Abstract
MeCP2, whose methylated DNA-binding domain (MBD) binds preferentially to DNA containing 5Me-CpG relative to linear unmethylated DNA, also binds preferentially, and with similar affinity, to unmethylated four-way DNA junctions through the MBD. The Arg133Cys (R133C) mutation in the MBD, a Rett syndrome mutation that abolishes binding to methylated DNA, leads to only a slight reduction in the affinity of the MBD for four-way junctions, suggesting distinct but partially overlapping modes of binding to junction and methylated DNA. Binding to unmethylated DNA junctions is likely to involve a subset of the interactions that occur with methylated DNA. High-affinity, methylation-independent binding to four-way junctions is consistent with additional roles for MeCP2 in chromatin, beyond recognition of 5Me-CpG.
INTRODUCTION
MeCP2 is an abundant vertebrate nuclear protein that binds preferentially to a symmetrically methylated CpG dinucleotide site in double-stranded DNA (1). It is abundant in pericentric heterochromatin, which has a high concentration of 5Me-CpG, but is also present throughout chromosomal arms (1). Two functional domains have been identified within MeCP2 (human, 486 amino acid residues), each of ∼100 residues. A methyl binding domain (MBD) confers specificity for methylated DNA (2) and has since been found in other members of the MBD family of proteins (3); the solution structures of the MBD of MeCP2 and of MBD1 have been determined (4,5) and show a very similar fold. A transcriptional repression domain (TRD) in MeCP2 (6) functions by recruitment of the co-repressor Sin3A and a histone deacetylase (7,8). Other transcriptional co-repressors, such as Ski and N-CoR, can also be recruited by MeCP2 and function similarly (9). MeCP2 can thus mediate repression of transcription through core histone deacetylation, which favours stable and transcriptionally inert chromatin, largely through effects at the level of higher-order structure (30 nm filament). This could provide a means of suppressing genome-wide transcriptional ‘noise’ (10) but also performs a critical role in repression at specific loci such as imprinted genes [e.g. (11)], and pro-viral (12) and other promoters, such as the brain-derived neurotrophic factor promoter (13,14). A third, less well-defined, domain that specifies binding to Group II WW domain splicing factors was mapped to the C-terminal region (residues 325–486) of MeCP2 (15). Mutations in all three domains have been found in Rett syndrome patients (InterRett-IRSA Phenotype Database http://www.ichr.uwa.edu.au/rett/irsa/).
Histone deacetylase inhibitors such as trichostatin A do not fully relieve the repressive effects of MeCP2 (8) suggesting another, deacetylation-independent, pathway for MeCP2-mediated repression (16,17). One possibility is that MeCP2 sequesters transcription factors or occludes their access to the polymerase, but it is also clear that MeCP2 is able to bind to unmethylated DNA in vitro, albeit with lower affinity than to methylated DNA [although only 2- to 3-fold fold lower in the case of mouse MeCP2 binding to oligonucleotides, measured by capillary electrophoresis (18)]. Another possibility, therefore, is that MeCP2 plays a methylation-independent structural role in chromatin, e.g. in stabilizing a repressive structure such as the 30 nm filament formed by folding of the 10 nm nucleosome filament, or even more stable structures based on this. Several (unrelated) observations support an uncoupling of repression by MeCP2 from DNA methylation. First, the chicken protein ARBP (attachment region binding protein), believed to be the chicken MeCP2 homologue (19), interacts preferentially in vitro with (unmethylated) nuclear matrix attachment region DNA (20) as well as with (unmethylated) chicken and mouse satellite DNA (19). A 25-residue sequence containing an ‘AT-hook’ motif (-GRGRGRPK-) located between the MBD and the TRD appears to be required (19). Second, MeCP2 has been shown recently to condense unmethylated, H1-deficient, reconstituted nucleosomal arrays, which then self-associate to form ‘oligomeric chromatin suprastructures’ (21). Third, the presence of MeCP2 and dimethylation of H3K9 at a promoter are sufficient to silence it, in the absence of DNA methylation (22).
Methylation-independent MeCP2-mediated repression of transcription from chromatin might occur by stabilization of nucleosomes and/or chromatin compaction (21), in a functionally similar manner to stabilization by H1. The detailed mode of interaction of H1 and MeCP2 might be different, although there is circumstantial evidence that they may have overlapping binding sites (6). H1 binds asymmetrically to the nucleosome, close to the dyad axis, contacting two DNA duplexes through its globular domain (23,24). It also binds in a structure-specific manner to four-way DNA junctions through its globular domain (25–27), most probably because a pair of junction arms mimics two juxtaposed duplexes in the nucleosome. Here we investigate the binding of MeCP2 and its MBD to four-way junction DNA, and compare this with binding to methylated DNA, as one approach towards assessing the potential of MeCP2 to mediate effects through chromatin-dependent, structure-specific, methylation-independent interactions.
