Abstract
Human Bex2 (brain expressed X-linked, hBex2) is highly expressed in the embryonic brain, but its function remains unknown. We have identified that LMO2, a LIM-domain containing transcriptional factor, specifically interacts with hBex2 but not with mouse Bex1 and Bex2. The interaction was confirmed both by pull-down with GST-hBex2 and by coimmunoprecipitation assays in vivo. Using electrophoretic mobility shift assay, we have demonstrated the physical interaction of hBex2 and LMO2 as part of a DNA-binding protein complex. We have also shown that hBex2 can enhance the transcriptional activity of LMO2 in vivo. Furthermore, using mammalian two-hybrid analysis, we have identified a neuronal bHLH protein, NSCL2, as a novel binding partner for LMO2. We then showed that LMO2 could up-regulate NSCL2-dependent transcriptional activity, and hBex2 augmented this effect. Thus, hBex2 may act as a specific regulator during embryonic development by modulating the transcriptional activity of a novel E-box sequence-binding complex that contains hBex2, LMO2, NSCL2 and LDB1.
INTRODUCTION
Interactions among transcriptional factors provide molecular mechanisms for the highly regulated temporal and spatial gene expression during embryonic development. Importantly, unveiling the function of a novel gene often relies on identifying its interactions with proteins of known function. We have identified a group of genes specifically expressed in human fetal brains (1) including human Bex2 (previously reported as human Bex1), a previously reported brain-expressed, X-linked protein of unknown function (2). The Bex gene family is composed of at least six members: Bex1/Rex3/EG2RVC/NADE4, Bex2, Bex3/NADE/HGR74 and three new Bexs, Bex4/5/6 (2–6). Two additional Bex orthologs, NADE2 and NADE3, were reported to be only expressed in human (7). Recently, the nomenclature for human Bex1 and Bex2 was revised since the previously designated human Bex1 has higher sequence identity with mouse Bex2 than with mouse Bex1 (74 and 68% identity, respectively) (8). With this revision in the nomenclature, the localization of Bex1–4 on the X chromosome from three species—human, rat and mouse—matches within the cluster (2,8). It has been shown that mouse Bex1 is transiently expressed during early development at the 2–8 cell stage, and then again expressed at the blastocyst stage (9). Murine Bex1 has also been suggested to play a role in neural development. For example, the expression of Bex1 was suppressed in F9 cells and PC12 cells upon retinoic acid or NGF treatment, respectively (4,10). Rat Bex1 was reported as one of the most commonly identified genes in a subtractive screen for ventral mesencephalic genes expressed at E10 (8). Additionally, the high expression profile of Bex1/2 in the adult brain supports a role of both proteins in the neural activity in mature neurons (2,6,8). However, the molecular mechanisms by which Bex proteins are involved in neurodevelopment remain elusive. Interestingly, both Bex1 and Bex2 were recently identified as binding partners for olfactory marker protein (OMP), a soluble protein possibly involved in olfactory signal transduction (11,12). Despite its interaction with OMP, Bex2's broader expression in the brain other than that restricted for OMP suggests that it may have a broader functional role in the brain (6). Moreover, mBex3, which has a relatively weaker sequence similarity with Bex1/2, was observed to interact with p75NTR in inducing apoptotic cell death (NADE) (5,7). Thus, the Bex family may play a dual role at the transcriptional and cytoplasmic signaling levels during embryonic development.
To determine the molecular and cellular mechanisms underlying the function of human Bex2 (previously named Bex1) (13), we have used a yeast two-hybrid system to screen for interacting proteins of hBex2, and identified LMO2 as a binding protein. LMO2, a member of the LIM-only class proteins and formerly named rhombotin-2 (rbtn-2 or Ttg-2), has been extensively reported to play a role in hematopoietic stem cell differentiation (14,15). The Drosophila homolog of LMO, dLMO, was also indicated in an evolutionarily conserved mechanism involved in patterning appendages (16). Interestingly, recent evidence indicates that dLMO controls the responsiveness of PDF-expressing ventral lateral neurons (principle circadian pacemaker cells) to cocaine, which in turn regulate the behavioral sensitivity of the flies to the drug (17). Furthermore, one of its distant relatives, LMO4, has been suggested to play a role in neurogenesis and neurodegenerative disease (18–20). As a highly expressed protein in the brain (21,22), LMO2 has been reported to be expressed in the hippocampus during development (23). This expression is up-regulated during seizure-induced responses, suggesting a role in neuronal regeneration after epileptic pathogenesis (24).
We next examined whether NSCL2, a neuronal form of Class II bHLH protein SCL, can interact with LMO2. It has been reported that LMO2 may interact with NSCL1 as an adaptor in addition to other transcription factors (25–27). However, this interaction failed to show direct regulation of transcriptional activity in a previous report (25). Here we have shown the functional interaction between LMO2 and NSCL2, suggesting the existence of a novel complex that is composed of hBex2, LMO2, NSCL2 and LDB1. The unveiling of this complex may help to understand the molecular mechanisms by which hBex2 plays a role in transcriptional regulation in the brain during neurodevelopment.
MATERIALS AND METHODS
cDNA plasmid construction
The full-length cDNAs for hBex1, hBex2, hBex3(NADE), mBex1, mBex2, mBex3 and various deletion mutants of hBex2 used in the yeast two-hybrid assay were cloned in frame with the GAL4 DNA-binding domain into vector pAS2-1 (BD_Clontech, CA). The Genbank accession numbers for the various Bex genes are: hBex1, AF251053 (for nomenclature change see Introduction); hBex2, AF237783; hBex3(NADE), AF187064; mBex1, AF097438; mBex2, AF097439; mBex3 and AF097440. The GAL4 AD fused LMO2 expressing plasmid was generated by inserting the LMO2 cDNA into pACT2 (BD Clontech, CA). The GST fusion protein constructs (pGEX-hBex2 and pGEX-LMO2) of hBex2 and LMO2 were constructed by in-frame insertion of hBex2 and LMO2 cDNAs into pGEX4T-1 and pGEX6P-1 (Amersham Pharmacia Biotech, Piscataway, NJ). hBex2-EGFP was constructed by in-frame insertion of the hBex2 cDNA with leading Kozak sequence into pEGFP-N1 vector (BD Clontech, CA). The chimeric constructs between hBex2 (H) and mBex1 (m) (mHm and HmH) were subcloned into pMRFP containing a monomer red fluorescent protein (R. Tsien, UCSD). pcDNA-LMO2 was constructed by inserting the LMO2 cDNA with leading Kozak sequence into pcDNA 3.1(+) vector. The same cDNA was used in constructing EGFP fused LMO2 expression plasmid pEGFP-LMO2. The PM3 (BD Clontech), PM3-LMO2, PM3-NSCL1 and PM3-NSCL2 are SV40 promoter-driven vectors expressing Gal4BD fused proteins. VP16 (BD Clontech), VP16-NSCL1 and VP16-NSCL2 are plasmids expressing VP16 fused proteins. The wild-type E-box-luciferase and the mutant form of E-box-luciferase were generated from pGAL-luc and were obtained as kind gifts from Dr Hakan Axelson.
Yeast two-hybrid screen
The full-length Bex2 was used as bait to screen a human fetal brain cDNA library as described in the user manual for the MATCHMAKER yeast two-hybrid system 2 (BD Clontech, CA). Yeast colonies with potential positive interactions were further tested for LacZ activity and survival on selective media. Plasmids containing cDNA for interacting proteins were extracted from the positive yeast clones and further sequenced with pACT2 specific primers.
