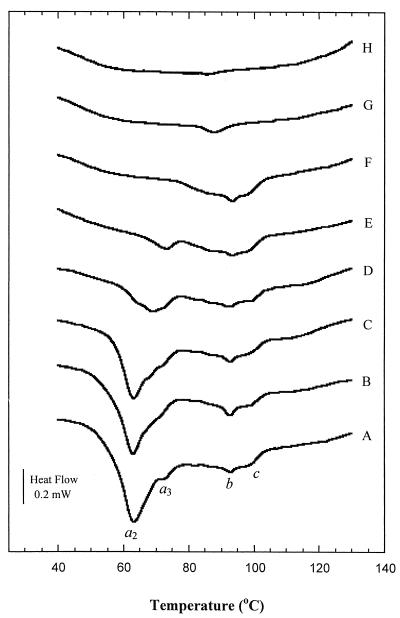
Pretreatment of bacteria at various temperatures was performed in the calorimeter with a partial scan to a predefined temperature. The reversibility of transitions after pretreatment was evaluated with a full second DSC scan (1 to 130°C) following the partial scan. Thermograms displaying changes in major conformational transitions of both organisms as a function of heat treatment are shown in Fig. 4 and 5. In general, pretreatments below 70°C resulted in significant changes in both the temperature and the area of the major peak observed in the thermograms of both bacteria, indicating an irreversible effect of pretreatment. It is apparent from Fig. 4 and 5 that the major peak in the DSC thermogram was obliterated after a pretreatment temperature to 85°C for E. coli (Fig. 4, thermogram F) and 75°C for L. plantarum (Fig. 5, thermogram F). Pretreatment resulted in changes in the shapes of existing peaks as well as in the appearance of new peaks. Thermograms of untreated bacteria are composed of several overlapping transitions. A new peak, which was previously partially obscured, may seem to appear if the overlapping transition disappears due to the heat treatment. For E. coli, peak a3, which was only partially visible due to the overlapping peak a2 in the control and 50 to 65°C pretreatment thermograms, became visible after pretreatment at 70°C (Fig. 4).
FIG. 4.
Effect of heat pretreatment on the thermogram of E. coli. (A) Control; (B to H) pretreatment temperatures of 50°C (B), 60°C (C), 65°C (D), 70°C (E), 85°C (F), 115°C (G), and 130°C (H). Thermograms are offset for clarity.
FIG. 5.
Effect of heat pretreatment on the thermogram of L. plantarum. (A) Control; (B to H) pretreatment temperatures of 50°C (B), 55°C (C), 60°C (D), 65°C (E) 75°C (F), 95°C (G), and 100°C (H). Thermograms are offset for clarity.
In Fig. 4 and 5, only the full scans following pretreatment which resulted in major irreversible changes are displayed. For E. coli, peak b, which is associated mainly with intracellular DNA denaturation (12), was reversible after heat treatment at 115°C. For L. plantarum, peak b shifted to a lower temperature following heat treatment at 95°C (Fig. 5, thermogram G) and was not reversible after the cell pellet was preheated to 100°C (thermogram H). After a partial scan to 115°C, the transition due to an outer cell wall component of E. coli (peak d) was absent. No evidence of native cellular components was observed in the thermogram of the 130°C heat-treated E. coli pellet (Fig. 4, thermogram H). However, for L. plantarum there was no evidence of native cellular components in thermograms of pellets heat treated at 100°C.
Comparison of DSC thermograms and viability of E. coli and L. plantarum after heat pretreatment.
