Abstract
Six bacteriophages active against Leuconostoc fallax strains were isolated from industrial sauerkraut fermentation brines. These phages were characterized as to host range, morphology, structural proteins, and genome fingerprint. They were exclusively lytic against the species L. fallax and had different host ranges among the strains of this species tested. Morphologically, three of the phages were assigned to the family Siphoviridae, and the three others were assigned to the family Myoviridae. Major capsid proteins detected by electrophoresis were distinct for each of the two morphotypes. Restriction fragment length polymorphism analysis and randomly amplified polymorphic DNA fingerprinting showed that all six phages were genetically distinct. These results revealed for the first time the existence of bacteriophages that are active against L. fallax and confirmed the presence and diversity of bacteriophages in a sauerkraut fermentation. Since a variety of L. fallax strains have been shown to be present in sauerkraut fermentation, bacteriophages active against L. fallax are likely to contribute to the microbial ecology of sauerkraut fermentation and could be responsible for some of the variability observed in this type of fermentation.
Industrial sauerkraut fermentation relies on indigenous bacterial populations initially present on raw cabbage (42). Several members of the lactic acid bacterium family are known to contribute to the complex sauerkraut fermentation, including Leuconostoc mesenteroides, Lactobacillus brevis, Pediococcus pentosaceus, and Lactobacillus plantarum (42). Leuconostoc species, including L. mesenteroides and L. fallax, are known to be present and to be predominant in the early heterofermentative stage of this fermentation (7, 24, 25, 36, 42).
Bacteriophage contamination is an important problem that is common in food fermentations, especially in the dairy industry. Among the bacteriophages that infect lactic acid bacteria, those specific for Streptococcus and Lactococcus species have been investigated most extensively (1). In contrast, little information is available on bacteriophages of Leuconostoc species. It was shown recently that phages active against lactic acid bacteria, including L. mesenteroides, L. plantarum, and P. pentosaceus, are present in pickle and sauerkraut fermentations (6, 57). Leuconostoc phages have also been identified as factors responsible for the failure of fermentations of several foods, including wine, coffee, and dairy products (4, 9, 18, 19, 27, 28, 39, 48, 49, 50).
Bacteriophages have the potential to control population levels and microbial diversity in natural bacterial communities (12). The interactions between bacterial and phage populations have been studied in dairy cultures (21), soil environments (17, 41), aquatic environments (8, 11, 12, 29, 54), and the phytosphere (5). To understand microbial succession and diversity in the complex sauerkraut fermentation, we must consider the roles and impact of bacteriophages.
The primary objective of this work was to investigate the presence of bacteriophages attacking L. fallax in a commercial sauerkraut fermentation. A comparative study of such phages was conducted in order to determine their morphologies, structural proteins, host ranges, growth characteristics, and genomes.
MATERIALS AND METHODS
Bacterial strains.
Cultures were stored at −80°C in MRS broth (Difco Laboratories, Detroit, Mich.) with 15% glycerol. Frozen cultures were propagated overnight prior to experiments. All bacterial strains were grown on MRS (20) plates or in MRS broth and incubated at 30°C aerobically. Bacterial strains and phages used in this study are shown in Table 1.
TABLE 1.
Bacterial strains and bacteriophages used in this study
| Organism or phage | Origin or reference |
|---|---|
| Bacterial strains | |
| Leuconostoc amelibiosum | ATCC 13146a |
| Leuconostoc citreum | ATCC 49370a |
| Leuconostoc fallax | ATCC 700006a |
| Leuconostoc lactis | ATCC 19256a |
| Leuconostoc mesenteroides subsp. cremoris | ATCC 19254a |
| Leuconostoc mesenteroides subsp. dextranicum | ATCC 19255a |
| Leuconostoc mesenteroides subsp. mesenteroides | ATCC 8293a |
| Weissella paramesenteroides | ATCC 33313a |
| Leuconostoc fallax LA 288b | 7 |
| Leuconostoc fallax LA 289b | 7 |
| Leuconostoc fallax LA 290b | 7 |
| Leuconostoc fallax LA 297b | 7 |
| Leuconostoc fallax LA 298b | 7 |
| Leuconostoc fallax LA 299b | 7 |
| Bacteriophages | |
| φR01 (host, LA 289) | This study |
| φR03 (host, LA 290) | This study |
| φR05 (host, LA 297) | This study |
| φR09 (host, LA 288) | This study |
| φR12 (host, LA 299) | This study |
| φR19 (host, LA 298) | This study |
American Type Culture Collection characteristic type strain.
Food Fermentation Laboratory, USDA Agricultural Research Service, Department of Food Science, North Carolina State University.
Phage isolation and preparation.