MATERIALS AND METHODS
Expression and purification of MeCP2 and its MBD
Plasmid pET17bMeCP2 (generated by D. Pate) contains the rat MeCP2 cDNA sequence (2) cloned into the BamH1/EcoR1 sites of pET17b; the recombinant MeCP2 expressed from this has an additional 20 amino acid residues (MASMTGGQQMGRDSSLVPSS) at its N-terminus, encoded by the vector. pET6HMBD (28) is pET-3c with an insert encoding the MBD (residues 78–164) of rat MeCP2; the MBD expressed from this has an N-terminal His6 tag. Escherichia coli BL21(DE3)pLysS cells transformed with plasmid pET17bMeCP2 or pET6HMBD were grown at 37°C in 2× YT medium to an OD600 = 0.4–0.6 and expression of MeCP2 or the MBD was induced with 0.5 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 1 h. The cells were collected by centrifugation, resuspended in buffer A (10 mM Tris–HCl, pH 7.5, 1 mM EDTA, 1 M NaCl and 0.5 mM phenylmethylsulphonyl fluoride) containing protease inhibitors (100 µg/ml benzamidine and 100 µg/ml 1-chloro-3-tosylamido-7-amino-l-2-heptanone) and lysed with a French press. The supernatant after centrifugation was diluted 10-fold in cold buffer B (buffer A without 1 M NaCl), containing protease inhibitors in the case of MeCP2, and applied to a S-Sepharose Fast Flow (Fast S) FPLC column (Pharmacia) pre-equilibrated with buffer B. Bound proteins were eluted with a gradient of 10 column volumes from buffer B to buffer A for MeCP2, and isocratically with buffer B alone for MBD, which binds much less tightly.
In the case of MeCP2, the peak fractions were brought to 35% ammonium sulphate saturation at 4°C, and the supernatant after centrifugation was applied to a PhenylSepharose FPLC column (Pharmacia) equilibrated with buffer C (35% saturated ammonium sulphate, 10 mM Tris–HCl, pH 7.5 and 1 mM EDTA). MeCP2 was eluted with a gradient from buffer C to buffer B, dialyzed against buffer B, applied to a Resource S column (Pharmacia) pre-equilibrated with buffer B, and eluted with a gradient from buffer B to buffer A. Peak fractions were pooled, dialyzed against buffer B or 10 mM sodium phosphate, pH 7.0, and stored at −80°C.
In the case of the MBD, the Fast S peak fractions were pooled and applied to a Ni2+–Sepharose Fast Flow column (Sigma) pre-equilibrated with buffer A containing 20 mM imidazole. The MBD was eluted with buffer A containing 300 mM imidazole, dialyzed against buffer B, and applied to a Resource S column (Pharmacia) as described for MeCP2, except that buffers also contained 3 mM dithiothreitol (DTT). Fractions containing the MBD were dialyzed twice against 10 mM Tris–HCl, pH 7.5 and 3 mM DTT, and stored at −80°C. The molecular mass was checked by MALDI-TOF mass spectrometry and the protein concentration was calculated from the amino acid composition determined by automated analysis (both procedures carried out in the Protein and Nucleic Acid Chemistry Facility, Department of Biochemistry, University of Cambridge).
Expression and purification of MBD R133C
The R133C mutation was introduced into pET6HMBD using the Stratagene QuikChange kit according to the manufacturer's instructions and verified by automated DNA sequencing (DNA Sequencing Facility, Department of Biochemistry, University of Cambridge). E.coli BL21(DE3) pLysS cells containing the mutant MBD plasmid were grown at 37°C in 2× YT medium to an OD600 of 0.4–0.6, cooled to 23°C and induced with 0.5 mM IPTG. After 5 h the cells were harvested and lysed as for the MBD. The cleared, filtered lysate was incubated with Ni2+–Sepharose 6B Fast Flow beads (Sigma) for 1 h at 4°C, and the beads were washed with buffer D (10 mM Tris–HCl, pH 7.5, 20 mM imidazole and 300 mM NaCl), and then with buffer E (10 mM Tris–HCl, pH 7.5, 75 mM imidazole and 300 mM NaCl). Bound proteins were eluted with 10 mM Tris–HCl, pH 7.5 and 300 mM imidazole and dialyzed overnight against 10 mM Tris–HCl, pH 7.5 and 75 mM NaCl. The supernatant after centrifugation was applied to a Resource S column (Pharmacia) pre-equilibrated with 10 mM Tris–HCl, pH 7.5, 75 mM NaCl. MBD(R133C) was eluted with a linear gradient from 75 mM to 1 M NaCl in 10 mM Tris–HCl, pH 7.5, dialyzed twice against 10 mM Tris–HCl, pH 7.5 or 10 mM sodium phosphate, pH 7.0, and stored at −80°C. The molecular mass and protein concentration were determined as described above.
Gel-retardation assays
Gel-retardation assays were carried out with three 32P-end-labelled probes: two 17 bp oligonucleotides formed by annealing 5′-CAGTGCGACACGGCATGCCTG-3′ and its complementary strand, with the central CpG (bold) either methylated or not on both strands, and a 32P-end-labelled four-way DNA junction with 15 bp arms formed by annealing four synthetic 30mer oligonucleotides (29), all of which were 5′-end-labelled. Binding of MeCP2 and the MBD to 32P-labelled DNA was carried out in 10 µl of 10 mM Tris–HCl, pH 7.5, 150 µg/ml BSA, 0.1% Nonidet-P40, 3 mM DTT, 5% glycerol and 50 or 125 mM NaCl, as specified, for MBD, or 150 mM NaCl for MeCP2, with or without unlabelled 17 bp oligonucleotide (methylated or not as specified) or sonicated E.coli DNA (Sigma) as competitors. The DNA (∼0.27 nM; added last, with competitor where applicable) was incubated with increasing concentrations of protein for 30 min at room temperature (incubation at 37°C did not affect the results; data not shown) and loaded on to 6.5 or 7% polyacrylamide gels containing 0.5× TB (45 mM Tris and 45 mM boric acid), which were run at 150 V at room temperature. Gels were vacuum-dried and exposed overnight at −80°C to pre-flashed X-ray film.