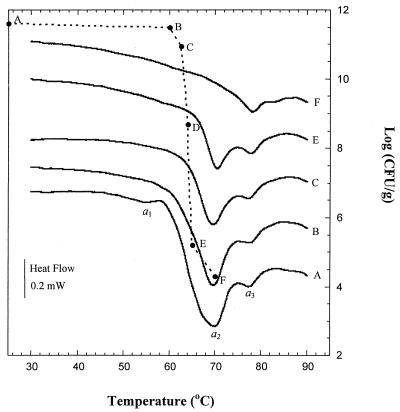
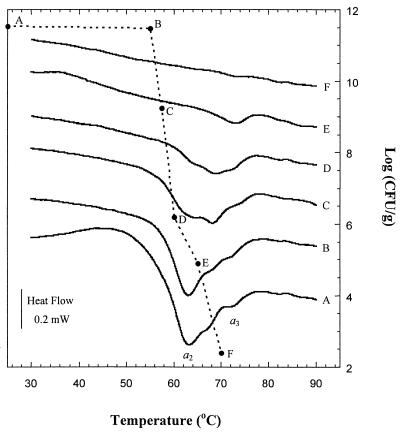
DSC thermograms of pellets of each microorganism were compared to each control thermogram after heat treatment at different temperatures (Fig. 6 and 7). The area under the curve of the second scan (apparent enthalpy, Joules per gram) was evaluated by integration. Significant reductions in the apparent enthalpy occurred with heat treatment up to 65°C (∼53%) for E. coli and up to 60°C (∼58%) for L. plantarum. The viability of each microorganism treated in the calorimeter under conditions identical to the corresponding DSC experiment was determined and plotted in Fig. 6 and 7. For E. coli, the viable-cell counts of the culture pellet displayed a slight change with heat treatment up to 60°C. Heat treatment of E. coli to higher temperature resulted in 6-log10-unit reductions for 65°C treatments and 7-log10-unit reductions for 70°C treatments. These reductions were accompanied by a decrease in the area of the peak (a2) corresponding to the denaturation of ribosomes in the thermogram (Fig. 6, thermograms E and F). For L. plantarum, irreversible denaturation of ribosomes was observed in the thermogram following a 57.5°C heat treatment (Fig. 7, thermogram C), with a viability loss of 2.3 log10 units.
FIG. 6.
Viable counts (--·--) and DSC thermograms of E. coli for control (A) and after heat pretreatment at 60°C (B), 62.5°C (C), 64°C (D, thermogram not shown), 65°C (E), and 70°C (F). Thermograms are offset for clarity.
FIG. 7.
Viable counts (--·--) and DSC thermograms of L. plantarum for control (A) and after heat pretreatment at 55°C (B), 57.5°C (C), 60°C (D), 65°C (E), and 70°C (F). Thermograms are offset for clarity.
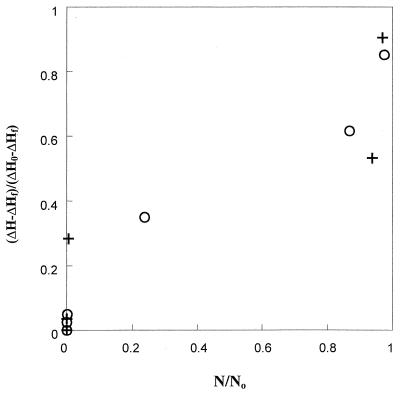
As reported by Lee and Kaletunç (10), by assuming that the apparent enthalpy is proportional to the number of viable cells after correction for residual apparent enthalpy, the fractional viability can be defined as the reduced apparent enthalpy, (ΔH − ΔHf)/(ΔH0 − ΔHf), where ΔH is the apparent enthalpy after a pretreatment, ΔHf is the residual apparent enthalpy after treatment resulting in no viability, and ΔH0 is the apparent enthalpy of untreated cells. Fractional viability values calculated from calorimetric data, (ΔH − ΔHf)/(ΔH0 − ΔHf), and plate count data, N/N0, are plotted in Fig. 8. A linear relationship between the reduced apparent enthalpy and the fraction of survivors was observed, except for the points corresponding to high temperature treatment.
FIG. 8.
Correlation between fractional apparent enthalpy and fractional viability for E. coli (○) and L. plantarum (+).
DISCUSSION
This study aims to assess bacterial resistance to heat treatment by (i) comparing the thermal resistance of bacteria and the thermal stability of isolated intact ribosomes, (ii) evaluating the reversibility of thermal transitions of various cellular components following heat treatment, and (iii) developing a relationship between fractional viability calculated from plate count data and calorimetric data.