Sauerkraut brine samples were obtained from an industrial fermentation tank inoculated with a single-strain L. mesenteroides starter culture. All phages were isolated after either 2 or 3 days of fermentation. The sauerkraut brines were centrifuged (5,000 × g, 10 min), and the resulting supernatants were filter sterilized (0.45-μm-pore-size membrane) and checked for phages by the overlay agar method (2) by using L. fallax strains as potential hosts. The L. fallax strains used in this study were isolated previously from sauerkraut fermentation (7). Bacteriophages were purified by the method described by Chow et al. (16). Bacterial cells (100 μl from an overnight culture) were mixed with a solution containing 100 μl of sauerkraut brine, 100 μl of 300 mM CaCl2 (52), and 3 ml of soft agar (0.7%, wt/vol) stored at 50°C before it was overlaid onto MRS agar plates. Phage lysates were prepared from plaques formed on the overlay plates. Single plaques were picked, mixed with 500 μl of bacterial cells in the early exponential growth phase (optical density at 600 nm, 0.2 to 0.3), 30 μl of CaCl2 (300 mM), and 500 μl of fresh MRS broth, and incubated at 30°C overnight. Double-layer plates containing each phage were prepared to obtain confluent lysis of the hosts. The plates were then flooded and mixed with 3 ml of MRS broth, and the liquid part was recovered. Liquid stocks of bacteriophages were prepared by infecting early-exponential-phase L. fallax cells. Each infected culture was incubated at 30°C for 3 to 5 h, which resulted in complete lysis of the culture. Cell debris was removed by centrifugation (5,000 × g, 10 min), and the phage lysates were sterilized by membrane filtration (pore size, 0.45 μm). The bacteriophages were precipitated at 4°C overnight in the presence of polyethylene glycol 8000 (10%, wt/vol) and NaCl (2.9%, wt/vol). Rapid bacteriophage sedimentation in the presence of polyethylene glycol was used for phage purification by phase partitioning (56). Phage pellets were collected by centrifugation (8,000 × g, 20 min). The bacteriophage pellets were resuspended in deionized water (1 to 2 ml), which allowed 50- to 100-fold concentration of the original lysate. DNase I and RNase A were each added to a final concentration of 1 μg/ml, and the preparations were incubated for 30 min at 35°C. Bacteriophage particles were purified further by ultracentrifugation in a three-step CsCl gradient (1.4, 1.5, and 1.7 g/cm3) (47). After centrifugation at 600,000 × g for 7 h with an S100 AT6 rotor in an RC M150 GX ultracentrifuge (Sorvall, Newtown, Conn.), the visible band of bacteriophage particles was extracted with a needle (47) and dialyzed with a 8,000-Da-molecular-size-cutoff membrane (Baxter Diagnostics Inc., McGaw Park, Ill.) against 500 ml of deionized water for 6 h, with five changes of water. Purified bacteriophage suspensions were stored at 4°C.
Electron microscopy.
Aliquots of a bacteriophage sample obtained by ultracentrifugation were subjected to electron microscopy for morphological analysis. Purified phage particles were negatively stained with 2% (wt/vol) uranyl acetate, deposited on carbon-coated grids (Ladd Research Industries Inc., Burlington, Vt.), and examined with a JEOL 100 CX II electron microscope (JEOL, Peabody, Mass.) at an accelerating voltage of 80 kV.
Bacteriophage host range.
Phage host range was established by using the spot test method (15). The plate inoculum consisted of 3 ml of soft agar mixed with 100 μl of an overnight culture and 100 μl of CaCl2 (300 mM). This mixture was briefly vortexed and spread inoculated onto the surface of an MRS agar plate (2). Single drops (serial dilutions of 5 μl) of each phage lysate were spotted onto the inoculated MRS agar plates, and the plates were incubated overnight at 30°C. Bacterial sensitivity to a bacteriophage was established by bacterial lysis at the spot where the phage lysate drop was deposited. Phage typing of each strain was done three independent times. Positive spot tests were confirmed by titration assays by using diluted phage preparations.
SDS-PAGE analysis of phage proteins.
Bacteriophage structural proteins were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Aliquots of a bacteriophage sample obtained by ultracentrifugation were subjected to electrophoresis with a 10 to 20% Tris-Tricine continuous-gradient precast Novex gel (Novex, San Diego, Calif.). A 20-μl aliquot of phage from a CsCl-purified high-titer phage lysate was mixed with 7 μl of 4× sample buffer and 3 μl of 10× NuPAGE sample reducing buffer (Novex). The mixture was heat treated at 100°C for 3 min. The resulting sample was loaded onto the polyacrylamide gel, and electrophoresis was carried out at 200 V for 40 min. Proteins were visualized on gels and were stained with Coomassie brilliant blue.
Bacteriophage DNA isolation.
A modification of the procedure of Durmaz and Klaenhammer (22) was used for bacteriophage DNA isolation and purification. One hundred milliliters of phage lysate was incubated for 1 h at 37°C after addition of DNase I and RNase A (2 mg/ml). Polyethylene glycol 8000 and NaCl were added to final concentrations of 10% (wt/vol) and 3% (wt/vol), respectively. After gentle mixing, the samples were incubated overnight at 4°C. The phages were pelleted by centrifugation at 8,000 × g for 20 min, and the supernatants were discarded. The phage pellets were resuspended in 1 ml of 50 mM Tris (pH 8.0). Six 500-μl phage suspensions were extracted two or three times with 500 μl of phenol and then twice with phenol-chloroform-isoamyl alcohol (25:24:1). The nucleic acids were precipitated with 50 μl of 3 M sodium acetate and 1 ml of 70% (vol/vol) ethanol and resuspended in 50 μl of Tris-EDTA buffer.
Genome fingerprinting by restriction fragment length polymorphism (RFLP) analysis.
