Abstract
Topoisomerases are enzymes of quintessence to the upkeep of superhelical DNA, and are vital for replication, transcription and recombination. An atomic-resolution model for human topoisomerase I in covalent complex with DNA is simulated using molecular dynamics with external potentials that mimic torque and bias the DNA duplex downstream of a single-strand cut to rotate around the intact strand, according to the prevailing enzymatic mechanism. The simulations reveal the first dynamical picture of how topoisomerase accommodates large-scale motion of DNA as it changes its supercoiling state, and indicate that relaxation of positive and negative supercoils are fundamentally different. To relax positive supercoils, two separate domains (the ‘lips’) of the protein open up by about 10–14 Å, whereas to relax negative supercoils, a continuous loop connecting the upper and lower parts (and which was a hinge for opening the lips) stretches about 12 Å while the lips remain unseparated. Normal mode analysis is additionally used to characterize the functional flexibility of the protein. Remarkably, the same combination of low-frequency eigenvectors exhibit the dominant contribution for both rotation mechanisms through a see-saw motion. The simulated mechanisms suggest mutations to control the relaxation of either type of supercoiling selectively and advance a hypothesis for the debated role of the N-terminal domain in supercoil relaxation.
INTRODUCTION
Topoisomerases are enzymes with crucial role in maintaining the proper topology and physical integrity of DNA superhelical structures. They remove knots and catenanes generated by unwinding at the replication fork, relieve torsional stress caused by supercoiling during DNA transcription or chromosome disentanglement, and are involved in recombination and repair (1–3). A precise understanding of their mechanism is important mainly for two reasons.
Firstly, because topoisomerase inhibition cancels cell proliferation, human topo I is a major anti-cancer target (4,5). A novel class of topo I inhibitors (the camptothecin (CPT) analogues topotecan and CPT-11) are some of the most potent anti-cancer agents to date (6).
Secondly, the chemical reaction performed by topoisomerases induces movement of DNA segments several orders of magnitude larger than the size of the protein. For this reason, topoisomerases are prime case studies for understanding the triggering of large-scale mechanical motions induced by biomolecular motors, and in particular the effect that external forces or torques have in tuning such machines at the single-molecule level (7,8).
Computer simulations of DNA–protein complexes under external forces (9), as virtual single-molecule experiments, are in a position suitable to suggest and explain actual single-molecule measurements on supercoiled DNA (10–15). This is particularly important since the two techniques (simulation and experiment) are complementary both in terms of spatial resolution and time scales.
Four distinct subfamilies of topoisomerases have been identified, topo IA (16), IB (17), IIA (18), IIB (19), and subsequently classified according to their structure and functionality [recently reviewed in (3)]. Human topoisomerase I is a IB enzyme, an eukaryotic type discovered by Champoux and Dulbecco in the early 1970s (17). Topoisomerases (topos) of type I eliminate the supercoiled DNA stress by nicking one strand of the helix and passing the other strand through the nick. During strand passage, the enzyme attaches covalently to a DNA end at the nick by forming a phosphodiester bond with a tyrosine at the active site. Two models have been proposed for strand passage in type I topos: (i) the enzyme-bridging model (20) (in which the enzyme stabilizes a gate through the nick), and (ii) the strand-rotation model (21) (in which one end of the nick is driven to rotate around the intact strand by the supercoiling free energy). For the type IA subfamily, evidence in favor of the enzyme-bridging model, suggested by structural data (22,23), was confirmed by recent single-molecule manipulations (11,12). For the type IB subfamily, structural evidence points toward the strand-rotation model (see below). While an exciting recent single-molecule work (appeared during the submission phase of this manuscript) on vaccinia topo I (a smaller topo IB enzyme) found that its activity is torque-dependent (24), no molecular dynamical details of the actual mechanism involving the protein are currently available.
Moreover, while topo IB enzymes relax, unlike topo IA, both positive and negative supercoils (i.e. overwound or underwound), no attempt to discriminate between the two types of DNA substrates has been made for human topoisomerase. Here, we present the first dynamical model that includes the protein response to the large-scale motion of the two supercoiled DNA states by atomistic simulations.
Available data on human topoisomerase I
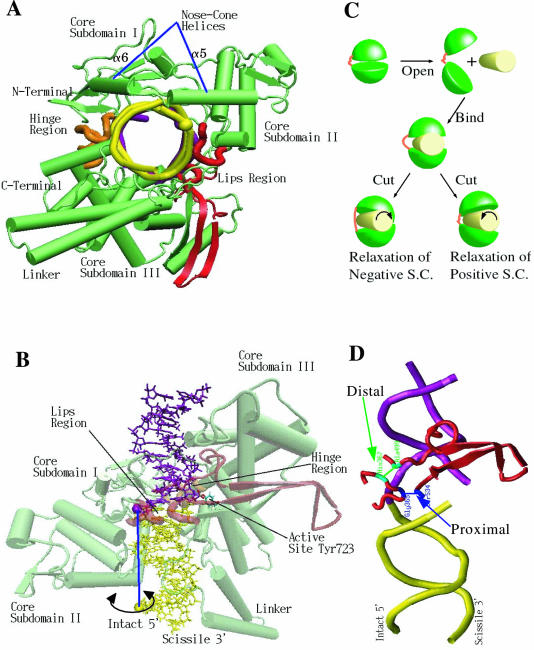
In a crystallographic tour de force, Holl, Champoux and coworkers (25–27) have obtained both covalent (i.e. after DNA nicking) and non-covalent (i.e. with intact DNA) complexes between human topoisomerase I and a 22 bp DNA duplex. Human topoisomerase I is a monomeric protein of 765 amino acids and is composed (2,25,28) of four major regions: the N-terminal, core, linker and the C-terminal domains. As seen in Figure 1A and B, the core subdomains I and II constitute an ‘upper cap’, which is connected by a flexible hinge to a ‘lower cap,’ containing the C-terminal domain, the core subdomain III, and the anti-parallel coiled-coil linker domain. Two contiguous α-helices (α5 and α6 in Figure 1A) form a V-shaped ‘nose cone,’ believed to be important in the topoisomerization mechanism (21), and belong to the upper-cap core subdomains II and I, respectively. When closed, the upper and lower caps bring together two opposable ‘lips’. These regions are positioned diametrically opposite the hinge region. The upper lip is part of the core subdomain I; the lower lip belongs to core subdomain III (see Figure 1A and B). The protein is believed (21) to open up its lips widely (hinging on the hinge region) in order to bind the incoming DNA duplex; this is then to be followed by the closing of the lips around the DNA strand (see binding step in Figure 1C). The steps of the cycle of activity of human topo I continue with cleavage of one of the two DNA strands through nucleophilic attack by the active site tyrosine (Tyr723 in Figure 1B). This results in covalent attachment of the protein to the 3′ phosphate at the site of cleavage. The resulting nick allows rotation of the DNA duplex downstream of the nick (see Figure 1B and C). This strand-rotation mechanism explains why topo IB enzymes can relax multiple supercoils, of either sign, at a time: the number of times the downstream part rotates equals the change in DNA's linking number [i.e. the number of times the two strands cross, a topological invariant under continuous deformations (29)]. The driving torque required for DNA rotation is thought to derive from the supercoiling energy; the direction of rotation depends on whether DNA is overwound (positively supercoiled) or underwound (negatively supercoiled). After a number of rotations occur [number believed to be, at least for vaccinia topos, torque-dependent (24)], a religation reaction sutures back the nick and the enzyme opens up to release a less-supercoiled DNA product.
Figure 1.
