Abstract
For the detection and identification of predominant bacteria in human feces, 16S rRNA-gene-targeted group-specific primers for the Bacteroides fragilis group, Bifidobacterium, the Clostridium coccoides group, and Prevotella were designed and evaluated. The specificity of these primers was confirmed by using DNA extracted from 90 species that are commonly found in the human intestinal microflora. The group-specific primers were then used for identification of 300 isolates from feces of six healthy volunteers. The isolates were clearly identified as 117 isolates of the B. fragilis group, 22 isolates of Bifidobacterium, 65 isolates of the C. coccoides group, and 17 isolates of Prevotella, indicating that 74% of the isolates were identified with the four pairs of primers. The remaining 79 isolates were identified by 16S ribosomal DNA sequence analysis and consisted of 40 isolates of Collinsella, 24 isolates of the Clostridium leptum subgroup, and 15 isolates of disparate clusters. In addition, qualitative detection of these bacterial groups was accomplished without cultivation by using DNA extracted from the fecal samples. The goal for this specific PCR technique is to develop a procedure for quantitative detection of these bacterial groups, and a real-time quantitative PCR for detection of Bifidobacterium is now being investigated (T. Requena, J. Burton, T. Matsuki, K. Munro, M. A. Simon, R. Tanaka, K. Watanabe, and G. W. Tannock, Appl. Environ. Microbiol. 68:2420-2427, 2002). Therefore, the approaches used to detect and identify predominant bacteria with the group-specific primers described here should contribute to future studies of the composition and dynamics of the intestinal microflora.
The human intestinal tract harbors a large, active, and complex community of microbes. The intestinal microflora plays several significant roles in the digestion of food, metabolism of endogenous and exogenous compounds, immunopotentiation, and prevention of colonization by pathogens in the gastrointestinal tract and hence is involved in maintaining human health (8, 36). The gut microflora has been investigated in great detail by using anaerobic culture techniques (5, 21, 23-25). The predominant genera in the large bowel are reported to be Bacteroides, Eubacterium, Clostridium, Ruminococcus, Peptococcus, Peptostreptococcus, Bifidobacterium, and Fusobacterium. Thus, intensive investigations have provided significant information concerning the flora. However, the classical culture methods are labor-intensive and time-consuming. Moreover, classification and identification based on phenotypic traits do not always provide clear-cut results and are sometimes unreliable.
For some years, molecular techniques based on 16S rRNA sequences have attracted attention as reliable methods for detection and identification of bacterial species (26, 42). Techniques such as the clone library method (35, 41) and denaturing gradient gel electrophoresis pattern analysis (33, 43) have allowed analysis of bacteria that are difficult to culture but represent a significant population. Methods involving 16S rRNA-targeted hybridization probes or PCR primers enable rapid and specific detection of a wide range of bacterial species and have become key procedures for detection of microorganisms (3, 7, 16, 19, 32, 39). Depending on the primers used, the hybridization method and the PCR technique can be used to detect bacteria at different phylogenetic levels. For complex mixed populations, 16S rRNA-targeted oligonucleotide probes have been used with fluorescent in situ hybridization as a culture-independent method (7, 17, 31). Franks et al. developed and used eight 16S rRNA-targeted probes for major species and groups of anaerobic intestinal bacteria to enumerate the bacterial population in fresh feces of healthy volunteers (7). According to their estimates, members of the genus Bacteroides and the Clostridium coccoides group constituted one-half of the fecal flora examined (7, 18).
On the other hand, PCR techniques with specific 16S ribosomal DNA (rDNA)-based oligonucleotide primers have been developed as powerful methods for detecting target bacteria in complex ecosystems (39). So far, specific oligonucleotide primers have been designed for many bacterial species which are known to be present in the intestinal tract, and these primers have been used successfully (14, 19, 20, 29, 34, 38-40). However, the complex microflora of the human gut is difficult to study with only primers that are specific at the species level due to the diversity of this ecosystem. Therefore, it is more convenient to have primers which are specific for major genera and groups present in the gut. Genus-specific primers have been designed for Bifidobacterium and have been extensively tested (15, 16). However, the number of such group-specific primers is still limited, in spite of a number of 16S rRNA-targeted group-specific hybridization probes which have been prepared (7, 11, 17, 32).
In this study, we designed 16S rRNA-gene-targeted group-specific primers for the Bacteroides fragilis group, Bifidobacterium, the C. coccoides group, and Prevotella. The specificity of these primers was tested with a range of reference strains that are the predominant bacteria in the human intestinal tract. After this validation, the specific primers were used for identification of colonies obtained by culture methods and for specific PCR detection with human fecal DNA.