Cell culture and transfection
All cells were maintained in 5% CO2 at 37°C in medium containing penicillin and streptomycin unless otherwise noted. HeLa and COS7 cells were maintained in DMEM (Invitrogen) with 10% cosmic calf serum (Hyclone, Logan, UT) and 2 mM l-glutamine, 1 mM sodium pyruvate and 0.1 mM nonessential amino acids. For transfection, cells were first plated on poly-d-lysine and Matrigel double-coated glass coverslips in a 24-well dish in culture medium without antibiotics the day before transfection. Cells on coverslips were then transfected by using LipofectAMINE 2000 according to the manufacturer's instructions (Invitrogen, San Diego, CA). After 12 h, transfected cells were fixed for immunofluorescence.
GST fusion protein and antiserum generation
pGEX-hBex2 and pGEX-LMO2 plasmids were transformed into Escherichia coli strain BL21(DE3). The protein expression and purification were performed according to the manufacturer's user manual (Amersham Pharmacia Biotech, Piscataway, NJ). Two New Zealand white female rabbits were immunized with 600 µg purified protein in complete Freund's adjuvant (Sigma, St Louis, MO) and were boosted three times with 300 µg protein in incomplete Freund's adjuvant at 3 week intervals. The titer and specificity of antisera were then determined by ELISA after each boost until the titer reached 1:1 × 106. The antisera were further tested by western blot against either the recombinant or expressed proteins.
GST fusion protein mediated pull-down assay
GST or GST-hBex2 fusion protein was expressed in E.coli strain BL21(DE3) and purified with Sepharose CL-4B resin (Amersham Pharmacia Biotech, Piscataway, NJ). The cell lysates from either pEGFP- or pEGFP-LMO2-transfected HeLa cells were then incubated with GST or GST-hBex2 bound resin at 4°C for 4 h. The pellets were washed three times with phosphate-buffered saline (PBS), separated by 10% SDS–PAGE and analyzed with western blot.
Immunofluorescent microscopy
HeLa cells transfected with the FLAG-tagged hBex2 and pEGFP-LMO2 were grown on 25 mm glass coverslips. The cells were fixed with 4% PFA and permeabilized with 0.1% Triton X-100 in PBS (pH 7.4). Cells were then incubated with anti-FLAG antibody (M2, Sigma, St Louis, MO), followed by incubation with TRITC conjugated secondary antibody. Confocal microscopic analysis was performed as described previously (28).
Electrophoretic mobility shift assay (EMSA)
EMSAs were performed with 32P-labeled double-strand oligonucleotides as described previously (29). Oligonucleotides containing the E-box motif (underlined sequences) were synthesized as follows: 5′-CTGGCTAGCGGGTCGCAGCTGCGTCCCGGGTCGCAGCTGCGTCCCGGGTCGCAGCTGCGTCCCCTCGAGATC-3′ and 5′-GATCTCGAGGGGACGCAGCTGCGACCCGG GACGCAGCTGCGACCCGGGACGCAGCTGCGACCCGCTAGCCAG-3′ (underlined is the hexameric sequence of E box). Nuclear extracts from M17 cells were prepared and incubated with labeled oligonucleotide for 20 min at room temperature, followed by incubation with pre-immune serum, anti-LMO2 anti-serum or anti-hBex2 anti-serum. The gel was dried, exposed to Kodak film overnight and then processed.
Transfection of cDNA plasmid into mammalian cells and luciferase assay
Cos-7 cells growing in 12-well plates with 90% confluence were transfected with 1 µg cDNA plasmid per well using LipofectAMINE 2000 as described in the manufacturer's user manual (Invitrogen, Carlsbad, CA). After 24 h, cell extracts were prepared and luciferase reporter assays were performed using the Dual-Luciferase Reporter Assay System (Catalog no. E1910, Promega, Madison, WI). The luciferase activity was measured using the LB9507 Luminometer (EG&G Berthold, Wellesley, MA). The transcriptional activity of the reporter gene was measured by relative luciferase activity value, which is the value of firefly luciferase activity divided by the value of renilla luciferase activity.
Coimmunoprecipitation
Fetal brain tissue (50 g) was homogenized in 250 ml of buffer A [10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EGTA, 0.5 mM phenylmethlysulfonyl fluoride (PMSF), 1 mM DTT, and 10 mg/ml for each of the following protease inhibitors: leupeptin, aprotinin and pepstatin]. The homogenate was then treated with NP-40 (0.2% final) for 15 min on ice and centrifuged for 15 min at 15 000 g, 4°C. The nuclear pellet was resuspended in 20 ml of buffer B (10 mM HEPES, pH 7.9, 0.1 mM EGTA, 0.4 M NaCl, 0.5 mM PMSF, 1 mM DTT and protease inhibitors) and incubated with rotation at 4°C for 15 min. The extract was then centrifuged, and the supernatant was dialyzed against 500 ml of buffer C (10 mM Tris–HCl, pH 7.9, 120 mM NaCl, 0.3% NP-40, 0.25% BSA and 1 mM DTT) at 4°C. Nuclear extracts (∼100 mg protein) were incubated with polyclonal anti-hBex2, anti-LDB1 or control antisera for 6 h at 4°C. The complexes were precipitated with protein A or G agarose, washed four times in RIPA buffer (50 mM Tris, pH 7.5, 150 mM NaCl, 5 mM EDTA, 1% Nonidet P-40, 1% deoxycholate and 0.1% SDS), and then detected by western blot using antisera α-LMO2 (Santa Cruz Biotechnology, Santa Cruz, CA) or α-NSCL2, (CeMines, Inc., Golden, CO), as indicated.
RESULTS
Identification of LMO2 as an interacting partner for hBex2
To gain insight into the function of hBex2 in development, we used a human fetal brain cDNA library in a Gal4 yeast two-hybrid system (BD Clontech, CA) to identify interacting proteins. Using the full-length hBex2 as bait, we screened ∼1 × 107 transformants and acquired ∼59 His and β-galactosidase positive yeast colonies. Following cDNA plasmid extraction and sequencing analysis, we chose to focus on a candidate cDNA clone corresponding to the complete coding sequence of LMO2, a LIM containing a transcriptional regulator involved in neurogenesis (24). The interaction between two proteins was further verified by retransformation into yeast strain SFY-526 to exclude the possibility of a false positive as a result of self-activation.