Purified bacteriophage DNA samples were subjected to restriction enzyme digestion with AluI, BamHI, EcoRI, HindIII, MboI, RsaI, and Sau3AI as suggested by the manufacturer (Promega Corp., Madison, Wis.). The restriction digests were separated on a 0.8% agarose gel and stained with ethidium bromide.
Genome fingerprinting by RAPD analysis.
The method used for randomly amplified polymorphic DNA (RAPD) analysis (54, 55) was derived from the method of Johansson et al. (33). Nine-mers with a G+C content of 80% were designed. The following primers were used in this study: primer A (5′ACGCGCCCT3′) and primer B (5′CCGAGTCCA3′) (Genosys Biotechnologies Inc., The Woodlands, Tex.). Each 100-μl reaction mixture for RAPD PCR analysis of bacteriophage DNA contained 66 μl of water, 10 μl of thermophilic DNA polymerase 10× PCR buffer, 10 μl of 25 mM MgCl2, 1 μl of a deoxynucleoside triphosphate mixture, 100 pmol of primer (Genosys Biotechnologies Inc.), 0.2 μg of DNA template, and 1 μl of Taq DNA polymerase (Promega). An initial denaturation step (95°C for 3 min) was performed with the reaction mixture prior to addition of Taq polymerase. DNA amplification was performed with a Gradient 96 Robocycler (Stratagene, La Jolla, Calif.) thermal cycler programmed as follows: 10 min at 94°C; four cycles of 45 s at 94°C, 2 min at 30°C, and 45 s at 72°C; 36 cycles of 15 s at 94°C, 30 s at 36°C, and 45 s at 72°C; and 10 min at 72°C. The DNA banding patterns were examined by 5% acrylamide gel electrophoresis, and a 1-kb ladder (Gibco-BRL, Grand Island, N.Y.) was used to provide size standards.
Disclaimer. Mention of a trademark or proprietary product does not constitute a guarantee or warranty of the product by the U.S. Department of Agriculture or North Carolina Agricultural Research Service, nor does it imply approval to the exclusion of other products that may be suitable.
RESULTS
Morphological diversity.
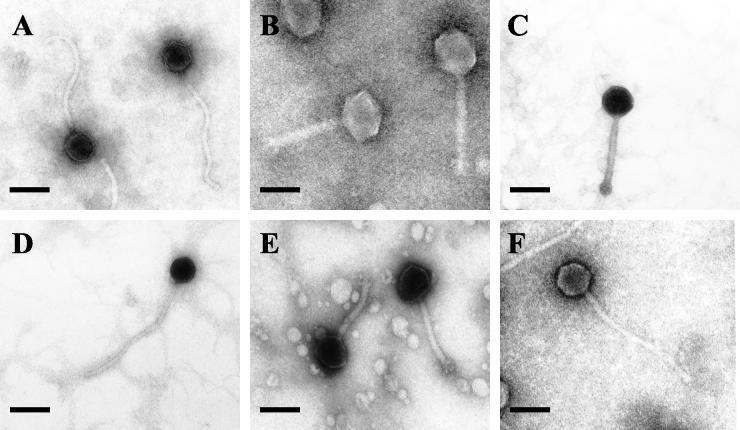
Six bacteriophages were examined by electron microscopy (Fig. 1). All the phages examined had tails and thus belonged to the order Caudovirales. The phages were assigned to two virus families on the basis of their morphological features. Phages φR01, φR09, and φR19 had isometric heads and long flexible noncontractile tails, consistent with the family Siphoviridae or Bradley's group B1 (1, 10). The other bacteriophages, φR03, φR05, and φR12, had isometric heads, visible collars, and shorter contractile tails with terminal base plates, which are characteristics of the family Myoviridae or Bradley's group A1 (1, 10). The morphological characteristics of these bacteriophages are summarized in Table 2.
FIG. 1.
Electron micrographs of L. fallax bacteriophages. (A) φR01; (B) φR03; (C) φR05; (D) φR09; (E) φR12; (F) φR19. Bars = 100 nm.
TABLE 2.
Morphological features of L. fallax phagesa
| Phage | Family | Head diam (nm) | Tail length (nm) | Tail diam (nm) | Additional features
|
|||
|---|---|---|---|---|---|---|---|---|
| Collar | Base plate | Tail pins | Terminal bulb | |||||
| φR01 | Siphoviridae | 68 (3) | 345 (12) | 13 (2) | − | − | − | + |
| φR03 | Myoviridae | 110 (6) | 203 (14) | 25 (2) | + | + | + | − |
| φR05 | Myoviridae | 78 (7) | 188 (12) | 16 (2) | + | + | + | − |
| φR09 | Siphoviridae | 69 (3) | 370 (6) | 13 (2) | − | − | − | + |
| φR12 | Myoviridae | 81 (2) | 174 (9) | 22 (2) | + | + | + | − |
| φR19 | Siphoviridae | 78 (5) | 356 (16) | 13 (2) | − | − | − | + |
The values are the means of six independent measurements for different phage particles. For phage φR05, only three phage particles were measured. The values in parentheses are standard deviations.
Host ranges.
All of the phages isolated from the sauerkraut brines were selectively lytic against L. fallax strains. The phages were not observed to lyse other Leuconostoc species included in this study. Within L. fallax, each of the six phages exhibited a distinctive host range, as shown in Table 3.
TABLE 3.