MATERIALS AND METHODS
Reference strains and culture conditions.
The strains listed in Table 1 were obtained from the American Type Culture Collection (Rockville, Md.) (ATCC), the Japan Collection of Microorganisms (Wako, Japan) (JCM), and the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany) (DSM). Most of the strains were cultured anaerobically in GAM broth (Nissui Seiyaku, Tokyo, Japan) supplemented with 1% glucose at 37°C overnight; the only exception was Escherichia coli, which was cultured aerobically in Trypticase soy broth (Difco, Detroit, Mich.) at 37°C overnight. When required, the number of bacteria was determined microscopically by the method of Jansen et al. (13). Vectashield with 4′,6′-diamidino-2-phenylindole (DAPI) (Vector Laboratories, Burlingame, Calif.) was used for DNA staining and mounting. Microscopic counts were determined from 10 images, and a minimum of 100 cells were counted per image.
TABLE 1.
Specificity tests with the group-specific primers
| Species | Strain | Reactions with the following group-specific primersb:
|
|||
|---|---|---|---|---|---|
| g-Bfrac | g-Prevod | g-Bifide | g-Ccocf | ||
| Bacteroides fragilis | NCTC 9343T | + | − | − | − |
| Bacteroides ovatus | JCM 5824T | + | − | − | − |
| Bacteroides thetaiotaomicron | JCM 5827T | + | − | − | − |
| Bacteroides uniformis | JCM 5828T | + | − | − | − |
| Bacteroides vulgatus | ATCC 8482T | + | − | − | − |
| Bacteroides distasonis | JCM 5825T | − | − | − | − |
| Bacteroides putredenis | ATCC 29800T | − | − | − | − |
| Prevotella buccae | DSM 20615 | − | + | − | − |
| Prevotella intermedia | JCM 7365T | − | + | − | − |
| Prevotella melaninogenica | JCM 6321 | − | + | − | − |
| Prevotella oralis | JCM 6330 | − | + | − | − |
| Bifidobacterium adolescentis | ATCC 15703T | − | − | + | − |
| Bifidobacterium angulatum | ATCC 27535T | − | − | + | − |
| Bifidobacterium bifidum | ATCC 29521T | − | − | + | − |
| Bifidobacterium breve | ATCC 15700T | − | − | + | − |
| Bifidobacterium catenulatum | ATCC 27539T | − | − | + | − |
| Bifidobacterium pseudocatenulatum | JCM 1200T | − | − | + | − |
| Bifidobacterium longum | ATCC 15707T | − | − | + | − |
| Bifidobacterium infantis | ATCC 15697T | − | − | + | − |
| Clostridium clostridioformea | JCM 1291T | − | − | − | + |
| Clostridium coccoidesa | JCM 1395T | − | − | − | + |
| Clostridium nexilea | DSM 1787T | − | − | − | + |
| Clostridium oroticuma | JCM 1429T | − | − | − | + |
| Clostridium sphenoidesa | JCM 1415T | − | − | − | + |
| Ruminococcus gnavusa | ATCC 29149T | − | − | − | + |
| Ruminococcus lactarisa | ATCC 29176T | − | − | − | + |
| Ruminococcus productusa | ATCC 27340T | − | − | − | + |
| Clostridium baratii | JCM 1385T | − | − | − | − |
| Clostridium celatum | JCM 1394T | − | − | − | − |
| Clostridium perfringens | JCM 1290T | − | − | − | − |
| Clostridium sporogenes | JCM 1416T | − | − | − | − |
| Clostridium limosum | JCM 1427T | − | − | − | − |
| Clostridium bifermentans | JCM 1386T | − | − | − | − |
| Clostridium glycolicum | JCM 1401T | − | − | − | − |
| Clostridium sordellii | JCM 3814T | − | − | − | − |
| Fusobacterium varium | ATCC 8501T | − | − | − | − |
| Fusobacterium gonidiaformans | ATCC 25563T | − | − | − | − |
| Fusobacterium prausnitzii | ATCC 27766T | − | − | − | − |
| Escherichia coli | ATCC 11775T | − | − | − | − |
| Enterococcus faecalis | ATCC 19433T | − | − | − | − |
| Enterococcus faecium | ATCC 19434T | − | − | − | − |
| Porphyromonas asaccharolytica | ATCC 25260T | − | − | − | − |
| Porphyromonas gingivalis | JCM 8525 | − | − | − | − |
| Collinsella aerofaciens | ATCC 25986T | − | − | − | − |
| Eubacterium biforme | ATCC 27806T | − | − | − | − |
| Propionibacterium acnes | ATCC 6919T | − | − | − | − |
| Lactobacillus acidophilus | ATCC 4356T | − | − | − | − |
| Peptostreptococcus prevotii | ATCC 9321T | − | − | − | − |
These species belong to the C. coccoides group.