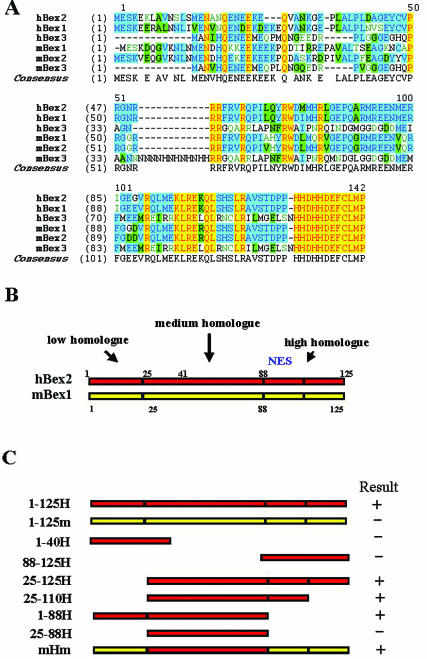
To determine the specificity of the interaction between LMO2 and hBex2 in the yeast two-hybrid system, we tested whether LMO2 could interact with other members of the Bex family, including hBex3, mBex1, mBex2 and mBex3. For a positive control, we also used the TAX protein of human T-cell leukemia virus type1 because of its self-association (28) (Table 1). Among the five Bex homolog genes tested in the yeast Gal4-BD construct, only hBex2 interacted with LMO2 (Table 1). More specifically, although it shares 74% amino acid sequence identity with hBex2 (Figure 1), mBex2 failed to show its interaction with LMO2 in our assay. Similarly, mBex1 also showed no interaction with LMO2 although it also shares high sequence identity (68%) with hBex2. Thus, the interaction of human Bex2 and LMO2 might be a human species-specific event.
Table 1.
Identification of LMO2 as an interaction partner for hBex2
| AD plasmid | BD plasmid | Result |
|---|---|---|
| pAS2-1-TAX | pACT2-TAX | + |
| pAS2-1 | pACT2 | − |
| pAS2-1 | pACT2-LMO2 | − |
| pAS2-1-hBex2 | pACT2 | − |
| pAS2-1-hBex2 | pACT2-LMO2 | + |
| pAS2-1-hBex3 | pACT2-LMO2 | − |
| pAS2-1-mBex1 | pACT2-LMO2 | − |
| pAS2-1-mBex2 | pACT2-LMO2 | − |
| pAS2-1-mBex3 | pACT2-LMO2 | − |
GAL4 yeast two-hybrid system was used to evaluate the interaction of genes in the Bex family with LMO2. TAX protein of human T-cell leukemia virus type 1 was used as a positive control. The interactions of listed GAL4–BD and GAL4–AD fusion pairs were indicated only as positive or negative for β-galactosidase activity tested in the yeast strain SFY526.
Figure 1.
(A) Multiple alignment of members of the Bex family. The C-terminals of the Bexs (hBex2 88–125) are conserved throughout the family, and the last 12 amino acids (114–125) are identical. hBex2 shares 74% amino acid sequence identity with mBex2 and 68% identity with mBex1. (B) Schematic diagram of hBex2 deletion constructs that were tested for interaction with LMO2 in yeast two-hybrid assay. hBex2 can be divided into three regions according to similarity analysis among members of the Bex family: the N-terminal low homolog region (1–25), the middle medium homolog region (25–88) and the C-terminal high homolog region (88–125). Deletion mutations were constructed accordingly. (C) Mapping of the LMO2-binding domain in hBex2. The interactions of the truncation mutation of hBex2 with full-length LMO2 were examined by a β-galactosidase activity assay in the yeast two-hybrid system. mHm indicates the chimeric construct containing the middle part of human Bex2 flanked by N- and C-termini of mouse Bex1. The TAX gene was also included as a positive control. The plus symbol indicates the positive binding between the transformed constructs.
To identify the binding domain within hBex2 responsible for interaction between hBex2 and LMO2, we generated several deletion and chimeric hBex2 mutants for yeast two-hybrid analysis. As shown in Figure 1A, members of the Bex family share sequence similarity mainly in the middle (58% identity) and C-terminal (95% identity) regions. Indicated by the yeast two-hybrid assay, the N-terminal region with relatively lower sequence similarity between hBex2 and mBex1 (1–26 amino acid sequence, 48% identity) was not required for binding to LMO2, nor was the C-terminal region (88–125), which shares high sequence homology among Bex family members (Figure 1C). In contrast, all constructs containing the middle region of hBex2 (26–88) expressed strong interaction (Figure 1C). However, the middle-region fragment alone does not bind to LMO2, suggesting that the sequence itself is insufficient for the binding. Interestingly, the chimeric mBex1 containing a swapped middle fragment (26–88) of hBex2 (mHm) showed positive interaction with LMO2 (Figure 1C), further supporting the specific requirement of this fragment within the hBex2 sequence for protein–protein interaction.
Protein–protein interaction between hBex2 and LMO2
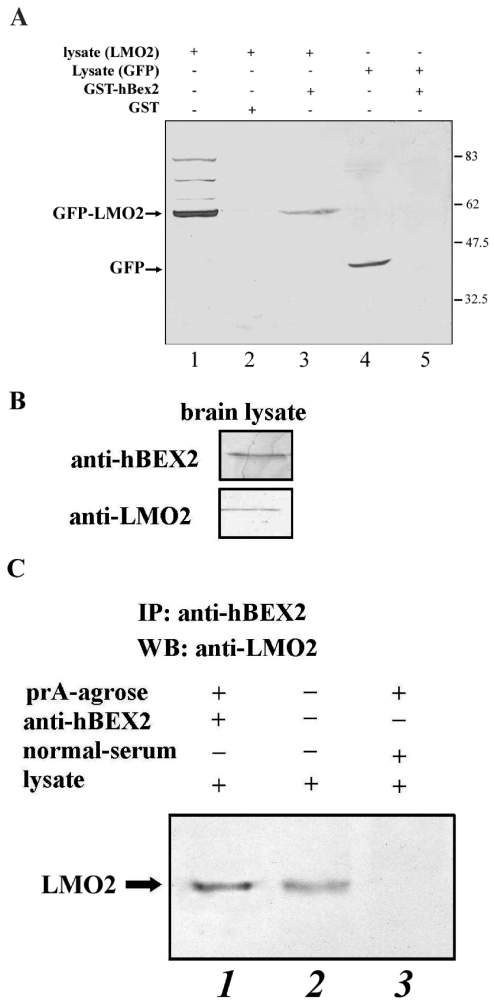
To further characterize the interaction between hBex2 and LMO2, we performed GST pull-down assays. GST and GST-hBex2 fusion proteins were used to test their interaction with EGFP-tagged LMO2 from cell extracts from transiently transfected HeLa cells. As shown in Figure 2A, EGFP-LMO2 bound to GST-hBex2 (lanes 3) but not to GST alone, as indicated by the antibody to the LMO2 fusion protein (lanes 2). We further tested whether human Bex2 interacts with LMO2 in vivo. As shown in Figure 2B, both proteins were expressed in human fetal brain extracts. Coimmunoprecipitation of hBex2 and LMO2 from these extracts using anti-hBex2 Ab indicates that their interaction may occur during development (Figure 2C).
Figure 2.
Interaction between hBex2 and LMO2 in vitro and in vivo. (A) pEGFP-LMO2 or pEGFP (short for pEGFP–LMO2 lysate or pEGFP lysate) were transiently expressed in HeLa cells. Cell lysates were used to perform pull-down assay by incubating with GST-hBex2 or GST bound to glutathione beads. Western analysis was performed with anti-GFP antibody. Lane 1, pEGFP-LMO2 cell lysate only; lane 2, GST pull-down from pEGFP-LMO2 lysate; lane 3, GST-hBex2 pull-down of pEGFP-LMO2 lysate; lane 4, pEGFP cell lysate only; lane 5, GST-hBex2 pull-down of pEGFP lysate. (B) Endogenous expression of hBex2 and LMO2 in human fetal brain to confirm the specificity of the antibodies. (C) Interaction of hBex2 and LMO2 in human fetal brain tissue. The fetal brain tissue was used to immunoprecipitate the human Bex2 with a polyclonal Ab. Precipitate was blotted with an anti-LMO2 Ab.