The primary low-temperature features (40 to 80°C) of whole-cell DSC profiles of bacteria are thought to correspond to the thermal unfolding of ribosomes (12). Using ribosomes isolated from E. coli, Mackey et al. (12) showed that an endotherm with three overlapping peaks appearing between 47 and 85°C for E. coli whole cells is associated with ribosome denaturation. In our study, although E. coli whole cells displayed a ribosomal denaturation endotherm consisting of three peaks, two peaks (a2 and a3) were observed for ribosomal denaturation in the L. plantarum thermogram (Fig. 1). It has been reported that the 30S ribosomal subunit is less thermally stable than the larger ribosomal subunit (3, 12, 21), suggesting that peak a1 may be attributed to denaturation of the 30S ribosomal subunit. It is also apparent from Fig. 1 that the peaks a2 and a3 are shifted to lower temperatures in comparison to the corresponding peaks for E. coli. The lower peak temperatures of peaks a2 and a3 of the L. plantarum thermogram suggest that the relative stabilities of the L. plantarum ribosomes are lower than those of the E. coli ribosomes. The pH of the L. plantarum medium is reduced due to lactic acid production during L. plantarum growth. It is possible that the increased acidity in the medium of L. plantarum may influence the stability of the ribosomal subunits directly or indirectly. Another factor that may influence ribosome stability is altered intracellular cation concentrations; in particular, Mg2+ is required for ribosome integrity (6). A loss of the peak (a1) is observed in the thermogram of acid-treated E. coli (unpublished results). Mohacsi-Farkas et al. (16) reported that the ribosomal denaturation peak of L. plantarum shifted to lower temperatures as the pH of the suspending medium decreased below 5. Furthermore, their results show that while a low-temperature (peak temperature, 57°C) endothermic transition appears when cells are suspended in buffer at pH 6.8 and 5, this transition is not observed on the thermograms for whole cells suspended in buffer at pH 4.6 and lower. Because the pH of the growth medium for L. plantarum was measured to be 4.4 in the present study, our results are in agreement with the previous data showing an absence of peak a1 and lower peak temperatures for peaks a2 and a3 induced by low pH. Ribosomes also were reported to be destabilized by the loss of Mg2+ from cells (2, 7, 19). The lack of a visible peak, a1, in the L. plantarum thermogram may indicate denaturation of the 30S ribosomal subunit as a result of Mg2+ loss (6, 24). Alternatively, peak a1 may be present but obscured by the other ribosomal peaks a2 and a3 because their transition are shifted to lower temperatures.
The comparison of ribosomal denaturation for each bacterium at pH 7.5 shows that both the peak temperature and the apparent enthalpy for E. coli are higher than those for L. plantarum, indicating a higher thermal stability and a greater energy requirement to disrupt the structure of the E. coli ribosome (Fig. 2). Similar behavior is observed for whole cells, suggesting that both the thermal stability and the energy required to inactivate the bacterial cells are higher for E. coli.
E. coli ribosomes show similar thermal stabilities, in terms of peak temperatures of corresponding transitions, in whole cells (75.1°C) and when isolated (74.3°C). However, for L. plantarum, the transition shapes and the thermal stabilities of whole cells washed with HEPES buffer (pH 7.5) and of isolated ribosomes are similar only when the ribosomes are suspended in potassium phthalate buffer (pH 4). Although an effect on ribosomes is indicated in both cases, the strong resemblance in the shape of observed peaks for whole cells and isolated ribosomes may have different causes and must be explored further. The higher thermal stability of isolated L. plantarum ribosomes when suspended in HEPES buffer (pH 7.5) containing 6 mM MgCl2 (Fig. 3b, thermogram B) than that of isolated ribosomes suspended in potassium phthalate buffer without Mg2+ (pH 4) (thermogram C) may be attributed to stabilization of ribosomes by magnesium ions in vitro (18). Similar behavior also was observed with E. coli whole cells, where the peak temperature of the major peak was 70°C following a water wash (Fig. 1) but 75.1°C following a HEPES buffer wash (Fig. 3a, thermogram A). Anderson et al. (2) note that the ionic composition and concentration of the buffer affect the thermal stability and shape of the ribosomal denaturation peak, which in turn may affect the thermal resistance of bacteria.
In the whole-cell thermograms, there are probably more transitions than are observable as discrete peaks. Some of these transitions may occur within the same temperature range and may be obscured by the larger ribosome denaturation peaks. Comparison of thermograms of ribosomal denaturation and whole cells (Fig. 1 and 2) shows that the difference in heat capacity between the native and denatured states, as shown by the difference in pre- and post-transition baselines, is 1.5 times greater for whole cells. A positive heat capacity is typically observed for denaturation of proteins. For a typical globular protein of ∼15 kDa, the change in the heat capacity is on the order of 0.4 to 0.67 J g−1 K−1 (5). Given the larger heat capacity change observed for denaturation of whole cells relative to ribosomes, it is probable that other cellular components contribute to the endothermic transitions attributed to the denaturation of ribosomes. Anderson et al. (2) indicate that the small number of peaks observed in whole-cell thermograms can be due to a larger number of transitions including protein unfolding and denaturation.