(A) DNA-human topoisomerase I covalent complex, seen from linker side of protein and along DNA helical axis. DNA downstream of the nick (see text) in yellow, upstream DNA in purple, protein is in green, (nose-cone helices pointed at by blue lines); lips region, including the 35 residue difference between the distal and proximal clamps (see text and Figure 1D), are in red, hinge region is in orange. (B) Different perspective of DNA-human topoisomerase complex focusing on the DNA duplex. Protein shown in transparent colors to make DNA visible. Active site residue (Tyr723) and the phosphate group at the nick point are represented by ball and stick model to show covalent attachment. The phosphate groups of −1 base (Ade) and and the +10 base (Ade), defining the rotation axis (which is in blue) are shown as large spheres, yellow for the +10 Ade and purple for the −1 Ade. The remaining coloring scheme is the same as in part A. In our molecular dynamics simulations, we rotate downstream DNA duplex (in yellow) around the blue rotation axis [mimicking the rotation driven by the supercoiling torque(24)] as indicated by the double-headed black circular arrow. (C) Caricature of supercoil relaxation, as revealed by simulations. View perspective as in Figure 1A; DNA: yellow cylinder, protein: two green hemispheres (upper cap and bottom part) connected by an orange thread-like hinge (see panels A, B of this figure and text); lips are on the right, at opposite side of the hinge. To bind DNA, the enzyme opens upper and lower ‘jaws’ and closes them around DNA. Following arrows to the left or right describes relaxation of negative or positive DNA supercoils, respectively. To relax negative supercoils, DNA needs to rotate clockwise in the enzyme's grip; torque applied in our MD simulation in that direction causes stretching of the (orange) hinge region. Counter-clockwise DNA rotation relaxes positive supercoils and produces opening of the lips. (D) The two clamps used in Carey et al. and Woo et al. Downstream DNA: yellow; upstream DNA: purple. The proximal clamp (Woo et al.) was produced by Cys mutations in Gly365 and Ser534, and shown in blue. The distal clamp (Carey et al.) was engineered by mutations in His367 and Ala499, and is colored green. The remaining lips of the protein is in red.
Based on the crystal structures of the covalent and noncovalent complex, Champoux and coworkers (21) have proposed what is now the prevalent model for human topo I activity, the so called ‘controlled-rotation’ mechanism: the downstream part of the DNA scissile strand, rather than being driven to rotate freely, rotates under the control of the surrounding protein. An important role in this process is thought to be played by charged residues of the nose cone and linker interacting electrostatically with the DNA downstream part. Eight additional crystal structures (26) exhibited significant crystal-to-crystal non-isomorphism and sizable differences in the trace of the backbone, documenting the remarkable flexibility needed by the closed enzyme to accommodate duplex rotation inside a cylinder-like space of large radius inside an enzyme maintaining its grip on the DNA. This indication of large-scale dynamical motions, together with available studies for related type IB topos (24,30–33), provided an incentive basis for our studies of the dynamical mechanisms important for the function of human topo I. However, it is very important to note that our simulations and the resultant findings are all based on the previously proposed ‘controlled-rotation’ mechanism (21), i.e., rotation of the downstream DNA around the intact strand. Therefore, if the DNA relaxes through another mechanism, our results should not be regarded as physiological.
MATERIALS AND METHODS
Torque-applied molecular dynamics
Constant-temperature molecular dynamics using the Nose–Hoover thermostat (34,35) and taking 2 fs steps by utilizing the SHAKE algorithm (36) were performed, with the non-bonded interactions (force) shifted to zero over 8–12 Å. The system was biased half-harmonically, i.e., only when it moved away from the target (37), with an external potential of the form
| 1 |
in which α is chosen to be 1000 kcal/mol/A4, (value adjusted so that full 360° rotations can be performed within the ns time scale). The reaction coordinate ρ leading from the initial state to the final state is given by
| 2 |
where rij = |ri − rj| is the distance between atoms i and j, R labels the coordinates of the final reference structure, and N represents the total number of atoms biased.
In this set-up, downstream DNA experienced forces from 0 to 2 nN, yielding an average effective torque of about 8.4 nN·Å. The reference structures towards which DNA was biased were obtained by applying a rotation transformation to the backbone atoms of DNA duplex in 10° angular increments until a full 360° turn (clock- or counter-clockwise) was obtained. Each simulation was 1.8 ns long, and took around 18 days on a 4 dual-node cluster of 8 AMD-Opteron-1.7GHz-64Bit CPUs.
The rotation axis is chosen to be a hypothetical axis that passes through the phosphate of the −1 adenine and the phosphate of the +10 adenine on the intact strand. The choice of the −1 Ade phosphate is motivated by the fact that, on one hand, it is exactly opposite the phosphate on the scissile strand which forms the phospodiester bond with the protein, and, on the other hand, it sits at the middle of the intact backbone part that connects the −1 Ade and +1 Ade. Choosing the rotation axis to pass through other atoms (or points in space) in this region will shift the rotation axis 1–2 Å on the helical curve, which is not expected to significantly affect the overall protein motion. The +10 Ade phosphate is chosen because it sits just below the −1 Ade phosphate in the 3D structure. Thus, the rotation axis becomes parallel with the helical axis of the rotating downstream duplex, which is needed for a proper (i.e. without base-pair opening) duplex rotation as hypothesized by the strand-rotation model. If our choice of the axis of rotation for DNA is incorrect, all of the described conformational changes in topo I should be regarded as artifacts of the simulations and one should not expect to observe them experimentally.
The CHARMM simulation package (38) was used for all calculations. A covalent model of the DNA–topoisomerase complex was set-up, with the position of the linker domain (missing in the covalent structure, PDB code 1a31) built using the non-covalent structure (PDB code 1a36).
The protein–DNA complex was completely immersed into a 60 Å-radius sphere of TIP3P (39) water molecules with at least 6 Å distance between the spherical boundary and the complex; waters overlapping by up to 2.8 Å were removed. A stochastic boundary potential was applied (40) and the system was equilibrated for 0.2 ns. Charge neutralization was performed by replacing 19 waters with sodium ions in the places where water oxygens had highest electrostatic energies. The systems was then equilibrated for an additional 100 ps without SHAKE (36) and 200 ps with the SHAKE algorithm. The final equilibrated structure has an 2.7 Å average RMS deviation from the original crystal structure. Six distinct simulations (corresponding to distinct ways to manipulate the rotating DNA) were set-up (see Table 1 in the main text) and analyzed in the light of each other.
Table 1.
Descriptions of the six systems used for MD simulations
| External potential is applied to: | Direction of the biased rotationa | Rotation increments | The system relaxes: | |
|---|---|---|---|---|
| System 1 | Both downstream and upstream parts | Downstream→Counterclockwise | 10° | Positive supercoils |
| Upstream→No rotation | 0° | |||
| System 2 | Both downstream and upstream parts | Downstream→Clockwise | 10° | Negative supercoils |
| Upstream→No rotation | 0° | |||
| System 3 | Both downstream and upstream parts | Downstream→Counterclockwise | 10° | Positive supercoils |
| Upstream→Clockwise | 10° | |||
| System 4 | Both downstream and upstream parts | Downstream→Clockwise | 10° | Negative supercoils |
| Upstream→Counterclockwise | 10° | |||
| System 5 | Only downstream part | Downstream→Counterclockwise | 10° | Positive supercoils |
| Upstream→No rotation | 0° | |||
| System 6 | Only downstream part | Downstream→Clockwise | 10° | Negative supercoils |
| Upstream→No rotation | 0° |
aWhen looking from the perspective in Figure 1A, i.e. from the linker side of the protein.
The resultant biasing forces mimic the effective torque arising from the twist energy contribution stored in the supercoiling free energy of the DNA substrate [estimated, based on measurements of the torsional modulus (15) to have the value, for an entire DNA molecule, on the average 4.5 cm long, at 5% supercoiled state, of around 12 kcal/mol]. However, the resultant torques in the simulation exceed by at least two orders of magnitude the physiologically-relevant torques expected in vivo. The strategy to apply forces or torques much larger than the actual ones is a necessity dictated by the limitations in the time scale accessible to molecular dynamics and has been used in several other studies (37,41–43). Given the large degree of forcing imposed to DNA on a short time scale (shorter than the intrinsic rotation time scale under physiological conditions), there exists, in principle, the possibility that protein motion was exaggerated because of the lack of relaxation time. Based on the reaction coordinate generated by the present study, calculations of the potential of mean force (in which the angle of DNA rotation is fixed and the protein is allowed to relax), as equilibrium calculations, surpass time scale limitations by restraining the system along a reaction pathway and are an obvious continuation of the work presented here. [While the method will be immune to a possible criticism of the shortness of reaction time scales, it will still have to address the issue of time to converge at the restraint points on the reaction path.]
Systems 1 and 2 represent the original strand rotation scheme where only the downstream part rotates. Although system 5 and system 6 may also be seen as representatives of a strand rotation scheme, they do not produce the desired motion (relative rotation of the downstream part) due to the fact that upstream DNA, together with the covalently attached protein, follow the downstream part when it is left free. The results of the simulations done on the system 5 and system 6 are found to be overall system rotations as a rigid body. This is because the upstream duplex is left free while the downstream part is subjected to a harmonic potential that drives the rotation. Such uneven treatment of the upstream part combined with the fact that the rotations are done on a rapid, ns time scale, causes the upstream duplex to rotate, unless external potentials are applied as in systems 1,2 and 3,4. Secondly, as explained above, applied potentials mimic the torque in the supercoiled DNA (although not quantitatively, but in what the resultant motions are concerned), and both the downstream and the upstream parts feel the same torque in opposite directions when DNA is nicked. Therefore, the upstream part should also be treated equally, and a restraining potential should be applied in some format. Therefore, we focused on the results of the simulation of these four systems, where both downstream and upstream duplexes are treated with external potentials.