RESULTS
MeCP2 interacts with four-way junction DNA, through its MBD
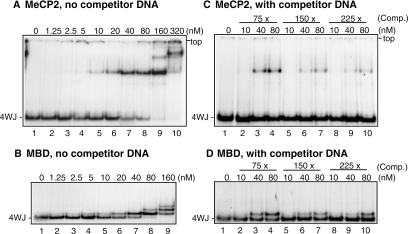
The ability of recombinant rat MeCP2 and the isolated MBD to interact with (unmethylated) 32P-labelled four-way junction DNA was investigated in gel-retardation assays. With MeCP2 a complex was first apparent at 5 nM protein (Figure 1A, lane 4); the apparent Kd for the interaction (in 150 mM NaCl) is 20–40 nM (lanes 6 and 7). The MBD (Figure 1B) behaved very similarly, with an apparent Kd (in 125 mM NaCl) of 20–40 nM (lanes 5 and 6). The lower mobility bands observed at the two highest protein concentrations in the absence of competitor are most probably owing to weaker, non-structure-specific binding of MBD and MeCP2 to additional sites on the junction arms. The H1 and H5 globular domains, and HMGB1, which bind to a pair of juxtaposed arms and the cross-over in the four-way junction, respectively, show similar secondary binding (27,30,31). A 75-fold molar excess of sheared E.coli DNA reduced binding of both MeCP2 (compare Figure 1A, lanes 7 and 8, with Figure 1C, lanes 3 and 4) and the MBD (compare Figure 1B, lanes 7 and 8, with Figure 1D, lanes 3 and 4) by roughly 50%, but even in the presence of a 225-fold molar excess of competitor (Figure 1C and D, lanes 9 and 10) binding was not abolished. The very similar behaviour of MeCP2 and its MBD (Figure 1A–D) suggests that the interaction of MeCP2 with four-way junction DNA is mediated through high-affinity binding of the MBD, with little contribution from other regions of the protein.
Figure 1.
Binding of MeCP2 and its MBD to four-way junction DNA. 32P-labelled four-way junction DNA (∼0.27 nM) was incubated with increasing amounts of MeCP2 (A and C) or MBD (B and D) in the absence (A and B) or presence (C and D) of E.coli competitor DNA in 75-, 150- or 225-fold molar excess over four-way junction DNA. The incubations contained 150 mM NaCl (MeCP2) or 125 mM NaCl (MBD). Samples were analysed in 6% (MeCP2) or 7.5% (MBD) polyacrylamide gels; autoradiographs are shown [of the whole gel in (A and C); of the portions showing bands in (B and D)].
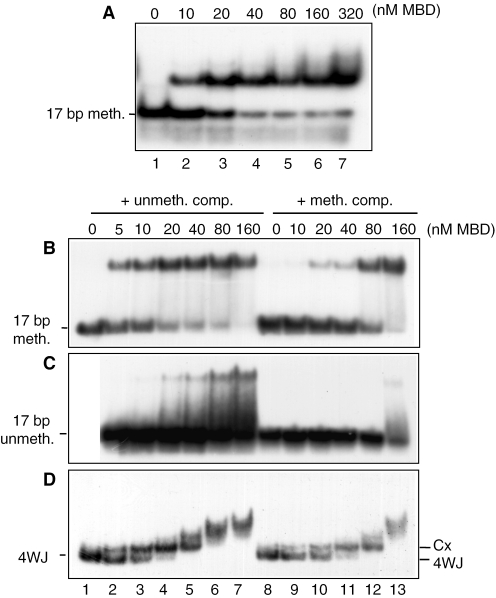
In gel-shift assays with a 17 bp singly methylated duplex, there was a single complex; the Kd was 10–20 nM in 125 mM NaCl (Figure 2A), similar to that of the MBD for four-way junctions under the same conditions (∼20–40 nM; Figure 1B). No additional lower mobility bands were observed at the higher protein concentrations in this case, further suggesting that the additional bands with four-way junctions are due to protein–DNA interactions with the junction arms, rather than protein–protein interactions between added and bound MBD [which, like MeCP2 (32), is monomeric when free in solution (data not shown)]. MBD binds to the corresponding unmethylated 17 bp duplex under the same conditions with much lower affinity, as expected, and the complexes are much less stable (data not shown). The DNA-binding properties of the MBD were further investigated by comparing its ability to bind 32P-labelled 17 bp DNA (both methylated and unmethylated) and four-way junction DNA in the presence of a 30-fold molar excess of 17 bp competitor DNA that was either unmethylated (Figure 2B–D, lanes 2–7) or methylated (lanes 8–13). Only the methylated competitor competed effectively for binding of the MBD to the radiolabelled methylated 17 bp DNA (Figure 2B) or to the four-way junction (Figure 2D), but even in the presence of the competitor strong binding persisted at the higher concentrations of MBD (80 nM and above). The weaker binding to the unmethylated probe was completely abolished when the competitor was methylated (Figure 2C) and largely abolished even by unmethylated competitor. Thus the MBD of MeCP2 interacts structure-specifically with (unmethylated) four-way junction DNA in conditions in which it does not bind to unmethylated linear DNA, and it binds four-way junction DNA with 15 bp arms and singly methylated DNA of 17 bp with similar affinity.