Bacteriophage host ranges
| Strain | Lysis by bacteriophagea:
|
|||||
|---|---|---|---|---|---|---|
| φR01 | φR03 | φR05 | φR09 | φR12 | φR19 | |
| L. amelibiosum ATCC 13146b | − | − | − | − | − | − |
| L. citreum ATCC 49370b | − | − | − | − | − | − |
| L. fallax ATCC 700006b | − | T | − | + | − | T |
| L. lactis ATCC 19256b | − | − | − | − | − | − |
| L. mesenteroides subsp. cremoris ATCC 19254b | − | − | − | − | − | − |
| L. mesenteroides subsp. dextranicum ATCC 19255b | − | − | − | − | − | − |
| L. mesenteroides subsp. mesenteroides ATCC 8293b | − | − | − | − | − | − |
| W. paramesenteroides ATCC 33313b | − | − | − | − | − | − |
| L. fallax LA 288c | + | − | + | + | − | + |
| L. fallax LA 289c | − | + | − | − | − | − |
| L. fallax LA 290c | + | − | + | + | − | + |
| L. fallax LA 297c | + | + | + | + | + | − |
| L. fallax LA 298c | + | + | + | + | − | + |
| L. fallax LA 299c | − | + | + | − | + | − |
+, plaque formation; −, no plaque formation; T, turbid plaque formation.
American Type Culture Collection type strain.
Food Fermentation Laboratory, USDA Agricultural Research Service, North Carolina State University.
Phage protein analysis.
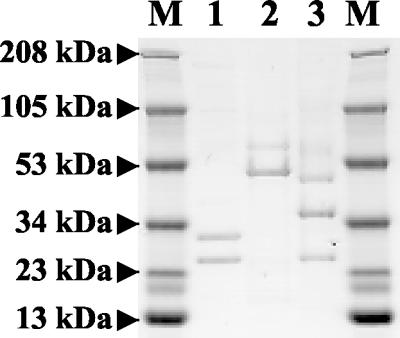
To further characterize the two phage families, which are distinguishable by morphology, the structural protein composition was analyzed by SDS-PAGE (Fig. 2). Two or three main structural proteins, as well as several minor proteins, were detected (data not shown). All three phages assigned to the Myoviridae family showed the same protein patterns as φR03, with two major structural proteins at 51 and 63 kDa. Two distinct patterns were observed for the Siphoviridae phages. Major 24- and 30-kDa structural proteins were detected for φR01, whereas φR09 and φR19 had three major proteins (24, 36, and 50 kDa). The profiles of the Myoviridae phages were different from those of the Siphoviridae bacteriophages. It has been observed previously that the major structural proteins of phages classified in the same DNA homology group are conserved (45). The protein patterns showed that members of the same morphotype have common structural proteins and that protein patterns can be used for phage morphotype characterization and differentiation.
FIG. 2.
SDS-PAGE patterns of phage structural proteins. Lanes M, molecular weight markers; lane 1, Siphoviridae phage φR01; lane 2, Myoviridae phage φR03; lane 3, Siphoviridae phage φR09.
Genome fingerprinting by RFLP analysis.
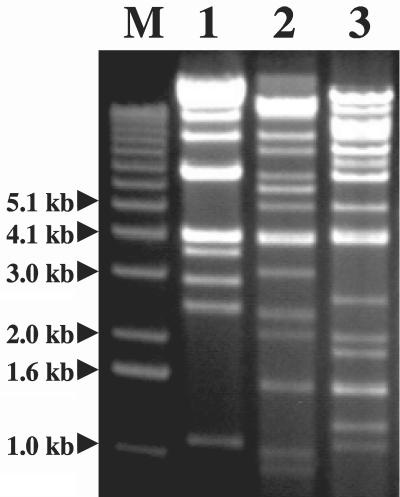
All the Myoviridae phage DNA samples were sensitive to BamHI, EcoRI, and HindIII and exhibited different but closely related restriction endonuclease patterns, and common bands appeared for the three phages. The EcoRI digestion patterns of Myoviridae phages are shown in Fig. 3. In contrast, the Siphoviridae phage DNA samples were highly refractory to digestion by several endonucleases, including AluI, BamHI, EcoRI, HindIII, RsaI, and Sau3AI (data not shown). Interestingly, only the Siphoviridae phage φR09 DNA sample was digested by MboI. The phage genome sizes estimated from RFLP patterns were approximately 59, 41, and 46 kb for φR03, φR05, and φR12, respectively. The DNA electrophoretic patterns of the phages resistant to endonucleases suggested that the other genomes are between approximately 40 and 55 kb long. The difference in sensitivity to restriction endonucleases provides further evidence that the L. fallax phages isolated from sauerkraut fermentation belong to two different phage families.
FIG. 3.
RFLP analysis of phage DNA: EcoRI digestion patterns of Myoviridae bacteriophages. Lane M, 1-kb DNA ladder; lane 1, φR03; lane 2, φR05; lane 3, φR12.
Genome fingerprinting by RAPD analysis.