+, positive; −, negative.
In addition, positive PCR results were obtained for Bacteroides heparinolytica ATCC 35895T, Bacteroides caccae ATCC 43185T, and Bacteroides stercoris ATCC 43183T.
In addition, positive PCR results were obtained for Prevotella buccalis DSM 20616T, Prevotella corporis JCM 8529T, Prevotella denticola JCM 8528T, Prevotella loescheii JCM 8530T, Prevotella oris JCM 8540T, Prevotella ruminicola JCM 8958T, and Prevotella veroralis JCM 6290T.
In addition, positive PCR results were obtained for Bifidobacterium angulatum ATCC 27535T, Bifidobacterium animalis ATCC 25527T, Bifidobacterium asteroides ATCC 25910T, Bifidobacterium boum JCM 1211T, Bifidobacterium choerinun ATCC 27686T, Bifidobacterium coryneforme ATCC 25911T, Bifidobacterium cunniculi ATCC 27916T, Bifidobacterium dentium ATCC 27534T, Bifidobacterium denticolens DSM 10105T, Bifidobacterium gallinarum JCM 6291T, Bifidobacterium indicum ATCC 25912T, Bifidobacterium inopinatum DSM 10107T, Bifidobacterium lactis DSM 10140T, Bifidobacterium magnum JCM 1218T, Bifidobacterium merycicum JCM 8219T, Bifidobacterium minimum ATCC 27538T, Bifidobacterium pseudolongum subsp. globosum ATCC 25864T, Bifidobacterium pseudolongum subsp. pseudolongum JCM 1205T, Bifidobacterium pullorum JCM 1214T, Bifidobacterium ruminantium JCM 8222T, Bifidobacterium saeculare DSM 6531T, Bifidobacterium subtile DSM 20096T, Bifidobacterium thermophilum ATCC 25866T, and Gardnerella vaginalis DSM 4944T.
In addition, positive PCR results were obtained for Clostridium aminovalericum DSM 1283T, Clostridium symbiosum JCM 1297T, Ruminococcus hansenii ATCC 27752T, Ruminococcus hydrogenotrophicus DSM 10507T, Ruminococcus schinkii DSM 10518T, and Ruminococcus torques ATCC 27756T.
Development of 16S rDNA-targeted species-specific primers.
By using 16S rRNA sequences obtained from the DDBJ, GenBank, and EMBL databases, multiple alignments of the target groups and reference organisms were constructed with the program Clustal X (37). After sequences unique to the group were compared with the sequences of a large number of reference strains, potential target sites for specific detection were identified (Tables 2 and 3). These oligonucleotide sequences were then checked by using the Check-Probe function of the Ribosomal Database Project software package (18). The primers were synthesized commercially by Greiner Japan (Tokyo, Japan).
TABLE 2.