Another visible difference between the E. coli and L. plantarum thermograms is a high-temperature endothermic transition (peak d) that is observed only in the DSC thermogram of E. coli whole cells. Mackey et al. (12) observed a peak corresponding to peak d in the thermogram of the cell envelope fraction and proposed this peak to be the result of cell envelope denaturation. These investigators hypothesized that a cell wall-associated thermostable protein may account for the appearance of this peak. Other DSC studies in our laboratory showed that the peak was observed in thermograms of Pseudomonas fluorescens but was absent from thermograms of S. aureus and Leuconostoc mesenteroides (unpublished results), suggesting that the origin of this peak is a cellular component of gram-negative bacteria. In most gram-negative bacteria, the outer cell wall layer exists as a true unit membrane. The outer cell wall membrane contains lipid, phospholipid, polysaccharide, and protein. The lipid and polysaccharide form a specific lipopolysaccharide layer. Rodriguez-Torres et al. (20), using DSC, reported that lipopolysaccharide shows endothermic transitions above 120°C, with the specific temperature depending on the linkage type. Reversibility studies demonstrate that this peak in E. coli is denatured by heat treatment above 110°C.
The peak temperatures associated with DNA denaturation are not significantly different for the two microorganisms, with that for L. plantarum is being slightly lower (93°C) than that for E. coli (94°C). Although the thermal stabilities of the DNA for both microorganisms are similar, their reversibility subsequent to heat treatment differs significantly. There is no indication of a DNA peak in the thermogram of the L. plantarum pellet preheated to 100°C (Fig. 5, thermogram H). The peak is preserved in the thermogram of the E. coli pellet heated up to 125°C (thermogram not shown), although the apparent enthalpy is reduced and the peak is shifted to lower temperatures as the heat pretreatment temperature is increased. The change in energy required for denaturation of DNA after heat treatment indicates partial refolding on cooling or folding to a different state (4). Furthermore, the appearance of a previously obscured, reversible peak (Tmax at 88.4°C) in the thermogram of L. plantarum preheated to 95°C may be due to partial reversibility of denatured DNA (Fig. 5, thermogram G). Mackey et al. (11) showed that there is a strong correlation between the G+C content of DNA and the Tmax of the putative DNA peak determined from a DSC scan of whole cells. The G+C content of L. plantarum (44 to 46 mol%) (9) is lower than that of E. coli (51.6 mol%) (11). Using the empirical relation between the G+C content and Tmax reported by Mackey et al. (11), we obtain a predicted DNA melting temperature of 92.4 to 93°C for L. plantarum and 94.8°C for E. coli. Mackey et al. (11) also reported a DNA melting temperature of 94.3°C determined by DSC for E. coli. Our experimental values for DNA melting are in close agreement with the literature data, including the expectation of a lower DNA peak temperature for L. plantarum.
DSC curves can be exploited further to determine the fractional viability of microorganisms based on calorimetric data, as described by Lee and Kaletunç (10). For both E. coli and L. plantarum, Fig. 4 and 5 reveal that as the severity of the heat treatment increases, the observed peak temperature of ribosomal denaturation increases, implying sequential damage to the ribosomal subunits and/or the existence of a range of thermal resistance in the microorganism population. It is apparent that a loss of viability of cells of both organisms occurs when the microorganisms are subjected to heat pretreatment in the range of 50 to 70°C. The viability loss is related to the apparent enthalpy change of ribosomal subunits monitored by DSC because preheating at 55 to 70°C affected the peaks associated with ribosome subunits but had no apparent influence on the thermally induced transitions of other cellular structures. Both the putative ribosomal peaks in the thermogram and cell viability of L. plantarum were noticeably reduced by preheating from 55 to 70°C (Fig. 7). A similar pattern was observed in the DSC profiles of E. coli, although the reductions in ribosomal peaks and cell viability occurred at higher temperature (Fig. 6). However, as discussed in detail by Lee and Kaletunç (10), the peak area corresponding to only the ribosome transition within the whole-cell thermogram cannot be determined accurately because the baseline is not well defined due to overlapping transitions. Instead, the total peak area corresponding to the total apparent enthalpy must be used. With increasing treatment temperature, the total apparent enthalpy (between approximately 50 and 130°C for E. coli and 50 and 110°C for L. plantarum) decreases gradually compared to the peak area for the untreated control. It is apparent from Fig. 4 and 5 that residual transitions remain even after the cells are inactivated, implying that the total area under the thermogram includes contributions related to both