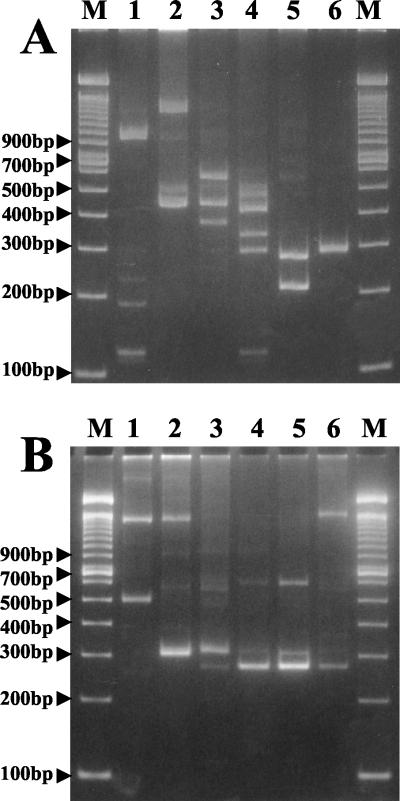
RAPD fingerprinting of the DNA of the six L. fallax bacteriophages is shown in Fig. 4. The primers used for RAPD characterization of bacteriophage DNA have been used previously for characterization of bacteria (7, 13, 33, 44). When primer B was used (Fig. 4A), all of the phages exhibited very different patterns, showing that all six phages are genetically unique and could be distinguished by RAPD fingerprinting. In contrast, when primer A was used (Fig. 4B), some similar bands were observed for phages belonging to the same family. Identical RAPD patterns were obtained in two different series of experiments (data not shown); however, the relative intensities of some of the bands were not consistent.
FIG. 4.
(A) RAPD fingerprinting of bacteriophage DNA with primer B. (B) RAPD fingerprinting of bacteriophage DNA with primer A. Lane 1, φR01; lane 2, φR03; lane 3, φR05; lane 4, φR09; lane 5, φR12; lane 6, φR19; lanes M, 100-bp DNA ladder.
DISCUSSION
This is the first report of bacteriophages active against L. fallax. In this study, we confirmed the presence of bacteriophages in a sauerkraut fermentation and demonstrated that individual L. fallax strains can be susceptible to more than one phage and members of more than one phage family. The six phages isolated from sauerkraut fermentation could be classified in two different families on the basis of morphology, structural proteins, and DNA fingerprinting. Bacteriophages are ubiquitous and likely to be prevalent in environments with high densities of metabolically active bacteria (40). Therefore, it was not surprising that L. fallax phages were isolated within 3 days after the initiation of fermentation, while Leuconostoc strains are thought to predominate (7, 24, 25, 42).
RAPD analysis of phage DNA provided a simple and reproducible method for differentiation of bacteriophages that were specific for L. fallax. Closely related phages in each family could be distinguished by using two primer sets. Use of RAPD for investigation of phage DNA relatedness also showed that bacteriophages with common morphotypes have common DNA bands. This method has been used extensively for microbial characterization, differentiation of closely related bacterial species, and identification of strain polymorphisms (31, 33, 58). Traditional polymorphism assays that are based on the PCR require target DNA sequence information for the design of amplification primers. In our case, no phage DNA sequence was available; therefore, amplification of phage DNA with arbitrary nucleotide sequence primers offers the advantage of detection of DNA polymorphisms in the absence of specific primers. RAPD analysis has several advantages over RFLP analysis, including no need for DNA purification, no need for prior knowledge of the molecular biology of the organism investigated (53), sensitivity, discriminatory power, and typing of DNA regions which are not accessible to RFLP analysis due to the presence of repetitive sequences (55). RAPD markers are well suited for DNA fingerprinting and provide an efficient assay for phage polymorphisms. In this study, RAPD phage fingerprinting results suggested that the six L. fallax phages were related but distinct. Phage genome fingerprinting with RAPD has been reported previously (43), and the results presented in this study show that RAPD analysis can be used as a rapid method for identification, typing, and discrimination of bacteriophages. Since several of the L. fallax phages produced similar bands, it should be possible to sequence these bands, determine their homologies, and design specific primers for direct detection of phage strains. DNA primers derived from RAPD fragments were recently used to specifically detect some members of the lactic acid bacteria (23, 38, 51). Also, a multiplex PCR protocol was developed for specific detection of three Lactococcus lactis phage species, showing that conserved regions of phage genomes can be used to design species-specific primers (35).
One interesting observation made during this study was the insensitivity of the Siphoviridae phages to restriction endonucleases. Bacteriophage resistance to restriction enzymes is common and has been reported previously (32, 33, 45, 46). Sixteen Campylobacter phages have been shown to be refractory to digestion by a number of commonly used restriction enzymes (46). Also, some L. lactis bacteriophages have been found to be highly refractory to digestion by several restriction enzymes (45). Several explanations have been proposed to explain phage DNA resistance to restriction enzymes, usually referred to as antirestriction mechanisms (37). Foremost among these explanations is the hypothesis that phage genomes adapt under the selection pressure of widespread restriction-modification systems (3, 26) and lose restriction sites naturally during evolution (37). Another explanation for the phage DNA insensitivity is integration of unusual bases in the viral DNA, such as hydroxymethyl uracil or hydroxymethyl cytosine, that make the DNA somewhat refractory to endonuclease cleavage (32). Alternatively, phage genomes may encode methyltransferases that modify specific nucleotides within the recognition site of one or more of the restriction endonucleases (30, 34, 45, 46). The varied distribution of restriction endonuclease sites in different phage species further supports the hypothesis that there is an adaptive mechanism involving loss of restriction sites (37) and the transfer of methylase genes in phage genomes (30). The frequency of restriction endonuclease sites and phage sensitivity to restriction enzymes have been shown to decrease with time, suggesting that there is a phage adaptive mechanism (37). In the present study of L. fallax phages, Siphoviridae phages exhibited little sensitivity to restriction enzymes, whereas Myoviridae phages were sensitive to all of the restriction enzymes tested, which might suggest that there was long-term evolution of the former phages. Whatever the mechanism, it is clear that the two families of L. fallax phages are different, and the data suggest that Siphoviridae phages exhibit clear distinctions in susceptibility to restriction enzymes and probably restriction-modification systems.