Figure 2.
Comparison of the binding of the MBD of MeCP2 to methylated DNA and four-way junctions. (A) Binding to methylated duplex DNA (no competitor DNA). 32P-labelled methylated 17 bp DNA duplex (∼0.27 nM) was incubated with increasing concentrations of MBD in the presence of 125 mM NaCl and the samples were analysed by 6.5% PAGE and autoradiography. (B–D) Binding of the MBD of MeCP2 to methylated DNA and four-way junction DNA in the presence of competitor. (B) 32P-labelled singly methylated, (C) unmethylated 17 bp duplex or (D) four-way junction DNA (all ∼0.27 nM) was incubated with increasing concentrations of MBD. Reactions contained a 30-fold molar excess of unlabelled 17 bp duplex DNA that was either unmethylated (lanes 2–7) or methylated (lanes 8–13) as competitor and 50 mM NaCl. Samples were analysed by 7.5% PAGE and autoradiography. Cx indicates the first protein–DNA complex with the four-way junction.
Overlapping binding sites for methylated and four-way junction DNA
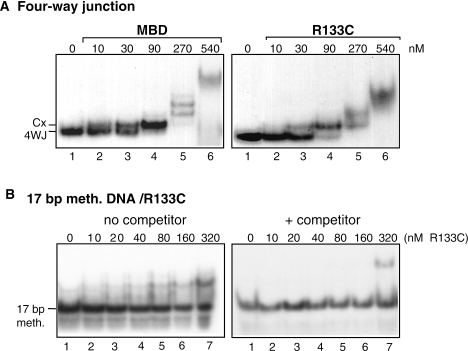
The Rett syndrome mutation R133C in MeCP2 disrupts binding of the MBD to methylated DNA in vitro (33–35). To test whether there was overlap between the interactions involved in methyl CpG-dependent binding of the MBD to linear DNA and structure-specific binding to four-way junctions, binding of the R133C MBD mutant to four-way junction DNA and methylated DNA was compared (Figure 3). The affinity of the R133C mutant was only slightly lower (∼2-fold) than that of wild-type MBD (Figure 3A). In contrast, binding to singly methylated 17 bp DNA was severely impaired and in the presence of a 20-fold excess of 17 bp methylated DNA was abolished (Figure 3B); there was no binding to unmethylated 17 bp DNA (data not shown). The binding site for interaction of the MBD with methylated duplex DNA must thus be largely distinct from the binding site(s) for four-way junctions, but there might be some overlap between the sites. Virtually identical near-UV CD spectra for the wild-type MBD and the R133C mutant (data not shown) indicate that the differences in methylated DNA binding are not a result of any significant conformational change introduced by the mutation, as indicated also by NMR spectroscopy (34).
Figure 3.
Effect of the Rett mutation R133C on binding of the MBD of MeCP2 to four-way junction DNA and methylated DNA. (A) 32P-labelled four-way junction DNA (∼0.27 nM) was incubated with increasing amounts of wild-type or R133C mutant MBD as indicated. (B) 32P-labelled 17 bp methylated DNA (∼0.27 nM) was incubated with increasing amounts of R133C mutant MBD in the absence (left) or presence (right) of a 20-fold excess of unlabelled 17 bp singly methylated DNA. All samples (A and B) contained 125 mM NaCl and were analysed by 6.5% PAGE and autoradiography. Cx indicates the position of the first protein–DNA complex with the four-way junction.
DISCUSSION
The main conclusion from the results reported here is that MeCP2, as well as its isolated MBD, binds to four-way junction DNA in a structure-specific, methyl-CpG-independent manner. The affinity for the junction is similar to that for methylated DNA (Figures 1B and 2A). Double-stranded competitor DNA, whether methylated or unmethylated, had only a small effect on junction binding (Figure 2D) but abolished binding to the unmethylated probe (Figure 2C), showing that the interaction with four-way junction DNA is specific. Mutation of a methyl-CpG binding residue [R133C, a Rett syndrome mutation that essentially abolishes binding to methylated DNA (33–35); Figure 3B] reduces binding to four-way junction DNA by only ∼2-fold (Figure 3A), suggesting largely distinct, but possibly overlapping, binding sites for the two DNA substrates. Formation of MeCP2-dependent ‘oligomeric chromatin suprastructures’ in vitro from reconstituted unmethylated nucleosomal arrays, like four-way junction binding, also appears not to be affected by the R133C mutation in MeCP2 (21). In other words, MeCP2 has a methyl-CpG-dependent binding mode and a methylation-independent chromatin binding mode which, in an array of MeCP2-containing nucleosomes, leads to chromatin condensation. Certain features of chromatin binding may be reflected in binding of MeCP2 to four-way junctions.