cell death and additional macromolecular transitions. After subtraction of the contributions due to enthalpy associated with inactive cells, a reduced apparent enthalpy value can be defined to determine the fraction of survivors in terms of calorimetric data. A plot of reduced apparent enthalpy against the fractional survivors from plate count data (Fig. 8) gives a linear relationship. As we have shown (10), these data can be interpreted in terms of D and z values of microorganisms which are subjected to linearly increasing temperature. In Fig. 8, the points close to a viability value of 1 represent low-temperature treatment while the points close to 0 represent high-temperature treatments. It is apparent that for both microorganisms at very low temperature, viabilities calculated from both plate count and calorimetric data are in close agreement. However, as the treatment temperature increases, a disparity appears between the plate count and calorimetric data for both microorganisms. The disparity between the viability data derived from the two methods is larger for L. plantarum than for E. coli. As the temperature of the treatment increases further, the disparity decreases. It is expected that as the temperature of the heat treatment increases, the number of injured microorganisms also increases. The injured microorganisms die during a complete DSC scan following the partial scan without having a chance to repair. However, the plate count method provides favorable conditions for injured cells to recover. We speculate that the disparity between viabilities calculated from calorimetric data and from plate count data at intermediate treatment temperatures may be due to injury of the microorganisms during pretreatment. L. plantarum may have a greater tendency to injury than E. coli. This speculation may also explain the lag period typically observed in the semilog survival curves of microorganisms and needs to be explored further.
In this study, the patterns of the temperature-induced changes in ribosomes, cell wall components, and DNA of E. coli (gram-negative) and L. plantarum (gram-positive) bacteria are compared by DSC. The results indicate that more intensive heat treatment is needed to inactivate E. coli than to inactivate L. plantarum. Mohacsi-Farkas et al. (15) also reported a higher heat inactivation temperature for E. coli than for L. plantarum.
The thermal tolerance of microorganisms may depend on the growth conditions as well as the cell structure. Both the thermal stability and enthalpy of ribosome denaturation are influenced by pH in vitro and in vivo for E. coli and L. plantarum. Work is in progress in our laboratory on the calorimetric evaluation of the thermal stability and apparent enthalpy change of ribosomes isolated from E. coli and L. plantarum as a function of pH.
We have demonstrated a correlation between viability calculated from calorimetric data and from plate count data. Calorimetric data provide unique information by direct measurement of the energy required to inactivate microorganisms. Evaluating and quantifying differences in thermograms of whole cells and isolated components permits us to rank the relative thermal stabilities of the various cellular components and identify those most susceptible to thermal disruption.
Acknowledgments
This work is supported by State and Federal Funds appropriated to the Ohio Agricultural Research and Development Center and The Ohio State University.
We thank the reviewers for insightful comments.
REFERENCES
- 1.Allwood, M. C., and A. D. Russel. 1967. Mechanism of thermal injury in Staphylococcus aureus. Appl. Microbiol. 15:1266-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, W. A., N. D. Hedges, M. V. Jones, and M. B. Cole. 1991. Thermal inactivation of Listeria monocytogenes studied in differential scanning calorimetry. J. Gen. Microbiol. 137:1419-1424. [DOI] [PubMed] [Google Scholar]
- 3.Bonincontro, A., S. Cinelli, M. Mengoni, G. Onori, G. Risuleo, and A. Santucci. 1998. Differential stability of E. coli ribosomal particles and free RNA towards thermal degradation studied by microcalorimetry. Biophys. Chem. 75:97-103. [DOI] [PubMed] [Google Scholar]
- 4.Cantor, C. R., and P. R. Schimmel. 1980. Biophysical chemistry, part III. The behavior of biological macromolecules, p. 1222-1223. W.H. Freeman & Co., San Francisco, Calif.
- 5.Gomez, J., V. J. Hilser, D. Xie, and E. Freire. 1995. The heat capacity of proteins. Proteins Struct. Funct. Genet. 22:404-412. [DOI] [PubMed] [Google Scholar]
- 6.Hurst, A. 1984. Reversible heat damage, p. 303-318. In A. Hurst and A. Nasim (ed.), Repairable lesions in microorganisms. Academic Press, Ltd., London, United Kingdom.
- 7.Hurst, A., and A. Hughes. 1978. Stability of ribosomes of Staphylococcus aureus S6 sublethally heated in different buffers. J. Bacteriol. 133:564-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kaletunç, G. 2001. Thermal analysis of bacteria using differential scanning calorimetry, p. 227-235. In F. Bozoglu, T. Deak, and B. Ray (ed.), Novel process and control technologies in the food industry. IOS Press, Amsterdam, The Netherlands.