Since diverse L. fallax strains have been shown to be present in sauerkraut fermentations (7), the presence of phages might interfere with natural fermentation and affect microbial ecology. Bacteriophage diversity is observed naturally in the environment (14), and it seems logical that phage variety reflects microbial variety and distribution in sauerkraut. In addition, natural L. fallax strain variety may prevent severe fermentation disturbance by bacteriophages. This study emphasizes the need for further investigations of phage ecology in sauerkraut fermentation in order to gain a more complete understanding of microbial interactions and succession. The heterofermentative stage of sauerkraut fermentation is very critical for establishing the proper conditions for microbial succession and ultimately the final quality of the product. L. fallax strains are clearly present during this stage (7) and, based on this study, are susceptible to lytic phage attack. None of the L. fallax strains isolated were sensitive to all of the bacteriophages, suggesting that sensitive and resistant populations may fluctuate during this stage of fermentation. The L. fallax-specific phages were diverse and thus should be expected to affect the course of events in this fermentation ecosystem. Efforts to control the initial stage of sauerkraut fermentation must consider the predominance of L. fallax and the involvement of lytic phages attacking this species. In this regard, use of L. fallax and L. mesenteroides starter cultures will likely face significant obstacles and require selection and development of phage-resistant varieties. The quantitative role of bacteriophages in the sauerkraut fermentation ecosystem and the influence of bacteriophages on population dynamics, microbial succession, microbial diversity, and genetic transfer remain to be investigated.
Acknowledgments
This study was supported by the U.S. Department of Agriculture, by Pickle Packers International, and by NRICGP under project 97-35503-4368.
We thank Janet Hayes for providing the bacterial strains, Evelyn Durmaz for providing phage DNA isolation methods, Valerie Knowlton for assistance with electron microscopy, and Eric Altermann and Michael Callanan for reviewing the manuscript.
Footnotes
Paper no. FSR02-5 of the Journal Series of the Department of Food Science, North Carolina State University, Raleigh.
REFERENCES
- 1.Ackermann, H. W. 2001. Frequency of morphological phage descriptions in 2000. Arch. Virol. 146:843-857. [DOI] [PubMed] [Google Scholar]
- 2.Adams, M. H. 1959. Bacteriophages. Interscience Publishers, New York, N.Y.
- 3.Allison, G. E., and T. R. Klaenhammer. 1998. Phage resistance mechanisms in lactic acid bacteria. Int. Dairy J. 8:207-226. [Google Scholar]
- 4.Arendt, E. K., and W. P. Hammes. 1992. Isolation and characterization of Leuconostoc oenos phages from German wines. Appl. Microbiol. Biotechnol. 37:643-646. [Google Scholar]
- 5.Ashelford, K. E., M. J. Day, M. J. Bailey, A. K. Lilley, and J. C. Fry. 1999. In situ population dynamics of bacterial viruses in a terrestrial environment. Appl. Environ. Microbiol. 65:169-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrangou, R. 2000. Leuconostoc fallax strains and their bacteriophages from industrial sauerkraut fermentations. M.S. thesis. North Carolina State University, Raleigh.