Normal mode analysis
The structure at the end of the equilibration phase of molecular dynamics is minimized until a root-mean-square gradient of 10−5 kcal/mol/Å is reached in a series of 7500 cycles of steepest descent, followed by 8000 steps of the adopted basis Newton–Raphson procedure. This number of steps is needed such that, after discarding the six rigid-body modes, no negative eigenvalues are obtained. The lowest 1500 modes have been determined on this minimized structure by a iterative diagonalization scheme of the Hessian, where repetitive reduced-basis diagonalizations have been performed. These reduced basis is constructed partially from the not yet converged eigenvectors in Cartesian coordinates. This provides very efficient large calculations on big systems (such as the one used here) by consuming less memory and will make possible repetitive calculations for a multitude of suggested mutants. The details of the procedure are given in (44). The projections of the lowest modes are done on the different reaction coordinates, chosen to be multidimensional vectors that correspond to the difference between the coordinates of the initial and the final structures. Ordinary dot product of each mode with the reaction coordinates were performed.
RESULTS
Molecular dynamics simulations were run on a covalent complex with external constraining potentials on DNA (mimicking torque on either positively or negatively supercoiled DNA) that pushed it harmonically towards a target position obtained by applying, in 10° increments, full-circle (clockwise or counter-clockwise) rotations to the scissile strand, around an axis passing through the intact one. Six distinct DNA-topo-solvent systems (containing 86 694 atoms each, including water and counterions, and differing in how and on which region the constraints were applied, see Table 1) were simulated for 1.8 ns each. Out of the six runs, only four of them (system 1–4) were adept at relaxing supercoils. For the other two, the upstream DNA part was allowed to move, and it followed (together with the covalently attached protein) to a large extent the rotation of the downstream part, resulting in no effective relaxation. (For computational details, see the Materials and Methods section.)
Although the ‘controlled-rotation’ mechanism suggests that the downstream duplex rotates around the intact strand (21), we have also tested an alternative model, in which DNA rotation is performed around the helical axis. This alternative model was however untenable due to the large underlying free energy profile relative to the strand rotation scheme (exceeding 70 kcal/mol for a mere sixth of a full rotation). Rotations around the helical axis were therefore ruled out.
Distinct protein motions involved in positive versus negative supercoil relaxation
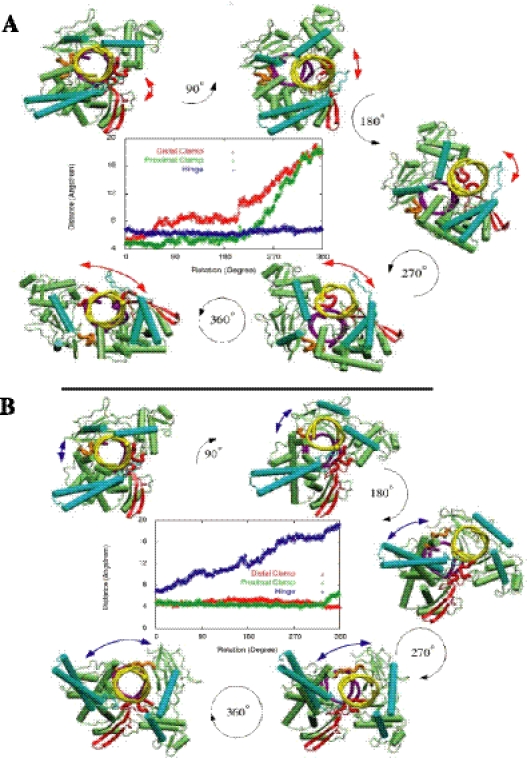
Whether the protein, during strand rotation, stays in the closed form or opens its lips has been a focus of two recent studies with seemingly contradictory conclusions. In 2003, Carey et al. (45) reported the engineering of two cysteines into the opposing lips that, after DNA-binding, formed a disulfide bond (a ‘distal clamp’, in reference to its relative distance from the active site, see Figure 1D) that sealed the two lips. They observed that the rate of DNA relaxation in the clamped form of the protein is comparable to the unclamped one, and concluded that relaxation occurs in a closed conformation. However, later in the same year, Woo et al. (46) published the design of a disulfide bond that clamped the two lips at a position that was closer to both the active site Tyr723 and to the bound DNA duplex; we refer to it as the ‘proximal clamp’ (see Figure 1D). Contrary to what was found for the distal clamp, they observed that DNA rotation is inhibited within the locked protein. Because the above studies showed that the two subtle alterations in the flexibility of the enzyme impacted differently on the enzyme's activity, it is of fundamental interest to study the extent of protein breathing (or opening) during strand rotation. Does the wild-type protein stay or not in the closed clamp conformation during DNA rotation? If it does open up, then how and how much? Previously, any possible opening was thought to occur by parting the two lips, so it was these regions that have been sealed in the clamped mutants. We stress that, importantly, both the Carey et al. and the Woo et al. clamped mutants used positively supercoiled DNA substrates.
We have set out to assess the opening of the protein by direct simulations of DNA rotation, in both the positive and negative direction, and have revealed unexpected structural dynamics (see the two movies in Supplementary Data). We have found that, while to relax positive supercoils the enzyme does tend to open up by a gradual separation of the two lips (see Figure 2A), the situation is quite different in the case of removal of negative supercoils (see Figure 2B). For this case, our simulations indicate that the upper and lower cap open up by stretching the hinge region (from Leu429 to Lys436, positioned diametrically opposite the lips across the DNA helical cross-section), while the two lips do not significantly change their relative positions. We find thus that negative supercoil rotations induce the reverse opening motion relative to the opening of the lips induced by rotations of positive supercoils (see Figure 1C). Moreover, the opening amplitudes imparted by the two types of supercoiling are similar; the hinge stretching and the lip opening distances are both within 10–14 Å. This result is reproduced in all four of our relaxation-competent runs (systems 1–4, see Table 1). The relevant distances are plotted, as a function of DNA rotation, in the insets to Figure 2A and B. It is of interest that the hinge region hypothesized here to be important for negative supercoil relaxation, is also believed (21) to be involved in the hinge-bending motion needed to achieve an open conformation of the DNA-free enzyme that can part the lip regions wide enough to allow the entry of the duplex strand (opening step in Figure 1C). This might imply that this region has been designed to be flexible both as a hinge and a stretch.
Figure 2.
(A) Positive supercoil relaxation, snapshots from the molecular dynamics results for system 1 in Table 1. Colors and perspective as in Figure 1A, except that nose-cone helices and linker domain are in darker green. Direction of downstream DNA rotation is counterclockwise. The two-sided red arrow points at the increasing separation of the lips. Notice that, in this case, the hinge does not stretch (see also inset). Distances in both figures measured between corresponding Cα atoms. Also note that in both cases in (A) and (B). DNA is still in the grip of the protein. (B) Negative supercoil relaxation: structural details from simulation (system 2 in Table 1). All details same as in panel A of this figure, except that direction of downstream DNA rotation is clockwise. Snapshots are at 90° intervals. Relative stretching of the hinge shown with two-sided blue arrows. Inset shows distances His367 to Ala499 (corresponding to distal clamp), Gly365 to Ser534 (proximal clamp), and Leu429 to Lys436 (hinge).
What are the interaction details that bring about the two distinct mechanisms? The nose-cone helices (in the upper cap) and the linker (in the lower cap) are thought (21) to control DNA rotation within the protein because these domains are positively charged. The two nose-cone helices sit oriented along the spiral groove of the rotating DNA duplex and, depending on the direction of rotation, impart a push to different domains of the surrounding protein, much like the way a nut around a screw is pushed in different parts of its inner lining when the screw rotates clock or anticlockwise. The DNA rotation that relieves positive supercoils affects mainly the right nose-cone helix α5 (i.e. on the right side of Figure 2A, see also movie 1), which pushes open the right side of the cap region. On the other hand, the rotation that relaxes negative supercoils moves first the left nose-cone helix α6 (in Figure 2B, see also movie 2), inducing motion of the left cap region, which in turn stretches the hinge.