Nuclear colocalization of hBex2 and LMO2
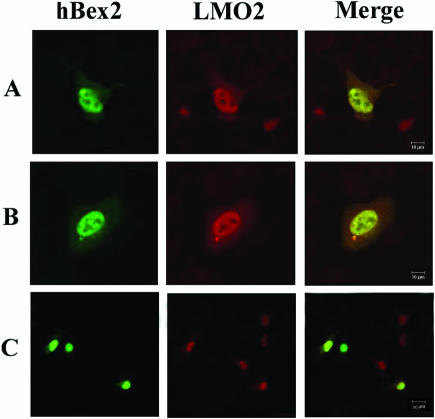
LMO2 is localized to the nucleus to play a role in transcriptional regulation (22). The interaction of hBex2 with LMO2 suggests their subcellular colocalization in the nucleus. Consistent with this prediction, evidence suggests that hBex2's nuclear export sequence (NES) may be insufficient for transporting the protein out of the nucleus (7). However, it has been suggested that hBex2 has a cytoplasmic localization pattern when it is overexpressed in HEK293 cells (8). To clarify this issue, we examined the subcellular localization of both endogenous and overexpressed hBex2 and LMO2 in cell lines. As shown in Figure 3, the endogenous Bex2 and LMO2 were primarily colocalized in the nuclei of human neuroblastoma M17 cells by double immunofluorescent staining. Similarly, the heterogeneously expressed FLAG-hBex2 and dsRed-LMO2 were also colocalized in the nuclei of transient transfected HeLa cells (Figure 3B). The nuclear localization of either protein does not seem to be facilitated by the overexpression of their binding partners (Figure 3C).
Figure 3.
Nuclear colocalization of hBex2 and LMO2. (A) Endogenous Bex2 and LMO2 were colocalized in the nuclei of M17 cells using specific antibodies for immunofluorescent staining. (B) FLAG-tagged hBex2 and dsRed-LMO2 were colocalized in the transient transfected HeLa cells. Anti-FLAG mAb and TRITC-conjugated secondary anti-mouse antibody were used sequentially to detect the expression of hBex2. The staining image was collected with a 60× objective. (C) Same as (B) with a 20× objective. Bars indicate 10 µm.
hBex2 increases the transcriptional activity of LMO2
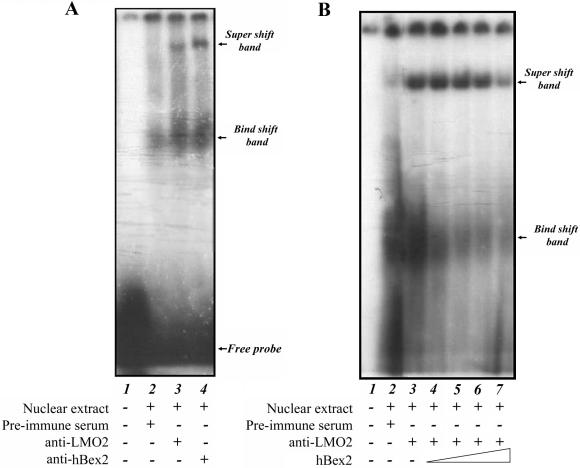
Previous studies have suggested that LMO2 can form a DNA-binding complex mediated by the E-box sequence within Class II bHLH transcription factors (29,30). We speculated that hBex2 might be involved in the complex formation and play a role in transcriptional regulation. To test this hypothesis, we performed EMSAs using a polyclonal antiserum against LMO2 and hBex2. As shown in Figure 4A, we found that anti-hBex2 antibody could generate a super-shift band similar to that seen in the lane with anti-LMO2 antibody. These data indicate that hBex2 participates in the formation of a LMO2/E-box complex without interfering in the interaction of LMO2 with other members of the complex that bind to the E-box motif. Furthermore, when the recombinant hBex2 was used in the assay, it clearly blocked the shift in a concentration-dependent manner (Figure 4B), supporting the specificity of the antibodies for the interactions.
Figure 4.
hBex2 is part of a complex with LMO2 that recognizes the E-box element. (A) EMSAs were performed with the γ32-ATP-labeled oligonucleotides of the E-box sequence using the nuclear extract from M17 cells. Labeled probe was incubated with nuclear extract for 20 min at room temperature (lanes 2–4), then further incubated with pre-immune serum (lane 2), anti-LMO2 anti-serum (lane 3) or anti-hBex2 anti-serum (lane 4). The super-shift band and bind-shift band are indicated by arrows. (B) The specificity of the formation of bind-shift band and super-shift band by interaction between hBex2 protein and its antibody. Purified hBex2 protein was added into the binding assay to compete with the anti-hBex2 Abs as follows: 0 µg (lane 3), 10 µg (lane 4), 20 µg (lane 5), 50 µg (lane 6) and 100 µg (lane 7). The competitive binding of the recombinant hBex2 reduced the intensity of both the bind-shift band and the super-shift band.
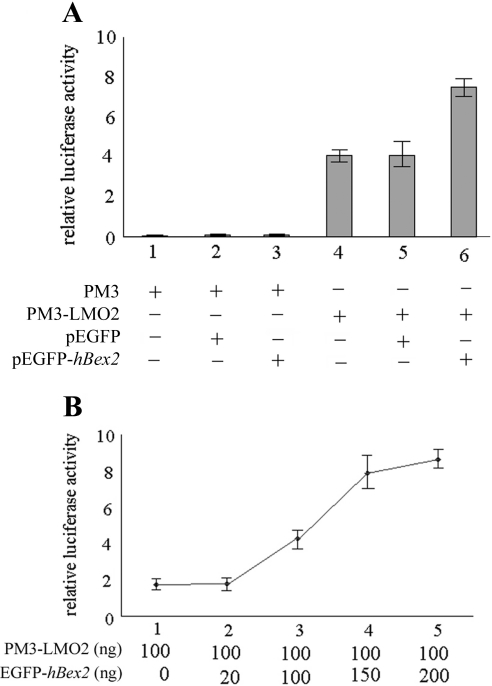
The transcriptional regulation activity of LMO2 has been detected by using reporter genes containing either the E-box motif (31,32) or Gal4-binding sequences in which LMO2 was a fusion protein with Gal4-BD domain (33). In order to examine whether the interaction between LMO2 and hBex2 affects the transcriptional activity of LMO2, we performed the luciferase reporter gene assay using the Gal4 system (25,33). cDNA plasmids for GFP-hBex2 and Gal4-BD-LMO2 were cotransfected into Cos-7 cells together with a plasmid for luciferase reporter gene driven by Gal4-binding enhancer. GFP-hBex2 alone had no effect on the luciferase activity (Figure 5A, column 3), while LMO2 activated the expression of the reporter gene (Figure 5A, column 4). When hBex2 and LMO2 were coexpressed, the luciferase activity was significantly increased compared with that in LMO2 expression alone (Figure 5A, column 6). The hBex2-mediated up-regulation of GAL4-BD-LMO2-dependent luciferase activity also correlates with the amount of hBex2 cDNA used in transfection (Figure 5B).