MeCP2 forms ‘tramline’ complexes with long DNA (21). Complexes of this type have been demonstrated previously for the linker histones H1 and H5 and their isolated globular domains (36,37) and in those cases, like binding of the proteins to the juxtaposed duplexes of four-way junctions (27), are taken to reflect the existence of two DNA-binding sites on opposite faces of the globular domain (38); the existence of two sites was supported by mutagenesis studies (25,39). In the nucleosome the two binding sites engage with two duplexes in the general vicinity of the dyad axis (24). Since MeCP2, like H1 and H5, assembles linear DNA into tramlines and binds selectively to four-way junctions, it seems possible that it, too, might bind two DNA duplexes, perhaps (but not necessarily) binding similarly to H1 and H5. However, if MeCP2 does contact two DNA strands, it remains to be determined whether the two binding sites are on the MBD, since the ability of the MBD to form tramlines was not reported (21).
The detailed mode of binding of MeCP2 to DNA and chromatin remains to be investigated. There is circumstantial evidence suggesting that MeCP2 and H1 may have overlapping binding sites in chromatin. Addition of MeCP2 to methylated plasmid DNA that had been assembled into chromatin in a Xenopus oocyte extract and reconstituted with H1 displaced some (40%) of the bound H1 (6), although if the nucleosome occupancy of the DNA was not complete, it is possible that the displaced H1 was bound not to nucleosomes but to stretches of naked DNA. However, if MeCP2 does bind at, or in such a way that it overlaps, a linker histone binding site, in contrast to H1 binding (40) it does not lead to protection of chromatosomes (containing 166 bp DNA) in vitro during micrococcal nuclease digestion (A. Sanderson and J. O. Thomas, unpublished data). It does confer protection from DNase I digestion on a methylated CpG at the dyad axis of a nucleosome assembled on methylated 5S rDNA (41), but this is presumably due to methylation-dependent binding rather than structure-specific recognition, since there was no protection against DNase I digestion when the DNA was not methylated. Moreover, despite tramline formation by MeCP2 it remains possible that the feature in four-way junctions that is recognized by MeCP2 and its MBD is not a pair of juxtaposed duplexes per se, but the distortion at the cross-over of the duplexes where bases are unstacked, e.g. as is the case for HMGB1 (29,42).
What might be the basis of the interaction of the MBD of MeCP2 with unmethylated DNA?
Molecular details of the interaction of the highly homologous (46% identical) MBD of MBD1 with methylated DNA are known from an NMR structure of the complex (43); similar interactions are inferred for the MBD of MeCP2, based on chemical shift changes in the protein on binding to methylated DNA (34). The MBD in both MeCP2 (5) and MBD1 (4) is a wedge-shaped α/β sandwich, with a C-terminal α-helix. In the complex of the MBD (of MBD1) with methylated DNA, the anti-parallel three-stranded β-sheet fits into the major groove (43). The buried interface is relatively small (810 Å2) (43), consistent with the ability of MeCP2 to bind to methylated nucleosomal DNA without nucleosomal disruption (41). Recognition of the two methyl CpGs takes place, asymmetrically, through hydrophobic and polar interactions in a pocket formed by six residues in β-strands 2 and 3 (β2 and β3) and loops 1 and 2 (L1 and L2). These residues are Val20 (in β2), Arg22 (L1), Asp32 (β3), Tyr34 (β3), Arg44 (L2) and Ser45 (L2). In addition there are extensive contacts with the sugar-phosphate backbone, including phosphate contacts with arginine or lysine residues in loops L1 and L2 and with the N-terminus of helix α1. Most of the residues involved in MBD1 are conserved in MeCP2. A subset of the interactions, e.g. involving the phosphates and sugars but possibly also those involving residues in the pocket that normally binds 5Me-CpG, is likely to account for the binding of MeCP2 to non-methylated DNA. However, the higher-affinity preferential binding to four-way DNA junctions relative to unmethylated linear DNA implies additional, structure-specific interactions with junction DNA (and possibly the ability to satisfy two DNA-binding sites intramolecularly), the nature of which is not yet clear.
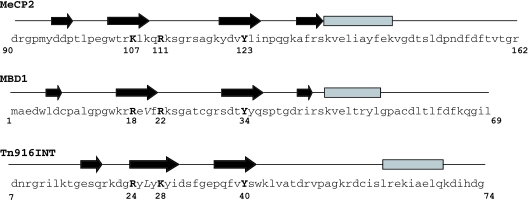
There are three unrelated β-sheet proteins/domains with a C-terminal α-helix that exhibit conserved features of DNA binding to the major groove of (unmethylated) DNA (44): the I-PpoI homing endonuclease (45), the DNA-binding domain of the Tn916 integrase [Tn916Int (46)], and the DNA-binding domain of the GCC-box binding protein AtERF1 (47). In these cases the β-sheet lies flat in the major groove, rather than being twisted as in the MBD of MBD1, as noted (43). Although the proteins have no overall sequence homology there are striking similarities between the MBD1 contacts with methylated DNA and the Tn916Int contacts with unmethylated DNA. Key protein–DNA contacts involve identical or similar amino acids at corresponding positions in the β-sheets and loops (Figure 4; see legend for details). We suggest that similar considerations are likely to apply to the binding of the MBD of MeCP2 (Figure 4, top line) to unmethylated DNA. The similarities in key residues between the methylated-DNA-binding MBD1 and the other proteins, whose DNA-binding sites are not methylated, suggests that the MBD of MeCP2 (and presumably of MBD1) could interact with unmethylated DNA using a subset of the contacts used to bind to methylated DNA.