Interaction energies along rotation coordinate
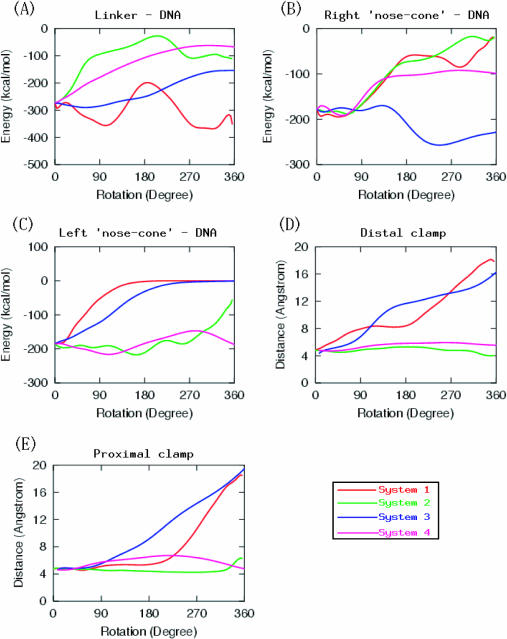
The energy of interaction of DNA with the right nose-cone helix, left nose-cone helix, as well as with the linker domain, is calculated along the rotations. In agreement with the above observation, they show that the linker domain and the right nose-cone control the rotation of the DNA predominantly for removal of positive supercoils, whereas the left nose-cone helix (α6) controls rotations that relax negative supercoils. This prompts at residue locations for charge-neutralization mutations with the potential to alter relaxation of one type of supercoiling, but not of the other.
As seen in Figure 3A, the interaction energy (95% of which was electrostatic) between the linker and DNA is conserved during rotations that relax positive supercoils (system 1 and system 3 in Table 1), whereas this interaction is lost during rotations that relax negative supercoils (systems 2 and 4 in Table 1). However, the situation is reversed for the interaction between DNA and the left nose-cone helix (see Figure 3C), in which the interaction quickly weakens for positive supercoil relaxation (system 1 and system 3), while it is conserved for negative relaxation (system 2 and system 4). [Due to finite sampling problems (common to all molecular dynamics simulations of large systems), the actual values of the energies presented here are to be taken in a qualitative sense. A more detailed energy analysis is to be done in future work using potential of mean force calculations.]
Figure 3.
(A) Interaction energies (locally-averaged, and in kcal/mol) of DNA with the linker domain (residues 636–712) as a function of the extent of rotation. (B) Interaction energy of DNA with the right nose-cone helix (α5, Thr303 to Gln318). (C) Interaction energy of DNA with left nose-cone helix (α6, Lys321 to Tyr338). (D) The variation of the distance (in angstroms) of the distal clamp (His367–Ala499) in all four systems, as a function of the extent of rotation of downstream DNA. (E) The corresponding plot for the proximal clamp (Gly365–Ser534). All distances are measured between the two Cα atoms. Legends in inset are the same for all panels.
For the interaction between DNA with the right nose-cone helix, the two simultaneous 180° rotations of both parts (system 3, see Table 1) produced conservation of the interaction as expected. However, a 360° rotation of just the downstream part (system 1 in Table 1) shows no such conservation (see Figure 3B). This can be a result of the fact that the interaction between the linker domain and the DNA is more stable in system 1 than in system 3, meaning that the linker is in control of positive supercoil relaxation. Again, the right nose-cone helix-DNA interaction is almost completely lost in systems 2 and 4 (see Figure 3B), showing that the left nose-cone helix, and not the right one, dominantly controls rotations that relax negative supercoils.
The above finding is in agreement with the recent single molecule experiment on vaccinia topo I (24), which revealed that friction between protein and DNA is important in the control of DNA rotation. As friction derives, on the atomic scale, from the interaction energy between the topoisomerase cavity and DNA, our aforementioned discussion on the preservation of the interaction supports the conclusion of the friction-based model proposed by Koster et al. (24). In addition to it, we have revealed specific sites in the protein that govern friction (e.g. the linker and nose-cone helices), and we surmise that, without the protein, DNA rotation might be faster, but certainly ‘uncontrolled’.
Analysis of clamping studies
The fact that the disulphide clamping studies of Carey et al. (45) and Woo et al. (46) produced opposite results (the proximal clamp inhibited relaxation, while the distal one did not) was interpreted to arise from the relative positioning of the clamps with respect to the rotating DNA. Although Carey et al. used an N-terminal truncated human topoisomerase I (topo70), while Woo et al. used a full length protein, the truncated topo70 (starting from residue 175) retained its activity. The fact that the first study used temperature changes and the second a DNA intercalator is not likely to be fundamentally significant, as both of the procedures have the same effect on the DNA substrate. The explanation of the observed differences is most likely that the proximal clamp strangles DNA more tightly than the distal one, impeding strand rotation. However, no time-resolved details of the entire rotation mechanism existed. Moreover, although both studies used only positively supercoiled DNA substrates, [For DNA intercalators, this requirement was due to the impossibility to assay negative supercoils because such studies induce DNA supercoiling after the formation of topo-DNA catenanes. A temperature shift method should, in principle, not have this problem.] their conclusions were extended to the general relaxation mechanism of both kinds of supercoils. This tacit assumption may have stemmed from the knowledge that human topo I can remove both positive and negative supercoils. However, in the light of the distinct opening motions found in the present rotation simulations, it is essential to differentiate between supercoiling signs. The actual simulations on the clamped enzyme systems are in progress, and more detailed explanations for the observed contradictory findings of Carey et al. (45) and Woo et al. (46) will be available at the atomic level. Meanwhile, we have investigated how the distances between the amino acids of the wild-type corresponding to the distal and proximal clamp Cys mutations change in our different molecular dynamics simulations. As mentioned, during rotations that remove negative supercoils we found, remarkably, that the two lips do not open up, and that both the proximal and distal clamp distances remain almost the same. This feature was reproducible, and both our runs that are adept at relaxing negative supercoils exhibited this feature. In system 2, the two clamp separations fluctuate around 5 ± 0.5 Å (see Figure 2B), except that the proximal clamp shows a 1.5 Å sudden increase during the last 40° rotations. In system 4, the fluctuations are within ±2.5 Å and the same for both clamps. The final changes (at the end of 360° rotations) are 0.5 Å decrease in the distal clamp and 1.3 Å increase in the proximal clamp for system 2, and 0.4 Å increase in distal clamp and 0.2 Å decrease in proximal clamp in system 4 (see Figure 3D and E). Therefore, these very small final changes can be regarded as negligible when compared to those obtained in rotations for removal of positive supercoils where the changes are observed around 10–14 Å (see Figure 3D and E). Given that (i) our simulations indicate that the behavior of the protein acting on negative supercoils is fundamentally distinct from the positive case, and that (ii) clamping experiments were performed on positive supercoils, it would be of great future interest to perform the two lip-clamping experiments with negatively supercoiled DNA substrates.
Possible implications of the N-terminal domain
Deletion of the first 190 amino acids from the N-terminus of human topo I (amino acids 1–190) is proved not to modify significantly human topo I's mechanism relative to the full length enzyme (47–49). However, three recent studies showed results that bring about remarkable insights when viewed from the point of the simulations reported here. Firstly, in 2001, Lisby et al. (50), by truncating amino acids from the N-terminal all the way down to position 206, proved that the ‘further-down’ residues 191–206 participate in topoisomerization either by binding DNA directly or by promoting DNA-binding to other regions of the enzyme. The truncated enzyme distinguished itself from the wild-type by exhibiting insensitivity to the anti-cancer drug CPT in relaxation and inability to ligate blunt end DNA fragments. This implicates the truncated region in the control of strand rotation. Secondly, in 2003, Christensen et al. (51) also reported that residues 190–210 of human topoisomerase I are required for enzyme activity in vivo (but not in vitro). Thirdly, while our work was in progress, Frøhlich et al. (52) strengthened the conclusion found by Lisby et al. (50) by studying the N-terminal residues more in detail, and hypothesized that Trp205 (among other amino acids positioned between 191–206) coordinates DNA rotation during topoisomerization.
From a structural point of view, Redinbo et al. (27) had resolved, in 2000, 12 additional residues at the N-terminal domain (positions 203–214). The observations that (i) these new residues were found to be in close proximity of the hinge region, and that (ii) our simulations indicate that relaxation of negative supercoils induces a stretch in the hinge (rather than causing the lips to open) can be corroborated to provide a possible explanation for the biochemical deletion studies presented above.