Partial 16S rDNA sequences of the reference organisms obtained with the group-specific primers
| Organism or sitea | Forward primer
|
Reverse primer
|
||
|---|---|---|---|---|
| Designation | Sequenceb | Designation | Sequenceb | |
| B. fragilis group-specific primers | g-Bfra-F | 5′ ATAGCCTTTCGAAA 3′ | g-Bfra-R | 3′ ATTTTAACGTCAACTATGACC 5′ |
| Target site | 5′ ATAGCCTTTCGAAA 3′ | 5′ TAAAATTGCAGTTGATACTGG 3′ | ||
| Bacteroides caccaec | .............. | ..................... | ||
| Bacteroides fragilisc | .............. | ....................T | ||
| Bacteroides ovatusc | .............. | ...............A..... | ||
| Bacteroides thetaiotaomicronc | .............. | ..................... | ||
| Bacteroides vulgatusc | .C............ | ..................... | ||
| Bacteroides distasonis | ...A..CGG..... | ..G......C.....A..... | ||
| Porphyromonas gingivalis | ...A..CG.T.... | .C.GCC...C.....A....C | ||
| Prevotella melaninogenica | ...A...GC..... | CTG........CGC....... | ||
| Bifidobacterium longum | .....TCC.G.... | .GG.TCC..GCCG.G...G.. | ||
| Clostridium coccoides | ...ACAG..AG... | C.GG.C....T.G..A....T | ||
| Collinsella aerofaciens | ......GCC..... | CCCG.AGC.CCCG..AC..CC | ||
| Prevotella-specific primers | g-Prevo-F | 5′ CACRGTAAACGATGGATGCC 3′ | g-Prevo-R | 3′ CCAGACGTTGGGCTGG 5′ |
| Target site | 5′ CACRGTAAACGATGGATGCC 3′ | 5′ GGTCTGCAACCCGACC 3′ | ||
| Prevotella buccalisc | .................... | ................ | ||
| Prevotella intermediac | .................... | ................ | ||
| Prevotella melaninogenicac | .................... | ................ | ||
| Prevotella oralisc | .................... | ................ | ||
| Bacteroides vulgatus | ..............A..A.T | A..............T | ||
| Porphyromonas gingivalis | .G............AT.A.T | A.........T....T | ||
| Bifidobacterium longum | .G.C.......G.......T | A.........T....T | ||
| Clostridium coccoides | .G.C..N...N...A..A.T | A......N..T....T | ||
| Collinsella aerofaciens | AG.C............C..T | ..GC..........C. | ||
| Bifidobacterium-specific primers | g-Bifid-F | 5′ CTCCTGGAAACGGGTGG 3′ | g-Bifid-R | 3′ ACATCTATAGCCCTTCTTGTGG 5′ |
| Target site | 5′ CTCCTGGAAACGGGTGG 3′ | 5′ TGTAGATATCGGGAAGAACACC 3′ | ||
| Bifidobacterium adolescentisc | ................. | ...................... | ||
| Bifidobacterium brevec | ................. | ...................... | ||
| Bifidobacterium catenulatumc | ................. | ...................... | ||
| Bifidobacterium longumc | ................. | ...................... | ||
| Propionibacterium acnes | ..T.A......T..G.C | C.C.......A...G....... | ||
| Bacteroides vulgatus | .CTTCT....G.AAGAT | CT........AC.......T.. | ||
| Clostridium coccoides | .AGT.A....T.AC..C | C........TA...G....... | ||
| Collinsella aerofaciens | .CG.CC....G.ACG.. | C.C..........TG....... | ||
| C. coccoides group-specific primers | g-Ccoc-F | 5′ AAATGACGGTACCTGACTAA 3′ | g-Ccoc-R | 3′ AAGCGTTCTTACTTTGAGTTTC 5′ |
| Target site | 5′ AAATGACGGTACCTGACTAA 3′ | 5′ TTCGCAAGAATGAAACTCAAAG 3′ | ||
| Clostridium clostridioformec | .................... | ...................... | ||
| Ruminococcus gnavusc | .................... | ...................... | ||
| Clostridium coccoidesc | ..................N. | ...................... | ||
| Eubacterium rectalec | T................... | ...................... | ||
| Clostridium perfringens | T............CA.GG.G | G........T.A.......... | ||
| Clostridium ramosum | G.G.........T.T.T.TT | ...................... | ||
| Bacteroides vulgatus | GNT..CAT....T.T.TG.. | CCG....CGG............ | ||
| Bifidobacterium longum | G.G...GTT....C.TTG.. | GC......GC.A.......... | ||
| Collinsella aerofaciens | TC.A...T.......CAG.. | GC......N-.A.......... | ||
The positions of the target sites for the forward primers are as follows (numbering based on the E. coli 16S rDNA sequence): B. fragilis group-specific primer, nucleotides 148 to 169; Prevotella-specific primer, nucleotides 803 to 827; Bifidobacterium-specific primer, nucleotides 153 to 169; and C. coccoides group-specific primer, nucleotides 477 to 496. The target sites for the reverse primers are as follows: B. fragilis group-specific primer, nucleotides 626 to 646; Prevotella-specific primer, nucleotides 1311 to 1335; Bifidobacterium-specific primer, nucleotides 699 to 720; and C. coccoides group-specific primer, nucleotides 895 to 916.
Only the nucleotides that are different from the nucleotides in the target sequences are shown.
Targeted organism.
TABLE 3.