- 9.Kandler, O., and D. Weiss. 1986. Regular, nonsporing, gram-positive rods, p. 1208-1234. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. The Williams & Wilkins Co., Baltimore, Md.
- 10.Lee, J., and G. Kaletunç. 2002. Calorimetric determination of inactivation parameters of microorganisms. J. Appl. Microbiol. 93:178-189. [DOI] [PubMed] [Google Scholar]
- 11.Mackey, B. M., S. E. Parsons, C. A. Miles, and R. J. Owen. 1988. The relationship between base composition of bacterial DNA and its intracellular melting temperature as determined by differential scanning calorimetry. J. Gen. Microbiol. 134:1185-1195. [DOI] [PubMed] [Google Scholar]
- 12.Mackey, B. M., C. A. Miles, S. E. Parsons, and D. A. Seymour. 1991. Thermal denaturation of whole cells and cell components of Escherichia coli examined by differential scanning calorimetry. J. Gen. Microbiol. 137:2361-2374. [DOI] [PubMed] [Google Scholar]
- 13.Mackey, B. M., C. A. Miles, D. A. Seymour, and S. E. Parsons. 1993. Thermal denaturation and loss of viability in Escherichia coli and Bacillus stearothermophilus. Lett. Appl. Microbiol. 16:56-58. [Google Scholar]
- 14.Miles, C. A., B. M. Mackey, and S. E. Parsons. 1986. Differential scanning calorimetry of bacteria. J. Gen. Microbiol. 132:939-952. [DOI] [PubMed] [Google Scholar]
- 15.Mohacsi-Farkas, C., J. Farkas, L. Meszaros, O. Reichart, and E. Andrassy. 1999. Thermal denaturation of bacterial cells examined by differential scanning calorimetry. J. Therm. Anal. Calor. 57:409-414. [Google Scholar]
- 16.Mohacsi-Farkas, C., J. Farkas, and A. Simon. 1994. Thermal denaturation of bacterial cells examined by differential scanning calorimetry. Acta Aliment. 23:157-168. [Google Scholar]
- 17.Niven, G. W., C. A. Miles, and B. M. Mackey. 1999. The effect of hydrostatic pressure on ribosome conformation in Escherichia coli: an in vivo study using differential scanning calorimetry. Microbiology 145:419-425. [DOI] [PubMed] [Google Scholar]
- 18.Noll, M., and H. Noll. 1976. Structural dynamics of bacterial ribosomes V. Magnesium-dependent dissociation of tight couples into subunits: measurement of dissociation constants and exchange rates. J. Mol. Biol. 105:111-130. [DOI] [PubMed] [Google Scholar]
- 19.Rheinberger, H., U. Geigenmuller, M. Wedde, and K. H. Neirhaus. 1988. Parameters for preparation of E. coli ribosomes and ribosomal sub-units active in tRNA binding. Methods Enzymol. 164:658-662. [DOI] [PubMed] [Google Scholar]
- 20.Rodriguez-Torres, A., M. C. Ramos-Sanchez, A. Orduna-Domingo, F. J. Martin-Gil, and J. Martin-Gil. 1993. Differential scanning calorimetry investigations on LPS and free lipids A of the bacterial cell wall. Res. Microbiol. 144:729-740. [DOI] [PubMed] [Google Scholar]
- 21.Stephens, P. J., and M. V. Jones. 1993. Reduced ribosomal thermal denaturation in Listeria monocytogenes following osmotic and heat shocks. FEMS Microbiol. Lett. 106:177-182. [DOI] [PubMed] [Google Scholar]
- 22.Teixeira, P., H. Castro, C. Mohacsi-Farkas, and R. Kirby. 1997. Identification of sites of injury in Lactobacillus bulgaricus during heat stress. J. Appl. Microbiol. 83:219-226. [DOI] [PubMed] [Google Scholar]
- 23.Tolker-Nielsen, T., and S. Molin. 1996. Role of ribosome degradation in the death of heat-stressed Salmonella typhimurium. FEMS Microbiol. Lett. 142:155-160. [DOI] [PubMed] [Google Scholar]
- 24.Tomlins, R. H., and Z. J. Ordal. 1976. Thermal injury and inactivation in vegetative bacteria, p. 153-190. In F. A. Skinner and W. B. Hugo (ed.), Inhibition and inactivation of vegetative microbes. Academic Press, Ltd., London, United Kingdom.