That is, taken together, our findings and the new N-terminus structural data thus suggest that the N-terminal region is, in fact, important for the removal of negative supercoils because some of its residues are packed against the hinge region. [This region would therefore be a hot spot for engineering mutations either in the hinge or N-linker to alter topo I activity.] More interestingly, Trp205 [found to be particularly important by Frøhlich et al. (52), see above] is the closest residue, among the 12 newly resolved residues, to the hinge region (Leu429 to Lys436): it has side chain atoms only around 4 Å away from those of Ser432 and Arg434. We cannot, at the moment, say more about the function of the N-terminal as it is absent from the covalent structure our studies were performed on. Importantly, however, all three biochemical studies (50–52) on the role of the N-terminal domain were performed on negatively supercoiled DNA. In the light of the different negative versus positive relaxation protein motions seen in our simulations, it is tempting to speculate that the N-terminal region can, in fact, control removal of only negative supercoils (through its close interactions with the hinge). Mutations in the N-terminus should not significantly affect positive supercoiling relaxation. [Although there could be ‘second-order’ effects due to the fact the opening motion of the lips hinges on the hinge region. Therefore, for an Cys-mutant experimental test of our prediction (i.e. that the hinge stretches upon relaxation of negative turns), it would be optimal if the disulfide bond is ‘locked’ after non-covalent binding of the protein (by pH change).] This is a testable hypothesis in future experiments that would compare hinge-mutated topo I activity on positively and negatively supercoiled DNA substrates.
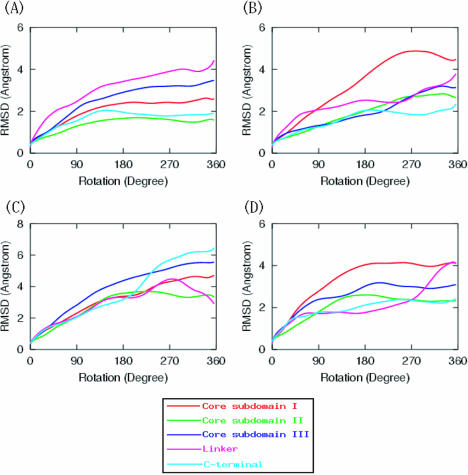
RMS deviations during MD simulations
In terms of the average root-mean-square deviations observed in all heavy protein atoms, system 1 produced the largest deformations in the linker part, while the core subdomain II shows the minimum RMS deviations, as seen in Figure 4A. This is consistent with the argument that the linker domain is in more control of the relaxation of positive supercoils than the right nose-cone helix (which is within the core subdomain II). This is because the DNA duplex is a double-stranded stiff chain, with persistence length of about 1000 Å, so even if we impose the rotation around the intact strand, the protein is capable of adapting to take the orientation of ‘minimal resistance’ during DNA rotation. Therefore, correlation between RMSD and the interaction energies with the DNA suggest that those domains that control the rotations are more flexible, and they adopt the necessary structural changes to conserve the interactions with the rotating DNA atoms. [Several equilibrium MD simulations on topoisomerases have been recently performed (53–55) and have explored additional correlations between domains; however, there was no attempt to model the actual relaxation mechanism (as was done here).] Similarly, both system 2 and 4 (which relax negative supercoils) exhibit a distinct RMSD in core subdomain I (see Figure 4B and D), which is larger than those observed for the other domains. This is also in agreement with the observation that the left nose-cone helix (which is within the core subdomain I) controls the rotation for relaxation of negative supercoils, more than the linker does. The study of Redinbo et al. (26) on protein flexibility concluded that the linker domain and the upper cap region (composed of subdomain I and II) show the maximal degree of flexibility, a result in good agreement with the above discussion. They observed that, at least in the crystal structures, the linker domain is the most flexible part of the protein exhibiting up to 4.6 Å non-isomorphic shifts, followed by the cap region with up to 3.6 Å deviations. These experimental findings are consistent with our calculated RMS deviations of the room-temperature solvated system, which are less than 4.5 Å, with the largest RMS deviations in the linker and the core-subdomain I (on the upper cap), followed by the C-terminal and core-subdomain III.
Figure 4.
Average RMS deviations for all four systems; (A) for system 1, (B) for system 2, (C) for system 3, and (D) for system 4. RMSDs were calculated on heavy atoms of protein's domains, and plotted as a function of the rotation angle. Legends in inset are the same for all panels. RMSDs were calculated by superimposing the coordinates of a domain (at every 100 MD steps) on the initial coordinates at the beginning of the rotation.
While we find that the RMS deviation of individual domains from their crystal structure reference stays below 4.5 Å (i.e. we do not artificially unfold domains by the rapid rotation), large, see-saw-like opening motions of the two caps as a whole due to the strand rotation are observed. Although large, these displacements are comparable to those reported in experiments on other topoisomerases. For Escherichia coli topo I, large conformational shifts of about 20 Å are also reported (22). Similarly, type IIA topoisomerases have been shown to undergo dramatic conformational changes that are larger than 20 Å (56,57). This paints a rather dynamic picture of the larger family of topoisomerase enzymes in general as they manipulate DNA by ample motions.
Same set of topoisomerase normal modes encodes both opening motions in a see-saw fashion
To gauge the inherent flexibility and large amplitude (low frequency) motions related to the conformational changes that the protein undergoes, and to complement the model provided by the molecular dynamics simulations, we have performed a normal mode analysis of human topoisomerase I on the MD-equilibrated structures. (This consists of the diagonalization of the second-derivative matrix of the potential energy, yielding the vibrational frequencies, as well as the directions of internal motions; see Materials and Methods for technical details). Such analyses have provided considerable insight into the nature of collective motions in many proteins (58–62) and have showed that, in most systems where an initial and a final structure are available, the first few low frequency modes are sufficient to describe the large-scale conformational changes involved in going from one structure to the other (63,64). This strategy has worked well, including for protein–DNA complexes (65), as well as for systems as large as the ribosome (66).
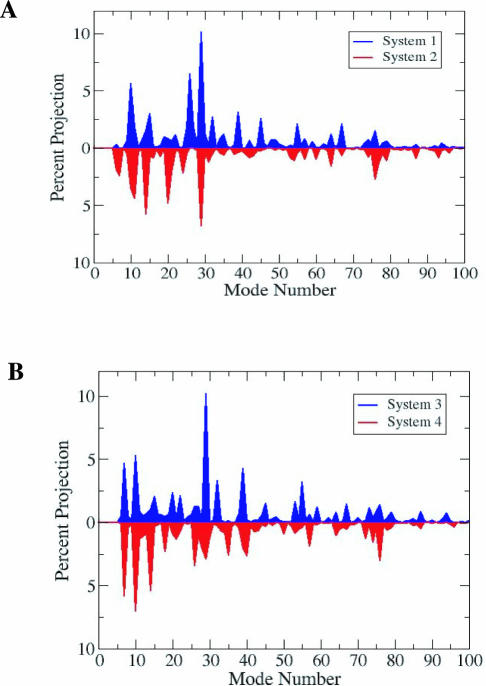
The difference vectors between the initial and the final (after a 360° DNA rotation) of the protein for the 4 relaxation-competent systems we have studied have been regarded as reaction coordinates, and the lowest 100 modes have been projected on the these reaction coordinates. The percent contributions to the reaction coordinate are plotted in Figure 5A (system 1 and 2), and the corresponding plot is presented in Figure 5B (for system 3 and 4). Significant contributions appear especially from the lowest 30 non-zero modes. As the reaction coordinate describes the breathing of the protein (whether this involves opening of the lips or stretching of the hinge), the significant contributions document the protein's large inherent flexibility which, perhaps by design, allows for flexing during DNA rotation. These implications are particularly attractive in the light of recently reported involvement of concerted protein motions in the activity of enzymes [see (67) and discussion below].
Figure 5.
(A) Projection of normal mode directions on the protein reaction coordinate corresponding to relaxation of positive (in blue) and negative (in red) supercoils. Reaction coordinate generated by fixing the upstream DNA and rotating the downstream part (see also Table 1) (B) Same as panel A except that the reaction coordinate of the protein was generated in two other independent runs, applying torque on both upstream and downstream DNA.
Interestingly, it is observed that the contributions to relaxation of positive supercoils are almost a mirror image of the contributions to relaxation of negative supercoils with respect to the mode number. This vividly demonstrates that the same modes are responsible for both positive and negative supercoil relaxations. The crystal structure ‘ground-state’ of the protein therefore seems to encode (and to almost equally favor) both lip opening and hinge stretching in a see-saw motion of the upper cap relative to the lower one.