Figure 5.
hBex2 enhances the transcriptional activity of LMO2. (A) pEGFP-hBex2 increases LMO2-induced luciferase activity in a mammalian GAL4 system. The reporter plasmid contains a GAL4-BD-binding motif within the promoter sequence upstream of the luciferase gene, allowing the binding of the LMO2-GAL4-BD chimeric protein. Cos-7 cells were transiently transfected with pGAL-LUC (firefly luciferase reporter gene plasmid) and TK (renilla luciferase expression plasmid) as reporter gene plasmids and expression gene plasmids including pM3-LMO2 and pEGFP-hBex2. pM3 and pEGPF were used as control vector DNAs. The transcriptional activity of the reporter gene was measured by relative luciferase activity value, which is the value of firefly luciferase activity divided by the value of renilla luciferase activity. All indicated relative luciferase activity values represent the means of three independent transfections, and error bars indicate the standard deviations. (B) The effect of hBex2 on LMO2 activity is correlated with the amount of hBex2 cDNA plasmid. Increased amounts of cDNA plasmids were used in the luciferase system as indicated from 0 to 200 ng.
LMO2 interacts with NSCL1 and NSCL2
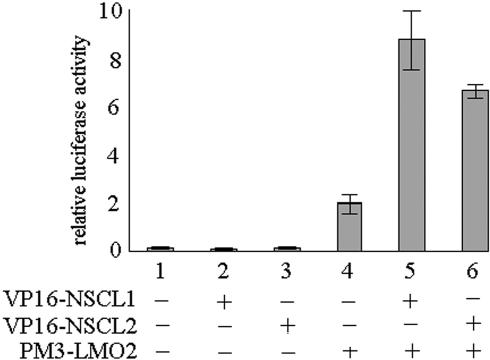
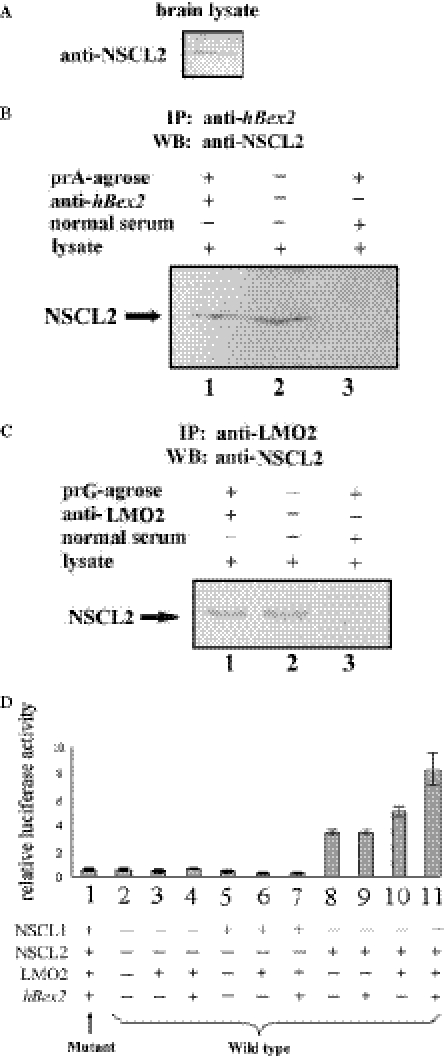
To further elucidate the mechanisms by which hBex2 regulates LMO2 function at the transcriptional level, we examined the interaction of LMO2 with class II bHLH proteins during the formation of a DNA-binding complex. There are two class II bHLH proteins, SCL (also known as TAL-1) and NSCL1, which are known to interact with LMO2 (25,29). Interestingly, LMO2 interacts with the brain specific SCL homologue NSCL1, but showed no effect on their transcriptional activity (25). To test whether LMO2 interacts with NSCL2, a NSCL1 homolog with high sequence similarity (34,35) that regulates its transcriptional function, we performed a mammalian two-hybrid assay. Both pGAL4-BD-LMO2 and pV16-NSCL2 fusion proteins were expressed in HeLa cells to examine their interaction as indicated by luciferase activity. As a control, pPM3-LMO2 and pVP16-NSCL1 were coexpressed and were found to induce luciferase activity, consistent with the previous report of their interaction (25) (Figure 6, column 5). Coexpression of GAL4-BD-LMO2 and VP16-NSCL2 significantly activated transcription of the reporter gene. This indicates that NSCL2 is also a binding partner for LMO2 (Figure 6, column 6). To test if NSCL2 is part of the complex of hBex2 and LMO2 in vivo, we performed coimmunoprecipitation with human fetal brain tissue. As shown in Figure 7A–C, NSCL2 was coexpressed and coimmunoprecipitated with hBex2 and LMO2 in human fetal brain extracts.
Figure 6.
Functional analysis of interaction between LMO2 and NSCL1 or NSCL2. A mammalian two-hybrid system was used to detect the protein–protein interaction as indicated by luciferase activity assay. Cos-7 cells were transiently transfected with pGALLUC and TK as reporter plasmids along with expression plasmids as indicated. The interactions were measured by relative luciferase activity. All experiments were repeated more than three times with triplication for each experiment. Error bars indicate standard deviations.
Figure 7.
hBex2 regulates NSCL2 activity through formation of transcriptional complex. (A) Endogenous expression of NSCL2 in human fetal brain to confirm the specificity of its polyclonal antibody. (B and C) Interaction of hBex2 and LMO2 with NSCL2, respectively, in human fetal brain tissue. The fetal brain tissue was used to immunoprecipitate the hBex2 (B) or LMO2 (C) with specific polyclonal Abs. Precipitate was blotted with anti-NSCL2 Ab. (D) hBex2 regulates E-box-dependent gene expression through LMO2 and NSCL2. The reporter system used here contains three tandem repeat sequences of wild-type E-box motif adjacent to the firefly luciferase. The mutant reporter gene contains an inactive form of E-box motif as a control. Cos-7 cells were transiently transfected with reporter gene plasmids along with expression plasmids as indicated. A renilla luciferase plasmid was cotransfected in each transfection to evaluate transfection efficiency. All experiments were repeated more than three times with triplication for each experiment. Error bars indicate standard deviations.
hBex2 influences the transcriptional activity of LMO2 and NSCL2
To further explore the physiological relevance of the interaction between LMO2 and NSCL2, and the potential regulatory role of hBex2 on the LMO2–NSCL2 complex, we generated a reporter construct containing three tandem repeated E-boxes adjacent to the firefly luciferase gene. In the absence of NSCL2, neither LMO2 alone, nor the coexpression of hBex2 and LMO2, had any effect on the basal luciferase activity (Figure 7D, columns 3–4). NSCL2, but not NSCL1, was able to activate transcription of the reporter gene (Figure 7D, columns 5 and 8), consistent with the previous report that NSCL1 was a relatively weak transcriptional activator (25). It has been reported that the enhancement or repression of class II bHLH proteins on transcriptional activity depends on both the target genes and the cellular context (32). Furthermore, LMO2 was able to up-regulate NSCL2's transcriptional activity (Figure 7D, column 10), but had no effect on NSCL1 (column 6). Importantly, coexpression of hBex2 augmented LMO2's effect on NSCL2 (column 11). In contrast, expression of hBex2 alone (data not shown) or together with LMO2 did not affect NSCL1-dependent transcription (Figure 7D, column 7). This result suggests that the ability of hBex2 to up-regulate the transcriptional activity of NSCL2 may be mediated by LMO2 within the complex.