- 7.Barrangou, R., S. S. Yoon, F. Breidt, T. R. Klaenhammer, and H. P. Fleming. 2002. Identification and characterization of Leuconostoc fallax strains isolated from industrial sauerkraut fermentation. Appl. Environ. Microbiol. 68:2877-2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bergh, O., Y. Borsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 9.Boizet, B., M. Mata, O. Mignot, P. Ritzenthaler, and T. Sozzi. 1992. Taxonomic characterization of Leuconostoc mesenteroides and Leuconostoc oenos bacteriophage. FEMS Microbiol. Lett. 90:211-216. [Google Scholar]
- 10.Bradley, D. E. 1967. Ultrastructure of bacteriophages and bacteriocins. Bacteriol. Rev. 31:230-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bratbak, G., M. Heldal, S. Norland, and T. F. Thingstad. 1990. Viruses as partners in spring bloom microbial trophodynamics. Appl. Environ. Microbiol. 56:1400-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bratbak, G., F. Thingstad, and M., Heldal. 1994. Viruses and the microbial loop. Microb. Ecol. 28:209-221. [DOI] [PubMed] [Google Scholar]
- 13.Brousseau, R., A. Saint-Onge, G. Prefontaine, L. Masson, and J. Cabana. 1993. Arbitrary primer polymerase chain reaction, a powerful method to identify Bacillus thuringiensis serovars and strains. Appl. Environ. Microbiol. 59:114-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bruttin, A., F. Desiere, N. d'Amico, J. P. Guerin, J. Sidoti, B. Huni, S. Lucchini, and H. Brussow. 1997. Molecular ecology of Streptococcus thermophilus bacteriophage infections in a cheese factory. Appl. Environ. Microbiol. 63:3144-3150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chopin, M. C., A. Chopin, and C. Roux. 1976. Definition of bacteriophage groups according to their lytic action on mesophilic lactic streptococci. Appl. Environ. Microbiol. 32:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chow, J. J., C. A. Batt, and A. J. Sinskey. 1988. Characterization of Lactobacillus bulgaricus bacteriophage ch2. Appl. Environ. Microbiol. 54:1138-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cresswell, N., P. R. Herron, V. A. Saunders, and E. M. H. Wellington. 1992. The fate of introduced streptomycetes, plasmid and phage populations in a dynamic soil system. J. Gen. Microbiol. 138:659-666. [Google Scholar]
- 18.Davey, G. P., L. J. H. Ward, and J. C. S. Brown. 1995. Characterization of four Leuconostoc bacteriophages isolated from dairy fermentations. FEMS Microbiol. Lett. 128:21-26. [Google Scholar]
- 19.Davis, C., N. F. A. Silveira, and G. H. Fleet. 1985. Occurrence and properties of bacteriophages of Leuconostoc oenos in Australian wines. Appl. Environ. Microbiol. 50:872-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.deMan, J. C., M. Rogosa, and M. E. Sharpe. 1960. A medium for the cultivation of lactobacilli. J. Appl. Bacteriol. 23:130-135. [Google Scholar]
- 21.Djordjevic, G. M., and T. R. Klaenhammer. 1998. Genes and gene expression in Lactococcus bacteriophages. Int. Dairy J. 7:489-508. [Google Scholar]
- 22.Durmaz, E., and T. R. Klaenhammer. 2000. Genetic analysis of chromosomal regions of Lactococcus lactis acquired by recombinant lytic phages. Appl. Environ. Microbiol. 66:895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Erlandson, K., and C. A. Batt. 1997. Strain-specific differenciation of lactococci in mixed starter culture populations using randomly amplified polymorphic DNA-derived probes. Appl. Environ. Microbiol. 63:2702-2707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fleming, H. P., R. F. McFeeters, and E. G. Humphries. 1987. A fermentor for study of sauerkraut fermentation. Biotechnol. Bioeng. 31:189-197. [DOI] [PubMed] [Google Scholar]
- 25.Fleming, H. P., K. H. Kyung, and F. Breidt. 1995. Vegetable fermentations, p. 631-661. In G. Reed and T. W. Nagodawithana (ed.), Bio/Technology, vol. 9, 2nd ed. VCH Publishing Co., Weinheim, Germany.
- 26.Forde, A., and G. F. Fitzgerald. 1999. Bacteriophage defense systems in lactic acid bacteria. Antonie van Leeuwenhoek. 76:89-113. [PubMed] [Google Scholar]
- 27.Henick-Kling, T., T. H. Lee, and D. J. D. Nicholas. 1986. Inhibition of bacterial growth and malolactic fermentation in wine by bacteriophage. J. Appl. Bacteriol. 61:287-293. [Google Scholar]
- 28.Henick-Kling, T., T. H. Lee, and D. J. D. Nicholas. 1986. Characterization of the lytic activity of bacteriophages of Leuconostoc oenos isolated from wine. J. Appl. Bacteriol. 61:525-534. [Google Scholar]
- 29.Hennes, K. P., and M. Simon. 1995. Significance of bacteriophages for controlling bacterioplankton growth in a mesotrophic lake. Appl. Environ. Microbiol. 61:333-340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill, C., L. A. Miller, and T. R. Klaenhammer. 1991. In vivo genetic exchange of a functional domain from a type IIA methylase between lactococcal plasmid pTR2030 and a virulent bacteriophage. J. Bacteriol. 173:4363-4370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holt, S. M., and G. L. Cote. 1998. Differentiation of dextran-producing Leuconostoc strains by a modified randomly amplified polymorphic DNA protocol. Appl. Environ. Microbiol. 64:3096-3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jensen, E. C., H. S. Schrader, B. Rieland, T. L. Thompson, K. W. Lee, K. W. Nickerson, and T. A. Kokjohn. 1998. Prevalence of broad-host-range lytic bacteriophages of Sphaerolitus natans, Escherichia coli, and Pseudomonas aeruginosa. Appl. Environ. Microbiol. 64:575-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johansson, M. L., M. Quednau, G. Molin, and S. Ahrne. 1995. Randomly amplified polymorphic DNA (RAPD) for rapid typing of Lactobacillus plantarum strains. Lett. Appl. Microbiol. 21:155-159. [DOI] [PubMed] [Google Scholar]
- 34.Kruger, D. H., and T. A. Bickle. 1983. Bacteriophage survival: multiple mechanisms for avoiding the deoxyribonucleic acid restriction systems of their hosts. Microbiol. Rev. 47:345-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Labrie, S., and S. Moineau. 2000. Multiplex PCR for detection and identification of lactococcal bacteriophages. Appl. Environ. Microbiol. 66:987-994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Martinez-Murcia, A. J., and M. D. Collins. 1991. A phylogenetic analysis of an atypical leuconostoc: description of Leuconostoc fallax sp. nov. FEMS Microbiol. Lett. 82:55-60. [DOI] [PubMed] [Google Scholar]
- 37.Moineau, S., S. Pandian, and T. R. Klaenhammer. 1993. Restriction/modification systems and restriction endonucleases are more effective on lactococcal bacteriophages that have emerged recently in the dairy industry. Appl. Environ. Microbiol. 59:197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moschetti, G., G. Blaiotta, F. Villani, and S. Coppola. 2000. Specific detection of Leuconostoc mesenteroides subsp. mesenteroides with DNA primers identified by randomly amplified polymorphic DNA analysis. Appl. Environ. Microbiol. 66:422-424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nel, L., B. D. Wingfield, L. J. Van der Meer, and H. J. J. Van Vuuren. 1987. Isolation and characterization of Leuconostoc oenos bacteriophages from wine and sugarcane. FEMS Microbiol. Lett 44:63-67. [Google Scholar]
- 40.Nowak, M. 1991. The evolution of viruses. Competition between horizontal and vertical transmission of mobile genes. J. Theor. Biol. 150:339-347. [DOI] [PubMed] [Google Scholar]
- 41.Pantastico-Caldas, M., K. E. Duncan, C. A. Istock, and J. A. Bell. 1992. Population dynamics of bacteriophage and Bacillus subtilis in soil. Ecology 73:1888-1902. [Google Scholar]
- 42.Pederson, C. S., and M. N. Albury. 1969. The sauerkraut fermentation. New York State Agricultural Experiment Station Technical Bulletin no. 824. New York State Agricultural Experiment Station, Geneva.