Normal mode analysis is a linear approximation of local dynamics, in distinction from molecular dynamics. It, nonetheless, is in good agreement with the MD results. Even if the normal modes were calculated in the absence of torque on DNA, torque was implicitly considered because it was applied to generate the reaction coordinates in the two directions. It therefore affected how the supercoiling sign of the DNA substrate directed protein motion. This is important because, for topo IV (a type II enzyme), studies of positive versus negative relaxation showed that for preferential cleavage of positively supercoiled DNA, substrate discrimination can take place before strand passage (14). It is of considerable interest to explore whether this strategy is used by type I enzymes.
DISCUSSION
Conclusions of computational studies
As a result of applications of MD simulations and of normal mode analysis, we have determined that:
The relaxation mechanisms for positive and negative supercoils are not the same. Removal of positive supercoils requires opening of the lips by 10–14 Å, while removal of negative supercoils needs the hinge region (from Leu429 to Lys436) to stretch about 12 Å.
The N-terminal part of the protein, a subject of ongoing debate, is likely to be important for the relaxation of negatively supercoiled DNA, and it may or may not be important for the relaxation of positively supercoiled DNA.
As suggested by structural data, the linker domain and the nose-cone helices are found to control the rotations needed to relax DNA. In additional insight, simulations reveal that the linker domain dominantly controls rotations for removal of positive supercoils, whereas the nose cones (particularly the left nose-cone helix) is the dominant controller of the rotations that remove negative supercoils.
The protein has large, and almost identical, inherent structural flexibility (as characterized by normal modes) towards both type of relaxations.
On the kinetic effects of clamping
Kinetic data are available for the related topo IB enzyme, vaccinia topoisomerase. For it, biochemical evidence shows that, on average, five strand rotations per binding event occur (68) [in contrast to a single-molecule estimate of about 19 rotations per cycle (24)], and that cleavage is slower than rotation and religation (32,69,70). However, no kinetic data are available to discern what the rate limiting step is for the human enzyme under processive conditions (i.e. when it relaxes several supercoils upon one binding event) and physiological DNA substrate concentrations. Equally frustrating is also that no data exist to bear on the question of whether or not rotation is slower in either the distal or proximal clamp states. It is certainly possible that in the closed clamp conformation, rotation is impeded relative to the unclamped state. What will be important to consider experimentally is whether rotation is faster than religation since, once cleavage occurs, these are the two competing reactions. These parameters have not been measured for the human enzyme and are certainly more difficult to measure than for the vaccinia topo (J. J. Champoux, private communication). Moreover, it is not clear if conclusions from vaccinia topo I can be extrapolated to human topo I, because the former has pronounced sequence specificity and, presumably, a distinct covalent DNA-binding pattern involving an extrahelical nucleotide position at the binding site (71). What will be important to address, in future computational studies of the Cys-clamped complex, is to which extent rotation is impeded by the distal or the proximal strand. The possibility that a difference between the seemingly contradictory results obtained when disulphide-clamping the lips by Carey et al. (45) and by Woo et al. (46) is due to the absence and, respectively, the presence of the N-terminus in the two constructs cannot formally be excluded. However, because of the distinct positioning of the two clamps, we expect that the clamped simulations will reveal significant dynamic and energetic differences of DNA interactions, in particular with the nose cone helices.
Simulated mechanisms suggest additional experiments
Our computational findings of the different mechanisms mentioned above can be used to engineer topoisomerases with inhibited activity towards, or that selectively relax only, one kind of supercoils. One way this can be achieved is by engineering the proximal clamp and therefore preventing relaxation of positive supercoils. As we show that the lips do not open significantly during relaxation of negative supercoils, this clamping should still permit relaxation of negative supercoils. In this case, one may observe a decrease in the rotation rate, as our MD simulations show that the distances corresponding to both the distal and the proximal clamps increase by 2 Å, and then decrease back to original values. However, this stretching barrier is small compared to the overall protein breathing motions (10–14 Å), and therefore relaxation of negative supercoils should not be inhibited. A protein that performs the reverse function, allowing relaxation of positive supercoils and preventing that of negative ones, may be obtained by engineering disulfide bonds in the hinge region. A possible pair of Cys mutants could involve, for example, Leu429 and Lys436, which have their Cαs only 6 Å away from each other in our equilibrium simulations. This hinge-clamping strategy is likely to allow relaxation of positive supercoils without much impediment, because the hinge region is unchanged during the relaxation of positive supercoils.
A fluorescence resonance energy transfer (FRET) experiment, with two fluorophores, one in the upper and one in the lower cap, could constitute another test of the proposed distinct conformational changes (72). To observe opening, the fluorophore pair would be placed next to the lips or next to the hinge. A technical issue to be resolved would be to find that pair of fluorophore labels for which the Förster distance of the donor-acceptor pair (at which FRET efficiency is 50%) would be around 20 Å to accommodate an extended hinge or the opening of the lips (see Figure 2).
FRET experiments could also possibly shed light in the following matter. The state of the protein upon a full DNA rotation [in both (+) and (−) cases] is not the same as the state of the protein at 0°. We have no rigorous proof to exclude the formal possibility that this is a consequence of the rapid time scale for the simulation (on which the protein might not have enough time to relax). Such a proof would require extending the simulation to times that are not available to current computer hardware. However, we believe that it is more likely that what we see at 360° is an intermediate state between open and closed. This intermediate would be the state in which rotation of the nicked DNA is allowed. It is important to note in this regard that DNA is thought to rotate several times before religation, as shown by recent single molecule (24) and bulk experiments (68). A transition from this metastable intermediate to the closed state would be followed by religation. Because such a transition can well take beyond microseconds, we were not able to observe it by direct molecular dynamics. It is also imaginable that another intermediate exists that would allow sliding along the DNA duplex, in-between two relaxation events. The conformations we have generated at 360° could also be representative of such a putative state (although if it exists, it should correspond to a non-covalent complex).
Another way to alter activity by altering flexibility and dynamics can be attempted in a systematic way by changing amino acids throughout the protein, gauging their flexibility by normal modes, and performing more detailed molecular dynamics studies on those particular mutants that show a significantly altered pattern of projections onto rotation coordinates. This is particularly important because recent evidence points to coupled networks of predominantly conserved residues that influence protein motion, which in turn points at an evolutionary selection of these coupled networks, of potential importance to protein engineering (67).
Topo I may be used as a biological machine that selectively relaxes only one type of supercoil, while preventing the other at specific-sites on the DNA chain. In an instance of a topo IB enzyme with specificity to the DNA sequence to which it binds, vaccinia topoisomerases, a possible ‘strangling machine’ could in principle be devised to alter preferentially the sign of supercoiling in a sequence-dependent fashion and possibly switch genes on and off.
Human topoisomerase I is the sole target of a novel class of potent anti-cancer drugs from the CPT family. Recent evidence (73) points to the fact that CPT, in addition to impeding religation, might also hinder rotation by intercalating at the nick site. Additional studies of the type presented here applied to the CPT-bound system should be important in understanding in detail the role of this anti-cancer drug.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online.