hBex2 complex contains NL1/CLIM2/LDB1
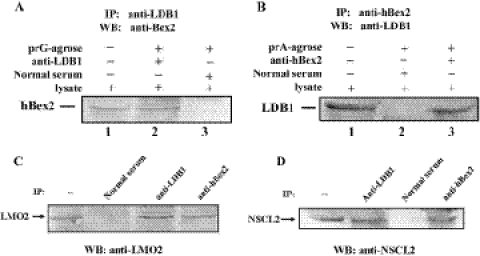
LDB1 is an important cofactor that is involved in the assembly of multiprotein complexes through interacting with the LIM-domain (hence LDB1—LIM domain-binding protein 1) (36). To examine if LDB1 is part of the hBex2 complex in vivo, we again performed coimmunoprecipitation with human fetal brain tissue. As shown in Figure 8A and B, both LDB1 and hBex2 were coexpressed in human fetal brain extracts and coimmunoprecipitated by either anti-hBex2 or anti-LDB1 antibody. Furthermore, we used antibodies to LDB1 and hBex2 for immunoprecipitation and confirmed that both LMO2 and NSCL2 are part of the complex now composed of at least hBex2, LMO2, NSCL2 and LDB1 (Figure 8C and D).
Figure 8.
Interaction of LDB1 with hBex2 mediated complex in vivo. Coimmunoprecipitation was performed using nuclear extracts from 4-month-old human fetal brain. Antibodies against either LDB1 (A) or hBex2 (B) were used to precipitate the protein complex, followed by western blotting conversely with two antibodies. Similar coimmunoprecipitation was performed to confirm the interaction of LMO2 (C) or NSCL2 (D) with LDB1 and hBex2.
DISCUSSION
In this report, we have presented evidence supporting the notion that human Bex2 (previously named human Bex1) can specifically bind to LMO2 and regulate the transcriptional activity of the E-box sequence-binding complex containing LMO2, NSCL2 and LDB1. Using a yeast two-hybrid system, we first identified LMO2 as a binding partner of hBex2. The interaction was characterized by both protein–protein interaction in vitro and nuclear colocalization. Deletion mutation analysis revealed that the interaction is a human Bex2 specific regulatory mechanism. Immunoprecipitation experiments with human fetal brain tissue further showed the interactions among the three proteins. Using EMSA and luciferase activity assay, we have demonstrated that hBex2 can physically interact with LMO2 and enhances its transcriptional activity through the formation of an E-box-binding complex. We further examined the interaction of LMO2 with the neuronal bHLH protein, NSCL2, and first showed that NSCL2 serves as functional binding partner for LMO2. Our work also suggests that hBex2 is able to augment the transcriptional activity of the target gene upon expression of all three proteins in vitro. Thus, hBex2 may act as a transcriptional regulator to modulate the activity of LMO2 and NSCL2, which play an important role in embryonic development.
Functional interaction between hBex2 and LMO2
The interaction of hBex2 with LIM domain-only protein, LMO2, was demonstrated in four independent assays: yeast two-hybrid analysis, in vitro affinity chromatography, nuclear colocalization and immunoprecipitation in vivo. As a ‘bridge molecule’, LMO2 plays a role in transcriptional regulation through its interaction with other proteins as a DNA-binding complex. There are two possible consequences of a direct interaction between hBex2 and LMO2. One is that hBex2 inhibits LMO2 from binding to other proteins. The second is that hBex2 facilitates the formation of an LMO2-containing protein complex. Our EMSA experiment supports the latter possibility (Figure 4). Furthermore, hBex2 up-regulates transcriptional activity of LMO2 through protein–protein interaction (Figure 5). Our work has further unveiled a novel binding partner for LMO2, NSCL2. hBex2 augmented NSCL2-dependent transcriptional activity, presumably through interacting with LMO2 (Figure 7). LMO2 contains two transactive domains in both the N- and C-terminus as well as two repressive LIM domains. The mechanisms by which hBex2 up-regulates LMO2 transcriptional activity may be a result of inhibiting the transcriptional activity of the complex through blocking both repressive LIM domains of LMO2, although our attempt in mapping the binding domain within LMO2 did not yield any conclusive information (data not shown).
Relevance of the novel LMO2 complex in neuronal function
Studies on the interaction between LMO2 and SCL in hematopoiesis has implicated a functional role of bHLH proteins in the LIM-modulation system (32,37). Since both hBex2 and LMO2 are expressed in areas of the brain where SCL is not detected, NSCL1 and NSCL2 became natural candidate bHLH proteins to examine their participation in the binding complex for the E-box motif (34,38,39). We have shown that LMO2 does not affect NSCL1-dependent transcription, consistent with a previous finding (25). However, we cannot exclude a potential regulatory role of NSCL1 in an LMO2 complex. The interaction between LMO2 and NSCL2 reported here suggests a neuronal-specific regulatory mechanism mediated by LMO2 and its binding partners. Furthermore, our work and that of others also provide evidence indicating a potential role for hBex2 complex in neurodevelopment. This evidence includes the high expression of hBex2, LMO2 and NSCL2 in brain; fetal brain tissue-derived yeast two-hybrid system used for detecting their interaction; and demonstration of interaction in vivo with human fetal brain tissue. However, further investigation should be performed to determine whether these proteins are colocalized in the same region or are differentiating neurons in embryonic brain tissue.
Nuclear translocation of hBex2 and its function
Consistent with bioinformatics predictions and physiological function, our study suggests that hBex2 is localized to the nucleus in order to play a role in transcriptional regulation. Interestingly, previous work has suggested that members of Bex family may differ in their translocation between the cytoplasm and nucleus. For example, the interaction between Bex and OMP may induce the nuclear localization of Bex (11). The overexpression of human Bex2 in HEK293 cells has shown a cytoplasmic localization pattern (8). The discrepancy between our work and this reported result may be due to the use of different cell lines. Future work on subcellular localization of hBex2 in neuronal tissue would be helpful to clarify this issue. In the case of hBex3 or NADE, it is predominantly distributed in the cytoplasm for its role in the P75NTR signaling pathway (5). It has been suggested that the protein translocation of Bex3 from the nucleus to the cytosol is required for this function (5). This translocation is determined by a NES motif, although mechanisms of regulating the subcellular trafficking remain elusive (5).
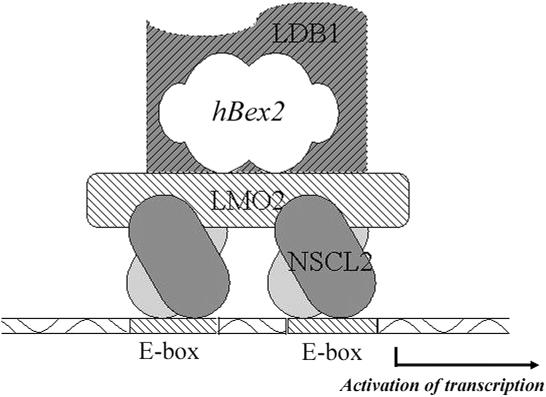
Finally, our study supports a working model in which hBex2 regulates the transcriptional activity of oligomeric DNA-binding complex containing LMO2 and NSCL2 (Figure 9). We suggest that human Bex2 binds to LMO2 as part of the oligomeric complex that binds to the E-box motif-containing sequence. This interaction may regulate the transcriptional activity of LMO2 through its binding to NSCL2 within the DNA-binding complex. The stoichiometry of the complex is unknown, but the E-box-binding modules of the oligomeric complex are provided by at least one NSCL2 molecule. Another half of an E-motif-binding protein could be either NSCL2 or another closely related bHLH protein. Therefore, the complex containing hBex2, LMO2, NSCL2 and LDB1 will be formed at the E-box site to regulate specific gene expression.