- 43.Perez, T., J. Albornoz, and A. Dominguez. 1998. An evaluation of RAPD fragment reproducibility and nature. Mol. Ecol. 7:1347-1357. [DOI] [PubMed] [Google Scholar]
- 44.Plengvidhya, V. 1999. Evaluation of molecular methods to follow the progress of starter cultures in sauerkraut fermentations. M.S. thesis. North Carolina State University, Raleigh.
- 45.Prevots, F., M. Mata, and P. Rizenthaler. 1990. Taxonomic differentiation of 101 lactococcal bacteriophages and characterization of bacteriophages with unusually large genomes. Appl. Environ. Microbiol. 56:2180-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sails, A. D., D. R. A. Wareing, F. J. Bolton, A. J. Fox, and A. Curry. 1998. Characterisation of 16 Campylobacter jejuni and C. coli typing bacteriophages. J. Med. Microbiol. 47:123-128. [DOI] [PubMed] [Google Scholar]
- 47.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 48.Santos, R., G. Vieira, M. A. Santos, and H. Paveia. 1996. Characterization of temperate bacteriophages of Leuconostoc oenos and evidence for two prophage attachment sites in the genome of starter culture strain PSU-1. J. Appl. Bacteriol. 81:383-392. [Google Scholar]
- 49.Saxelin, M. L., E. L. Nurmiaho-Lassila, V. T. Merilainen, and R. I. Forsen. 1986. Ultrastructure and host specificity of bacteriophages of Streptococcus cremoris, Streptococcus lactis subsp. diacetylactis, and Leuconostoc cremoris from Finnish fermented milk “Viili.” Appl. Environ. Microbiol. 52:771-777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sozzi, T., J. M. Poulin, R. Maret, and P. Pousaz. 1978. Isolation of a bacteriophage of Leuconostoc mesenteroides from dairy products. J. Appl. Microbiol. 44:159-161. [Google Scholar]
- 51.Tilsala-Timisjarvi, A., and T. Alatossava. 1998. Strain-specific identification of probiotic Lactobacillus rhamnosus with randomly amplified polymorphic DNA-derived PCR primers. Appl. Environ. Microbiol. 64:4816-4819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Watanabe, K., and S. Takesue. 1972. The requirement for calcium in infection with Lactobacillus phage. J. Gen. Virol. 17:19-30. [DOI] [PubMed] [Google Scholar]
- 53.Welsh, J., and M. McClelland. 1990. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 18:7213-7218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wichels, A., S. F. Biel, H. R. Gelderblom, T. Brinkhoff, G. Muyzer, and C. Schutt. 1998. Bacteriophage diversity in the North Sea. Appl. Environ. Microbiol. 64:4128-4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Williams, J. G. K., A. R. Kubelik, K. J. Livak, J. A. Rafalski, and S. V. Tingey. 1990. DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res. 18:6531-6535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamamoto, K. R., B. M. Alberts, R. Benzinger, L. Lawhorne, and G. Treiber. 1970. Rapid bacteriophage sedimentation in the presence of polyethylene glycol and its application to large-scale virus purification. Virology 40:734-744. [DOI] [PubMed] [Google Scholar]
- 57.Yoon, S. S., R. Barrangou, F. Breidt, T. R. Klaenhammer, and H. P. Fleming. 2002. Characterization of bacteriophages present in fermenting sauerkraut. Appl. Environ. Microbiol. 68:973-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zavaleta, A. I., A. J. Martinez-Murcia, and F. Rodriguez-Valera. 1997. Intraspecific genetic diversity of Oenococcus oeni as derived from DNA fingerprinting and sequence analyses. Appl. Environ. Microbiol. 63:1261-1267. [DOI] [PMC free article] [PubMed] [Google Scholar]