Supplementary Material
Acknowledgments
This work was supported from funds from the University of Michigan. Computational resources from the National Science Foundation (TeraGrid) are kindly acknowledged. We thank Professor J. J. Champoux for valuable suggestions and discussions. Funding to pay the Open Access publication charges for this article was provided by the University of Michigan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Wang J.C. DNA topoisomerases. Annu. Rev. Biochem. 1996;65:635–692. doi: 10.1146/annurev.bi.65.070196.003223. [DOI] [PubMed] [Google Scholar]
- 2.Champoux J.J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 3.Corbett K.D., Berger J.M. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophy. Biomol Struct. 2004;33:95–118. doi: 10.1146/annurev.biophys.33.110502.140357. [DOI] [PubMed] [Google Scholar]
- 4.Bjornsti M.A., Benedetti P., Viglianti G.A., Wang J.C. Expression of human DNA topoisomerase-I in yeast-cells lacking yeast DNA topoisomerase-I—restoration of sensitivity of the cells to the antitumor drug camptothecin. Cancer Res. 1989;49:6318–6323. [PubMed] [Google Scholar]
- 5.Pommier Y. Diversity of DNA topoisomerases I and inhibitors. Biochimie. 1998;80:255–270. doi: 10.1016/s0300-9084(98)80008-4. [DOI] [PubMed] [Google Scholar]
- 6.Li T.K., Liu L.F. Tumor cell death induced by topoisomerase-targeting drugs. Annu. Rev. Pharmacol Toxicol. 2001;41:53–77. doi: 10.1146/annurev.pharmtox.41.1.53. [DOI] [PubMed] [Google Scholar]
- 7.Bustamante C., Bryant Z., Smith S.B. Ten years of tension: single-molecule DNA mechanics. Nature. 2003;421:423–427. doi: 10.1038/nature01405. [DOI] [PubMed] [Google Scholar]
- 8.Strick T., Allemand J.F.O., Croquette V., Bensimon D. The manipulation of single biomolecules. Phys. Today. 2001;54:46–51. [Google Scholar]
- 9.Andricioaei I., Goel A., Herschbach D.R., Karplus M. Dependence of DNA polymerase replication rate on external forces: a model based on molecular dynamics simulations. Biophys. J. 2004;87:1478–1497. doi: 10.1529/biophysj.103.039313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Charvin G., Bensimon D., Croquette V. Single-molecule study of DNA unlinking by eukaryotic and prokaryotic type-II topoisomerases. Proc. Natl Acad. Sci. USA. 2003;100:9820–9825. doi: 10.1073/pnas.1631550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dekker N.H., Viard T., de La Tour C.B., Duguet M., Bensimon D., Croquette V. Thermophilic topoisomerase I on a single DNA molecule. J. Mol. Biol. 2003;329:271–282. doi: 10.1016/s0022-2836(03)00320-6. [DOI] [PubMed] [Google Scholar]
- 12.Dekker N.H., Rybenkov V.V., Duguet M., Crisona N.J., Cozzarelli N.R., Bensimon D., Croquette V. The mechanism of type IA topoisomerases. Proc. Natl Acad. Sci. USA. 2002;99:12126–12131. doi: 10.1073/pnas.132378799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Postow L., Ullsperger C., Keller R.W., Bustamante C., Vologodskii A.V., Cozzarelli N.R. Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. 2001;276:2790–2796. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- 14.Crisona N.J., Strick T.R., Bensimon D., Croquette V., Cozzarelli N.R. Preferential relaxation of positively supercoiled DNA by E.coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 2000;14:2881–2892. doi: 10.1101/gad.838900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bryant Z., Stone M.D., Gore J., Smith S.B., Cozzarelli N.R., Bustamante C. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 16.Wang J.C. Interaction between DNA and an Escherichia coli protein omega. J. Mol Biol. 1971;55:523–533. doi: 10.1016/0022-2836(71)90334-2. [DOI] [PubMed] [Google Scholar]
- 17.Champoux J.J., Dulbecco R. Activity from mammalian-cells that untwists superhelical DNA—possible swivel for DNA-replication. Proc. Natl Acad. Sci. USA. 1972;69:143–146. doi: 10.1073/pnas.69.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gellert M., Mizuuchi K., Odea M.H., Nash H.A. DNA gyrase—enzyme that introduces superhelical turns into DNA. Proc. Natl Acad. Sci. USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bergerat A., Gadelle D., Forterre P. Purification of a DNA topoisomerase-II from the hyperthermophilic archaeon sulfolobus-shibatae—a thermostable enzyme with both bacterial and eucaryal features. J. Biol. Chem. 1994;269:27663–27669. [PubMed] [Google Scholar]
- 20.Kirkegaard K., Wang J.C. Bacterial-DNA topoisomerase-I can relax positively supercoiled DNA containing a single-stranded loop. J. Mol. Biol. 1985;185:625–637. doi: 10.1016/0022-2836(85)90075-0. [DOI] [PubMed] [Google Scholar]
- 21.Stewart L., Redinbo M.R., Qiu X.Y., Hol W.G.J., Champoux J.J. A model for the mechanism of human topoisomerase I. Science. 1998;279:1534–1541. doi: 10.1126/science.279.5356.1534. [DOI] [PubMed] [Google Scholar]
- 22.Feinberg H., Lima C.D., Mondragon A. Conformational changes in E. coli DNA topoisomerase I. Nature Struct. Biol. 1999;6:918–922. doi: 10.1038/13283. [DOI] [PubMed] [Google Scholar]
- 23.Changela A., DiGate R.J., Mondragon A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–1081. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- 24.Koster D.A., Croquette V., Dekker C., Shuman S., Dekker N.H. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase ib. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 25.Redinbo M.R., Stewart L., Kuhn P., Champoux J.J., Hol W.G.J. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 26.Redinbo M.R., Stewart L., Champoux J.J., Hol W.G.J. Structural flexibility in human topoisomerase I revealed in multiple non-isomorphous crystal structures. J. Mol. Biol. 1999;292:685–696. doi: 10.1006/jmbi.1999.3065. [DOI] [PubMed] [Google Scholar]
- 27.Redinbo M.R., Champoux J.J., Hol W.G.J. Novel, insights into catalytic mechanism from a crystal structure of human topoisomerase I in complex with DNA. Biochemistry. 2000;39:6832–6840. doi: 10.1021/bi992690t. [DOI] [PubMed] [Google Scholar]
- 28.Stewart L., Ireton G.C., Champoux J.J. The domain organization of human topoisomerase I. J. Biol. Chem. 1996;271:7602–7608. doi: 10.1074/jbc.271.13.7602. [DOI] [PubMed] [Google Scholar]
- 29.Călugăreanu G. L'intégrale de Gauss et l'analyse des nœuds tridimensionnels. Rev. Math. Pures Appl. 1959;4:5–20. [Google Scholar]
- 30.Caserta M., Amadei A., Camilloni G., Dimauro E. Regulation of the function of eukaryotic DNA topoisomerase-I—analysis of the binding step and of the catalytic constants of topoisomerization as a function of DNA topology. Biochemistry. 1990;29:8152–8157. doi: 10.1021/bi00487a024. [DOI] [PubMed] [Google Scholar]
- 31.Camilloni G., Caserta M., Amadei A., Dimauro E. The conformation of constitutive DNA interaction sites for eukaryotic DNA topoisomerase-I on intrinsically curved DNAs. Biochim. Biophys. Acta. 1991;1129:73–82. doi: 10.1016/0167-4781(91)90214-7. [DOI] [PubMed] [Google Scholar]
- 32.Stivers J.T., Shuman S., Mildvan A.S. Vaccinia DNA topoisomerase-I—single-turnover and steady-state kinetic-analysis of the DNA strand cleavage and ligation reactions. Biochemistry. 1994;33:327–339. doi: 10.1021/bi00167a043. [DOI] [PubMed] [Google Scholar]
- 33.Tian L.G., Claeboe C.D., Hecht S.M., Shuman S. Remote phosphate contacts trigger assembly of the active site of DNA topoisomerase IB. Structure. 2004;12:31–40. doi: 10.1016/j.str.2003.11.025. [DOI] [PubMed] [Google Scholar]
- 34.Nosé S. A unified formulation of the constant temperature molecular-dynamics methods. J. Chem. Phys. 1984;81:511–519. [Google Scholar]
- 35.Hoover W.G. Canonical dynamics: equilibrium phase-space distributions. Phys. Rev. A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 36.Ryckaert J.P., Ciccotti G., Berendsen H.J.C. Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n-alkanes. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 37.Paci E., Karplus M. Forced unfolding of fibronectin type 3 modules: an analysis by biased molecular dynamics simulations. J. Mol. Biol. 1999;288:441–459. doi: 10.1006/jmbi.1999.2670. [DOI] [PubMed] [Google Scholar]
- 38.Brooks B.R., Bruccoleri R.E., Olafson B.D., States D.J., Swaminathan S., Karplus M. CHARMM: a program for macromolecular energy, minimization, and dynamics. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 39.Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. [Google Scholar]