Figure 4.
Sequences of the DNA-binding domains of MBD1, MeCP2 and Tn916Int, indicating key DNA-contacting residues in similar positions in MBD1 and Tn916Int, and (by extrapolation) MeCP2. The secondary-structure elements defined by NMR (5,43,46) are shown above each sequence (β-sheets shown as arrows, α-helices as bars). Secondary structure elements and loops of MBD1 complexed with DNA (43) are: β1 (residues 6–7), β2 (16–21), L1 (22–30), β3 (31–36), β4 (42–43), L2 (44–45), α1 (46–53). Conserved residues with similar DNA-binding roles in the MBD1 and Tn916Int protein–DNA complexes (43,46) are shown in boldface. In the MBD of MBD1 and DNA-binding domain of Tn916Int, Arg18 and Arg24, respectively, contact a phosphate group in the DNA backbone; Arg22 and Lys28, respectively, donate a hydrogen bond to a guanine and make a hydrophobic contact with the methyl group in a 5Me-cytosine (MBD1) or a thymine (Tn916Int); Tyr34 (MBD1) and Tyr40 (Tn916Int) form a hydrogen bond with the N4 of a methyl cytosine or cytosine, respectively. Val20 and Leu26, shown in italics, make van der Waals contacts with the methyl group of a methyl cytosine (MBD1) or a thymine (Tn916), respectively. The MBDs of MeCP2 (top line) and MBD1 are highly homologous; Arg18, Arg22 and Tyr34 in MBD1 all correspond to similar or identical residues in MeCP2 (Lys107, Arg111 and Tyr123). However, there is no hydrophobic residue in MeCP2 at the position (Lys109) corresponding to Val20 in MBD1.
Whatever the details of the interaction of the MBD of MeCP2 with four-way junction DNA, it represents high-affinity, methylation-independent binding to a DNA structure that is accepted as mimicking certain features of DNA architecture around the dyad axis of the nucleosome. This mode of binding may be related to the ability of MeCP2 to condense extended arrays of unmethylated nucleosomes (21). Further investigation of the interaction of MeCP2 with unmethylated DNA and chromatin is in progress in order to define any direct role it may play in chromatin structure and function.
Acknowledgments
We thank Adrian Bird for plasmids containing the rat MeCP2 and MBD cDNA inserts and David Pate for constructing p17bMeCP2. T.C.G. thanks CAPES (Brazil) for the award of a PhD Studentship, the Lundgren Fund of the University of Cambridge for additional financial support and Margaret Duggan for helpful discussion. The Open Access charge for this article is covered by a grant from the JISC to NAR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Lewis J.D., Meehan R.R., Henzel W.J., Maurer-Fogy I., Jeppesen P., Klein F., Bird A. Purification, sequence, and cellular localization of a novel chromosomal protein that binds to methylated DNA. Cell. 1992;69:905–914. doi: 10.1016/0092-8674(92)90610-o. [DOI] [PubMed] [Google Scholar]
- 2.Nan X., Meehan R.R., Bird A. Dissection of the methyl-CpG binding domain from the chromosomal protein MeCP2. Nucleic Acids Res. 1993;21:4886–4892. doi: 10.1093/nar/21.21.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hendrich B., Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol. Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ohki I., Shimotake N., Fujita N., Nakao M., Shirakawa M. Solution structure of the methyl-CpG-binding domain of the methylation-dependent transcriptional repressor MBD1. EMBO J. 1999;18:6653–6661. doi: 10.1093/emboj/18.23.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wakefield R.I., Smith B.O., Nan X., Free A., Soteriou A., Uhrin D., Bird A.P., Barlow P.N. The solution structure of the domain from MeCP2 that binds to methylated DNA. J. Mol. Biol. 1999;291:1055–1065. doi: 10.1006/jmbi.1999.3023. [DOI] [PubMed] [Google Scholar]
- 6.Nan X., Campoy F.J., Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 7.Jones P.L., Veenstra G.J., Wade P.A., Vermaak D., Kass S.U., Landsberger N., Strouboulis J., Wolffe A.P. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nature Genet. 1998;19:187–191. doi: 10.1038/561. [DOI] [PubMed] [Google Scholar]
- 8.Nan X., Ng H.H., Johnson C.A., Laherty C.D., Turner B.M., Eisenman R.N., Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 9.Kokura K., Kaul S.C., Wadhwa R., Nomura T., Khan M.M., Shinagawa T., Yasukawa T., Colmenares C., Ishii S. The Ski protein family is required for MeCP2-mediated transcriptional repression. J. Biol. Chem. 2001;276:34115–34121. doi: 10.1074/jbc.M105747200. [DOI] [PubMed] [Google Scholar]