Figure 9.
hBex2 regulates the transcriptional activity of oligomeric DNA-binding complex containing LMO2 and NSCL2. Human Bex2 binds to LMO2 as part of the oligomeric complex that binds to the E-box motif-containing sequence. This interaction may regulate the transcriptional activity of LMO2 through its binding to NSCL2 and LDB1 during the formation of DNA-binding complex. The stoichiometry of the complex is unknown, but the E-box-binding modules of the oligomeric complex are provided by at least one molecule of NSCL2. The other half-E-motif-binding protein may also be NSCL2, or other bHLH proteins. hBex2, LMO2, NSCL2 and LDB1 form a complex at the E-box site and regulate specific gene expression.
Acknowledgments
We thank Ms. Lesley Colgan for critical review of this manuscript. This work was supported by grants from the National Program for the Key Basic Research Project (‘973’-No. 2001CB510206, 2005CB522507, 2004CB518604, 2004CB518604 and 2005CB522400), the National High Technology Research and Development Program (2004AA221060) and the National Sciences Foundation of China (No. 30421003 and 30430200). Part of the work was also supported by grant from NIMH to YL (R01 MH62092-01A1). Funding to pay the Open Access publication charges for this article was provided by National Program for the Key Basic Research Project, China.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pan M., Yuan J., Chen B., Zhou Y., Qiang B. [Differential display analysis of human fetal brain mRNAs and isolation of brain-specific novel ESTs] Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 1997;19:93–99. [PubMed] [Google Scholar]
- 2.Brown A.L., Kay G.F. Bex1, a gene with increased expression in parthenogenetic embryos, is a member of a novel gene family on the mouse X chromosome. Hum. Mol. Genet. 1999;8:611–619. doi: 10.1093/hmg/8.4.611. [DOI] [PubMed] [Google Scholar]
- 3.Rapp G., Freudenstein J., Klaudiny J., Mucha J., Wempe F., Zimmer M., Scheit K.H. Characterization of three abundant mRNAs from human ovarian granulosa cells. DNA Cell Biol. 1990;9:479–485. doi: 10.1089/dna.1990.9.479. [DOI] [PubMed] [Google Scholar]
- 4.Faria T.N., LaRosa G.J., Wilen E., Liao J., Gudas L.J. Characterization of genes which exhibit reduced expression during the retinoic acid-induced differentiation of F9 teratocarcinoma cells: involvement of cyclin D3 in RA-mediated growth arrest. Mol. Cell. Endocrinol. 1998;143:155–166. doi: 10.1016/s0303-7207(98)00127-0. [DOI] [PubMed] [Google Scholar]
- 5.Mukai J., Hachiya T., Shoji-Hoshino S., Kimura M.T., Nadano D., Suvanto P., Hanaoka T., Li Y., Irie S., Greene L.A., et al. NADE, a p75NTR-associated cell death executor, is involved in signal transduction mediated by the common neurotrophin receptor p75NTR. J. Biol. Chem. 2000;275:17566–17570. doi: 10.1074/jbc.C000140200. [DOI] [PubMed] [Google Scholar]
- 6.Koo J.H., Saraswati M., Margolis F.L. Immunolocalization of Bex protein in the mouse brain and olfactory system. J. Comp. Neurol. 2005;487:1–14. doi: 10.1002/cne.20486. [DOI] [PubMed] [Google Scholar]
- 7.Mukai J., Suvant P., Sato T.A. Nerve growth factor-dependent regulation of NADE-induced apoptosis. Vitam. Horm. 2003;66:385–402. doi: 10.1016/s0083-6729(03)01011-2. [DOI] [PubMed] [Google Scholar]
- 8.Alvarez E., Zhou W., Witta S.E., Freed C.R. Characterization of the Bex gene family in humans, mice, and rats. Gene. 2005;357:18–28. doi: 10.1016/j.gene.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 9.Williams J.W., Hawes S.M., Patel B., Latham K.E. Trophectoderm-specific expression of the X-linked Bex1/Rex3 gene in preimplantation stage mouse embryos. Mol. Reprod. Dev. 2002;61:281–287. doi: 10.1002/mrd.10100. [DOI] [PubMed] [Google Scholar]
- 10.Lee N.H., Weinstock K.G., Kirkness E.F., Earle-Hughes J.A., Fuldner R.A., Marmaros S., Glodek A., Gocayne J.D., Adams M.D., Kerlavage A.R., et al. Comparative expressed-sequence-tag analysis of differential gene expression profiles in PC-12 cells before and after nerve growth factor treatment. Proc. Natl Acad. Sci. USA. 1995;92:8303–8307. doi: 10.1073/pnas.92.18.8303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behrens M., Margolis J.W., Margolis F.L. Identification of members of the Bex gene family as olfactory marker protein (OMP) binding partners. J. Neurochem. 2003;86:1289–1296. doi: 10.1046/j.1471-4159.2003.01940.x. [DOI] [PubMed] [Google Scholar]
- 12.Koo J.H., Gill S., Pannell L.K., Menco B.P., Margolis J.W., Margolis F.L. The interaction of Bex and OMP reveals a dimer of OMP with a short half-life. J. Neurochem. 2004;90:102–116. doi: 10.1111/j.1471-4159.2004.02463.x. [DOI] [PubMed] [Google Scholar]
- 13.Yang Q.S., Xia F., Gu S.H., Yuan H.L., Chen J.Z., Ying K., Xie Y., Mao Y.M. Cloning and expression pattern of a spermatogenesis-related gene, BEX1, mapped to chromosome Xq22. Biochem. Genet. 2002;40:1–12. doi: 10.1023/a:1014565320998. [DOI] [PubMed] [Google Scholar]
- 14.McGuire E.A., Hockett R.D., Pollock K.M., Bartholdi M.F., O'Brien S.J., Korsmeyer S.J. The t(11;14)(p15;q11) in a T-cell acute lymphoblastic leukemia cell line activates multiple transcripts, including Ttg-1, a gene encoding a potential zinc finger protein. Mol. Cell. Biol. 1989;9:2124–2132. doi: 10.1128/mcb.9.5.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boehm T., Foroni L., Kaneko Y., Perutz M.F., Rabbitts T.H. The rhombotin family of cysteine-rich LIM-domain oncogenes: distinct members are involved in T-cell translocations to human chromosomes 11p15 and 11p13. Proc. Natl Acad. Sci. USA. 1991;88:4367–4371. doi: 10.1073/pnas.88.10.4367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zeng C., Justice N.J., Abdelilah S., Chan Y.M., Jan L.Y., Jan Y.N. The Drosophila LIM-only gene, dLMO, is mutated in Beadex alleles and might represent an evolutionarily conserved function in appendage development. Proc. Natl Acad. Sci. USA. 1998;95:10637–10642. doi: 10.1073/pnas.95.18.10637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai L.T., Bainton R.J., Blau J., Heberlein U. Lmo mutants reveal a novel role for circadian pacemaker neurons in cocaine-induced behaviors. PLoS Biol. 2004;2:e408. doi: 10.1371/journal.pbio.0020408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leuba G., Vernay A., Vu D., Walzer C., Belloir B., Kraftsik R., Bouras C., Savioz A. Differential expression of LMO4 protein in Alzheimer's disease. Neuropathol. Appl. Neurobiol. 2004;30:57–69. doi: 10.1046/j.0305-1846.2003.00511.x. [DOI] [PubMed] [Google Scholar]