- 40.Brooks C.L., Brunger A., Karplus M. Active-site dynamics in protein molecules: a stochastic boundary molecular-dynamics approach. Biopolymers. 1985;24:843–865. doi: 10.1002/bip.360240509. [DOI] [PubMed] [Google Scholar]
- 41.Izrailev S., Stepaniants S., Balsera M., Oono Y., Schulten K. Molecular dynamics study of unbinding of the avidin-biotin complex. Biophys. J. 1997;72:1568–1581. doi: 10.1016/S0006-3495(97)78804-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bockmann R.A., Grubmuller H. Nanoseconds molecular dynamics simulation of primary mechanical energy transfer steps in F-1-ATP synthase. Nature Struct. Biol. 2002;9:198–202. doi: 10.1038/nsb760. [DOI] [PubMed] [Google Scholar]
- 43.Jin M., Andricioaei I., Springer T.A. Conversion between three conformational states of integrin I domains with a C-terminal pull spring studied with molecular dynamics. Structure. 2004;12:2137–2147. doi: 10.1016/j.str.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 44.Mouawad L., Perahia D. Diagonalization in a mixed basis—a method to compute low-frequency normal-modes for large macromolecules. Biopolymers. 1993;33:599–611. [Google Scholar]
- 45.Carey J.F., Schultz S.J., Sisson L., Fazzio T.G., Champoux J.J. DNA relaxation by human topoisomerase I occurs in the closed clamp conformation of the protein. Proc. Natl Acad. Sci. USA. 2003;100:5640–5645. doi: 10.1073/pnas.1031537100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woo M.H., Losasso C., Guo H., Pattarello L., Benedetti P., Bjornsti M.A. Locking the DNA topoisomerase I protein clamp inhibits DNA rotation and induces cell lethality. Proc. Natl Acad. Sci. USA. 2003;100:13767–13772. doi: 10.1073/pnas.2235886100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bronstein I.B., Wynne-Jones A., Sukhanova A., Fleury F., Ianoul A., Holden J.A., Alix A.J.P., Dodson G.G., Jardillier J.C., Nabiev I., Wilkinson A.J. Expression, purification and DNA-cleavage activity of recombinant 68-kDa human topoisomerase I target for antitumor drugs. Anticancer Res. 1999;19:317–327. [PubMed] [Google Scholar]
- 48.Stewart L., Ireton G.C., Champoux J.J. Reconstitution of human topoisomerase I by fragment complementation. J. Mol. Biol. 1997;269:355–372. doi: 10.1006/jmbi.1997.1056. [DOI] [PubMed] [Google Scholar]
- 49.Stewart L., Ireton G.C., Parker L.H., Madden K.R., Champoux J.J. Biochemical and biophysical analyses of recombinant forms of human topoisomerase I. J. Biol. Chem. 1996;271:7593–7601. doi: 10.1074/jbc.271.13.7593. [DOI] [PubMed] [Google Scholar]
- 50.Lisby M., Olesen J.R., Skouboe C., Krogh B.O., Straub T., Boege F., Velmurugan S., Martensen P.M., Andersen A.H., Jayaram M., Westergaard O., Knudsen B.R. Residues within the N-terminal domain of human topoisomerase I play a direct role in relaxation. J. Biol. Chem. 2001;276:20220–20227. doi: 10.1074/jbc.M010991200. [DOI] [PubMed] [Google Scholar]
- 51.Christensen M.O., Barthelmes H.U., Boege F., Mielke C. Residues 190–210 of human topoisomerase I are required for enzyme activity in vivo but not in vitro. Nucleic Acids Res. 2003;31:7255–7263. doi: 10.1093/nar/gkg927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Frohlich R.F., Andersen F.F., Westergaard O., Andersen A.H., Knudsen B.R. Regions within the N-terminal domain of human topoisomerase I exert important functions during strand rotation and DNA binding. J. Mol. Biol. 2004;336:93–103. doi: 10.1016/j.jmb.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 53.Chillemi G., Castrignano T., Desideri A. Structure and hydration of the DNA-Human topoisomerase I covalent complex. Biophys. J. 2001;81:490–500. doi: 10.1016/S0006-3495(01)75716-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chillemi G., Fiorani P., Benedetti P., Desideri A. Protein concerted motions in the DNA-human topoisomerase I complex. Nucleic Acids Res. 2003;31:1525–1535. doi: 10.1093/nar/gkg242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fiorani P., Bruselles A., Falconi M., Chillemi G., Desideri A., Benedetti P. Single mutation in the linker domain confers protein flexibility and camptothecin resistance to human topoisomerase I. J. Biol. Chem. 2003;278:43268–43275. doi: 10.1074/jbc.M303899200. [DOI] [PubMed] [Google Scholar]
- 56.Fass D., Bogden C.E., Berger J.M. Quaternary, M changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nature Struct. Biol. 1999;6:322–326. doi: 10.1038/7556. [DOI] [PubMed] [Google Scholar]
- 57.Cabral J.H.M., Jackson A.P., Smith C.V., Shikotra N., Maxwell A., Liddington R.C. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–906. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 58.Go N., Noguti T., Nishikawa T. Dynamics of a small globular protein in terms of low-frequency vibrational-modes. Proc. Natl Acad. Sci. USA. 1983;80:3696–3700. doi: 10.1073/pnas.80.12.3696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levitt M., Sander C., Stern P.S. Protein normal-mode dynamics—trypsin-inhibitor, crambin, ribonuclease and lysozyme. J. Mol. Biol. 1985;181:423–447. doi: 10.1016/0022-2836(85)90230-x. [DOI] [PubMed] [Google Scholar]
- 60.Brooks B., Karplus M. Normal-modes for specific motions of macromolecules—application to the hinge-bending mode of lysozyme. Proc. Natl Acad. Sci. USA. 1985;82:4995–4999. doi: 10.1073/pnas.82.15.4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ma J.P., Karplus M. Ligand-induced conformational changes in ras p21: a normal mode and energy minimization analysis. J. Mol. Biol. 1997;274:114–131. doi: 10.1006/jmbi.1997.1313. [DOI] [PubMed] [Google Scholar]
- 62.Cui Q., Li G.H., Ma J.P., Karplus M. A normal mode analysis of structural plasticity in the biomolecular motor F-1-ATPase. J. Mol. Biol. 2004;340:345–372. doi: 10.1016/j.jmb.2004.04.044. [DOI] [PubMed] [Google Scholar]
- 63.Tama F., Sanejouand Y.H. Conformational change of proteins arising from normal mode calculations. Protein Eng. 2001;14:1–6. doi: 10.1093/protein/14.1.1. [DOI] [PubMed] [Google Scholar]
- 64.Krebs W.G., Alexandrov V., Wilson C.A., Echols N., Yu H.Y., Gerstein M. Normal mode analysis of macromolecular motions in a database framework: developing mode concentration as a useful classifying statistic. Proteins-Struct. Func. Genetics. 2002;48:682–695. doi: 10.1002/prot.10168. [DOI] [PubMed] [Google Scholar]
- 65.Delarue M., Sanejouand Y.H. Simplified normal mode analysis of conformational transitions in DNA-dependent polymerases: the elastic network model. J. Mol. Biol. 2002;320:1011–1024. doi: 10.1016/s0022-2836(02)00562-4. [DOI] [PubMed] [Google Scholar]
- 66.Tama F., Valle M., Frank J., Brooks C.L. Dynamic reorganization of the functionally active ribosome explored by normal mode analysis and cryo-electron microscopy. Proc. Natl Acad. Sci. USA. 2003;100:9319–9323. doi: 10.1073/pnas.1632476100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Benkovic S.J., Hammes-Schiffer S. A perspective on enzyme catalysis. Science. 2003;301:1196–1202. doi: 10.1126/science.1085515. [DOI] [PubMed] [Google Scholar]
- 68.Stivers J.T., Harris T.K., Mildvan A.S. Vaccinia DNA topoisomerase I: evidence supporting a free rotation mechanism for DNA supercoil relaxation. Biochemistry. 1997;36:5212–5222. doi: 10.1021/bi962880t. [DOI] [PubMed] [Google Scholar]
- 69.Kwon K., Stivers J.T. Fluorescence spectroscopy studies of vaccinia type IB DNA topoisomerase—closing of the enzyme clamp is faster than DNA cleavage. J. Biol. Chem. 2002;277:345–352. doi: 10.1074/jbc.M109449200. [DOI] [PubMed] [Google Scholar]
- 70.Kwon K., Jiang Y.L., Song F.H., Stivers J.T. F-19 NMR studies of vaccinia type IB topoisomerase—conformational dynamics of the bound DNA substrate. J. Biol. Chem. 2002;277:353–358. doi: 10.1074/jbc.M109450200. [DOI] [PubMed] [Google Scholar]
- 71.Sekiguchi J., Shuman S. Covalent DNA binding by vaccinia topoisomerase results in unpairing of the thymine base 5′ of the scissile bond. J. Biol. Chem. 1996;271:19436–19442. doi: 10.1074/jbc.271.32.19436. [DOI] [PubMed] [Google Scholar]
- 72.Weiss S. Measuring conformational dynamics of biomolecules by single molecule fluorescence spectroscopy. Nature Struct. Biol. 2000;7:724–729. doi: 10.1038/78941. [DOI] [PubMed] [Google Scholar]
- 73.Staker B.L., Hjerrild K., Feese M.D., Behnke C.A., Burgin A.B., Stewart L. The mechanism of topoisomerase I poisoning by a camptothecin analog. Proc. Natl Acad. Sci. USA. 2002;99:15387–15392. doi: 10.1073/pnas.242259599. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.