- 10.Bird A.P. Gene number, noise reduction and biological complexity. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- 11.Gregory R.I., Randall T.E., Johnson C.A., Khosla S., Hatada I., O'Neill L.P., Turner B.M., Feil R. DNA methylation is linked to deacetylation of histone H3, but not H4, on the imprinted genes Snrpn and U2af1-rs1. Mol. Cell Biol. 2001;21:5426–5436. doi: 10.1128/MCB.21.16.5426-5436.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lorincz M.C., Schubeler D., Groudine M. Methylation-mediated proviral silencing is associated with MeCP2 recruitment and localized histone H3 deacetylation. Mol. Cell Biol. 2001;21:7913–7922. doi: 10.1128/MCB.21.23.7913-7922.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen W.G., Chang Q., Lin Y., Meissner A., West A.E., Griffith E.C., Jaenisch R., Greenberg M.E. Derepression of BDNF transcription involves calcium-dependent phosphorylation of MeCP2. Science. 2003;302:885–889. doi: 10.1126/science.1086446. [DOI] [PubMed] [Google Scholar]
- 14.Martinowich K., Hattori D., Wu H., Fouse S., He F., Hu Y., Fan G., Sun Y.E. DNA methylation-related chromatin remodeling in activity-dependent BDNF gene regulation. Science. 2003;302:890–893. doi: 10.1126/science.1090842. [DOI] [PubMed] [Google Scholar]
- 15.Buschdorf J.P., Stratling W.H. A WW domain binding region in methyl-CpG-binding protein MeCP2: impact on Rett syndrome. J. Mol. Med. 2004;82:135–143. doi: 10.1007/s00109-003-0497-9. [DOI] [PubMed] [Google Scholar]
- 16.Kaludov N.K., Wolffe A.P. MeCP2 driven transcriptional repression in vitro: selectivity for methylated DNA, action at a distance and contacts with the basal transcription machinery. Nucleic Acids Res. 2000;28:1921–1928. doi: 10.1093/nar/28.9.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu F., Thiesen J., Stratling W.H. Histone deacetylase-independent transcriptional repression by methyl-CpG-binding protein 2. Nucleic Acids Res. 2000;28:2201–2206. doi: 10.1093/nar/28.10.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fraga M.F., Ballestar E., Montoya G., Taysavang P., Wade P.A., Esteller M. The affinity of different MBD proteins for a specific methylated locus depends on their intrinsic binding properties. Nucleic Acids Res. 2003;31:1765–1774. doi: 10.1093/nar/gkg249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weitzel J.M., Buhrmester H., Stratling W.H. Chicken MAR-binding protein ARBP is homologous to rat methyl-CpG-binding protein MeCP2. Mol. Cell. Biol. 1997;17:5656–5666. doi: 10.1128/mcb.17.9.5656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Kries J.P., Buhrmester H., Stratling W.H. A matrix/scaffold attachment region binding protein: identification, purification, and mode of binding. Cell. 1991;64:123–135. doi: 10.1016/0092-8674(91)90214-j. [DOI] [PubMed] [Google Scholar]
- 21.Georgel P.T., Horowitz-Scherer R.A., Adkins N., Woodcock C.L., Wade P.A., Hansen J.C. Chromatin compaction by human MeCP2—assembly of novel secondary chromatin structures in the absence of DNA methylation. J. Biol. Chem. 2003;278:32181–32188. doi: 10.1074/jbc.M305308200. [DOI] [PubMed] [Google Scholar]
- 22.Zhao W., Soejima H., Higashimoto K., Nakagawachi T., Urano T., Kudo S., Matsukura S., Matsuo S., Joh K., Mukai T. The essential role of histone H3 Lys9 di-methylation and MeCP2 binding in MGMT silencing with poor DNA methylation of the promoter CpG island. J. Biochem. (Tokyo) 2005;137:431–440. doi: 10.1093/jb/mvi048. [DOI] [PubMed] [Google Scholar]
- 23.Thomas J.O. Histone H1: location and role. Curr. Opin. Cell Biol. 1999;11:312–317. doi: 10.1016/S0955-0674(99)80042-8. [DOI] [PubMed] [Google Scholar]
- 24.Zhou Y.B., Gerchman S.E., Ramakrishnan V., Travers A., Muyldermans S. Position and orientation of the globular domain of linker histone H5 on the nucleosome. Nature. 1998;395:402–405. doi: 10.1038/26521. [DOI] [PubMed] [Google Scholar]
- 25.Duggan M.M., Thomas J.O. Two DNA-binding sites on the globular domain of histone H5 are required for binding to both bulk and 5 S reconstituted nucleosomes. J. Mol. Biol. 2000;304:21–33. doi: 10.1006/jmbi.2000.4205. [DOI] [PubMed] [Google Scholar]
- 26.Varga-Weisz P., van Holde K., Zlatanova J. Preferential binding of histone H1 to four-way helical junction DNA. J. Biol. Chem. 1993;268:20699–20700. [PubMed] [Google Scholar]
- 27.Varga-Weisz P., Zlatanova J., Leuba S.H., Schroth G.P., van Holde K. Binding of histones H1 and H5 and their globular domains to four-way junction DNA. Proc. Natl Acad. Sci. USA. 1994;91:3525–3529. doi: 10.1073/pnas.91.9.3525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cross S.H., Charlton J.A., Nan X., Bird A.P. Purification of CpG islands using a methylated DNA binding column. Nature Genet. 1994;6:236–244. doi: 10.1038/ng0394-236. [DOI] [PubMed] [Google Scholar]