- 19.Vu D., Marin P., Walzer C., Cathieni M.M., Bianchi E.N., Saidji F., Leuba G., Bouras C., Savioz A. Transcription regulator LMO4 interferes with neuritogenesis in human SH-SY5Y neuroblastoma cells. Brain Res. Mol. Brain Res. 2003;115:93–103. doi: 10.1016/s0169-328x(03)00119-0. [DOI] [PubMed] [Google Scholar]
- 20.Hermanson O., Sugihara T.M., Andersen B. Expression of LMO-4 in the central nervous system of the embryonic and adult mouse. Cell Mol. Biol. (Noisy-le-grand) 1999;45:677–686. [PubMed] [Google Scholar]
- 21.Boehm T., Spillantini M.G., Sofroniew M.V., Surani M.A., Rabbitts T.H. Developmentally regulated and tissue specific expression of mRNAs encoding the two alternative forms of the LIM domain oncogene rhombotin: evidence for thymus expression. Oncogene. 1991;6:695–703. [PubMed] [Google Scholar]
- 22.Foroni L., Boehm T., White L., Forster A., Sherrington P., Liao X.B., Brannan C.I., Jenkins N.A., Copeland N.G., Rabbitts T.H. The rhombotin gene family encode related LIM-domain proteins whose differing expression suggests multiple roles in mouse development. J. Mol. Biol. 1992;226:747–761. doi: 10.1016/0022-2836(92)90630-3. [DOI] [PubMed] [Google Scholar]
- 23.Bulchand S., Subramanian L., Tole S. Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev. Dyn. 2003;226:460–469. doi: 10.1002/dvdy.10235. [DOI] [PubMed] [Google Scholar]
- 24.Hinks G.L., Shah B., French S.J., Campos L.S., Staley K., Hughes J., Sofroniew M.V. Expression of LIM protein genes Lmo1, Lmo2, and Lmo3 in adult mouse hippocampus and other forebrain regions: differential regulation by seizure activity. J. Neurosci. 1997;17:5549–5559. doi: 10.1523/JNEUROSCI.17-14-05549.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manetopoulos C., Hansson A., Karlsson J., Jonsson J.I., Axelson H. The LIM-only protein LMO4 modulates the transcriptional activity of HEN1. Biochem. Biophys. Res. Commun. 2003;307:891–899. doi: 10.1016/s0006-291x(03)01298-1. [DOI] [PubMed] [Google Scholar]
- 26.Wadman I.A., Osada H., Grutz G.G., Agulnick A.D., Westphal H., Forster A., Rabbitts T.H. The LIM-only protein Lmo2 is a bridging molecule assembling an erythroid, DNA-binding complex which includes the TAL1, E47, GATA-1 and Ldb1/NLI proteins. EMBO J. 1997;16:3145–3157. doi: 10.1093/emboj/16.11.3145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Retaux S., Bachy I. A short history of LIM domains (1993–2002): from protein interaction to degradation. Mol. Neurobiol. 2002;26:269–281. doi: 10.1385/MN:26:2-3:269. [DOI] [PubMed] [Google Scholar]
- 28.Zhou H.J., Wong C.M., Chen J.H., Qiang B.Q., Yuan J.G., Jin D.Y. Inhibition of LZIP-mediated transcription through direct interaction with a novel host cell factor-like protein. J. Biol. Chem. 2001;276:28933–28938. doi: 10.1074/jbc.M103893200. [DOI] [PubMed] [Google Scholar]
- 29.Grutz G.G., Bucher K., Lavenir I., Larson T., Larson R., Rabbitts T.H. The oncogenic T cell LIM-protein Lmo2 forms part of a DNA-binding complex specifically in immature T cells. EMBO J. 1998;17:4594–4605. doi: 10.1093/emboj/17.16.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bao J., Talmage D.A., Role L.W., Gautier J. Regulation of neurogenesis by interactions between HEN1 and neuronal LMO proteins. Development. 2000;127:425–435. doi: 10.1242/dev.127.2.425. [DOI] [PubMed] [Google Scholar]
- 31.Ono Y., Fukuhara N., Yoshie O. Transcriptional activity of TAL1 in T cell acute lymphoblastic leukemia (T-ALL) requires RBTN1 or -2 and induces TALLA1, a highly specific tumor marker of T-ALL. J. Biol. Chem. 1997;272:4576–4581. doi: 10.1074/jbc.272.7.4576. [DOI] [PubMed] [Google Scholar]
- 32.Vitelli L., Condorelli G., Lulli V., Hoang T., Luchetti L., Croce C.M., Peschle C. A pentamer transcriptional complex including tal-1 and retinoblastoma protein downmodulates c-kit expression in normal erythroblasts. Mol. Cell. Biol. 2000;20:5330–5342. doi: 10.1128/mcb.20.14.5330-5342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mao S., Neale G.A., Goorha R.M. T-cell oncogene rhombotin-2 interacts with retinoblastoma-binding protein 2. Oncogene. 1997;14:1531–1539. doi: 10.1038/sj.onc.1200988. [DOI] [PubMed] [Google Scholar]
- 34.Lipkowitz S., Gobel V., Varterasian M.L., Nakahara K., Tchorz K., Kirsch I.R. A comparative structural characterization of the human NSCL-1 and NSCL-2 genes. Two basic helix-loop-helix genes expressed in the developing nervous system. J. Biol. Chem. 1992;267:21065–21071. [PubMed] [Google Scholar]
- 35.Brown L., Espinosa R., III, Le Beau M.M., Siciliano M.J., Baer R. HEN1 and HEN2: a subgroup of basic helix–loop–helix genes that are coexpressed in a human neuroblastoma. Proc. Natl Acad. Sci. USA. 1992;89:8492–8496. doi: 10.1073/pnas.89.18.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matthews J.M., Visvader J.E. LIM-domain-binding protein 1: a multifunctional cofactor that interacts with diverse proteins. EMBO Rep. 2003;4:1132–1137. doi: 10.1038/sj.embor.7400030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wadman I., Li J., Bash R.O., Forster A., Osada H., Rabbitts T.H., Baer R. Specific in vivo association between the bHLH and LIM proteins implicated in human T cell leukemia. EMBO J. 1994;13:4831–4839. doi: 10.1002/j.1460-2075.1994.tb06809.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Begley C.G., Lipkowitz S., Gobel V., Mahon K.A., Bertness V., Green A.R., Gough N.M., Kirsch I.R. Molecular characterization of NSCL, a gene encoding a helix–loop–helix protein expressed in the developing nervous system. Proc. Natl Acad. Sci. USA. 1992;89:38–42. doi: 10.1073/pnas.89.1.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gobel V., Lipkowitz S., Kozak C.A., Kirsch I.R. NSCL-2: a basic domain helix–loop–helix gene expressed in early neurogenesis. Cell Growth Differ. 1992;3:143–148. [PubMed] [Google Scholar]