- 29.Webb M., Thomas J.O. Structure-specific binding of the two tandem HMG boxes of HMG1 to four-way junction DNA is mediated by the A domain. J. Mol. Biol. 1999;294:373–387. doi: 10.1006/jmbi.1999.3150. [DOI] [PubMed] [Google Scholar]
- 30.Lee K.B., Thomas J.O. The effect of the acidic tail on the DNA-binding properties of the HMG1,2 class of proteins: insights from tail switching and tail removal. J. Mol. Biol. 2000;304:135–149. doi: 10.1006/jmbi.2000.4206. [DOI] [PubMed] [Google Scholar]
- 31.Varga-Weisz P., van Holde K., Zlatanova J. Competition between linker histones and HMG1 for binding to four-way junction DNA: implications for transcription. Biochem. Biophys. Res. Commun. 1994;203:1904–1911. doi: 10.1006/bbrc.1994.2410. [DOI] [PubMed] [Google Scholar]
- 32.Klose R.J., Bird A.P. MeCP2 behaves as an elongated monomer that does not stably associate with the Sin3a chromatin remodeling complex. J. Biol. Chem. 2004;279:46490–46496. doi: 10.1074/jbc.M408284200. [DOI] [PubMed] [Google Scholar]
- 33.Ballestar E., Yusufzai T.M., Wolffe A.P. Effects of Rett syndrome mutations of the methyl-CpG binding domain of the transcriptional repressor MeCP2 on selectivity for association with methylated DNA. Biochemistry. 2000;39:7100–7106. doi: 10.1021/bi0001271. [DOI] [PubMed] [Google Scholar]
- 34.Free A., Wakefield R.I., Smith B.O., Dryden D.T., Barlow P.N., Bird A.P. DNA recognition by the methyl-CpG binding domain of MeCP2. J. Biol. Chem. 2001;276:3353–3360. doi: 10.1074/jbc.M007224200. [DOI] [PubMed] [Google Scholar]
- 35.Yusufzai T.M., Wolffe A.P. Functional consequences of Rett syndrome mutations on human MeCP2. Nucleic Acids Res. 2000;28:4172–4179. doi: 10.1093/nar/28.21.4172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Draves P.H., Lowary P.T., Widom J. Co-operative binding of the globular domain of histone H5 to DNA. J. Mol. Biol. 1992;225:1105–1121. doi: 10.1016/0022-2836(92)90108-v. [DOI] [PubMed] [Google Scholar]
- 37.Thomas J.O., Rees C., Finch J.T. Cooperative binding of the globular domains of histones H1 and H5 to DNA. Nucleic Acids Res. 1992;20:187–194. doi: 10.1093/nar/20.2.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramakrishnan V., Finch J.T., Graziano V., Lee P.L., Sweet R.M. Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature. 1993;362:219–223. doi: 10.1038/362219a0. [DOI] [PubMed] [Google Scholar]
- 39.Goytisolo F.A., Gerchman S.E., Yu X., Rees C., Graziano V., Ramakrishnan V., Thomas J.O. Identification of two DNA-binding sites on the globular domain of histone H5. EMBO J. 1996;15:3421–3429. [PMC free article] [PubMed] [Google Scholar]
- 40.Simpson R.T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978;17:5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- 41.Chandler S.P., Guschin D., Landsberger N., Wolffe A.P. The methyl-CpG binding transcriptional repressor MeCP2 stably associates with nucleosomal DNA. Biochemistry. 1999;38:7008–7018. doi: 10.1021/bi990224y. [DOI] [PubMed] [Google Scholar]
- 42.Pohler J.R.G., Norman D.G., Brahmam J., Bianchi M.E., Lilley D.M.J. HMG box proteins bind to four-way junctions in their open confirmation. EMBO J. 1998;17:817–826. doi: 10.1093/emboj/17.3.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohki I., Shimotake N., Fujita N., Jee J., Ikegami T., Nakao M., Shirakawa M. Solution structure of the methyl-CpG binding domain of human MBD1 in complex with methylated DNA. Cell. 2001;105:487–497. doi: 10.1016/s0092-8674(01)00324-5. [DOI] [PubMed] [Google Scholar]
- 44.Connolly K.M., Ilangovan U., Wojciak J.M., Iwahara M., Clubb R.T. Major groove recognition by three-stranded beta-sheets: affinity determinants and conserved structural features. J. Mol. Biol. 2000;300:841–856. doi: 10.1006/jmbi.2000.3888. [DOI] [PubMed] [Google Scholar]
- 45.Flick K.E., Jurica M.S., Monnat R.J., Jr, Stoddard B.L. DNA binding and cleavage by the nuclear intron-encoded homing endonuclease I-PpoI. Nature. 1998;394:96–101. doi: 10.1038/27952. [DOI] [PubMed] [Google Scholar]
- 46.Wojciak J.M., Connolly K.M., Clubb R.T. NMR structure of the Tn916 integrase-DNA complex. Nature Struct. Biol. 1999;6:366–373. doi: 10.1038/7603. [DOI] [PubMed] [Google Scholar]
- 47.Allen M.D., Yamasaki K., Ohme-Takagi M., Tateno M., Suzuki M. A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998;17:5484–5496. doi: 10.1093/emboj/17.18.5484. [DOI] [PMC free article] [PubMed] [Google Scholar]