Group-specific primers based on 16S rDNA sequences
| Target bacteria | Primera | Sequence (5′ to 3′) | Product size (bp) | Annealing temp (°C) |
|---|---|---|---|---|
| Bacteroides fragilis group | g-Bfra-F | ATAGCCTTTCGAAAGRAAGAT | 501 | 50 |
| g-Bfra-R | CCAGTATCAACTGCAATTTTA | |||
| Prevotella | g-Prevo-F | CACRGTAAACGATGGATGCC | 527-529 | 55 |
| g-Prevo-R | GGTCGGGTTGCAGACC | |||
| Bifidobacterium | g-Bifid-F | CTCCTGGAAACGGGTGG | 549-563 | 55 |
| g-Bifid-R | GGTGTTCTTCCCGATATCTACA | |||
| Clostridium coccoides group | g-Ccoc-F | AAATGACGGTACCTGACTAA | 438-441 | 50 |
| g-Ccoc-R | CTTTGAGTTTCATTCTTGCGAA |
The standardized primer names are as follows: g-Bfra-F, S-G-Bfra-0148-a-S-21; g-Bfra-R, S-G-Bfra-0626-a-A-21; g-Prevo-F, S-G-Prevo-0803-a-S-20; g-Prevo-R, S-G-Prevo-1311-a-A-16; g-Bifid-F, S-G-Bifid-0153-a-S-17; g-Bifid-R, S-G-Bifid-0699-a-A-22; g-Ccoc-F, S-G-Ccoc-0477-a-S-20; and g-Ccoc-R, S-G-Ccoc-0895-a-A-22 (1).
PCR amplification.
Each PCR mixture (25 μl) was composed of 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 2.5 mM MgCl2, each deoxynucleoside triphosphate at a concentration of 200 μM, each group-specific primer (Table 3) at a concentration of 0.25 μM, template DNA, and 0.45 U of Taq DNA polymerase (Perkin-Elmer, Norwalk, Conn.). The PCR was carried out with a Gene Amp PCR System 9600 (Perkin-Elmer). The amplification program consisted of one cycle of 94°C for 5 min; 40 cycles of 94°C for 20 s, 55 or 50°C for 20 s, and 72°C for 30 s; and finally one cycle of 72°C for 5 min. The amplification products were subjected to gel electrophoresis in 1% agarose, followed by ethidium bromide staining.
Fecal samples.
Fecal specimens from six healthy adult volunteers who were 28 to 52 years old (five males and one female) were collected, and serial 10-fold dilutions were prepared with prereduced dilution buffers in an anaerobic cabinet, after which 0.05-ml samples of the 107 to 109 dilutions were plated on nonselective Medium 10 agar (12). The plates were subsequently incubated at 37°C for 4 days under strictly anaerobic conditions with N2-CO2-H2 (88:5:7, vol/vol/vol) as the gas phase, and cultural counts (CFU) for total anaerobes were determined in duplicate. Total cell counts were also determined by using DAPI staining as described above.
Isolation of predominant bacteria.
Fifty colonies that appeared on the Medium 10 agar plates inoculated with the highest dilution were transferred with a sterile toothpick to 50 μl of 10 mM Tris-HCl-1 mM EDTA (pH 8.0) (TE buffer). One microliter of the suspension was smeared onto a glass slide for Gram staining. The remaining suspension was boiled for 15 min to lyse the cells and used as template DNA for the PCR.
16S rDNA sequence analysis.
Each PCR was performed with primers 926f (5′-AAA CTY AAA KGA ATT GAC GG-3′) and 1392r (5′-ACG GGC GGT GTG TRC-3′) to amplify 16S rDNA (positions 906 to 1406 in the Escherichia coli numbering system) directly from the transferred colonies. The PCR was performed under the following conditions: 94°C for 3 min; 35 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min; and finally 72°C for 5 min. The PCR products were purified with Microspin S-400 columns (Pharmacia Biotech, Uppsala, Sweden) as recommended by the manufacturer. The purified DNA was used for 16S rDNA sequence analysis performed with an ABI Prism dye terminator cycle sequencing Ready Reaction kit and primers 926f and 1392r as the sequencing primers. The sequences were automatically analyzed with an ABI model 373A DNA sequencer (Applied Biosystems, Foster City, Calif.). The assembled partial rDNA sequences were compared with sequences in the GenBank database (2).
DNA extraction from fecal samples.
Fecal samples (10 mg) were washed three times by suspending them in 1.0 ml of phosphate-buffered saline and centrifuging each preparation at 14,000 × g in order to reduce the PCR inhibitors. The fecal pellets were resuspended in 450 μl of extraction buffer (100 mM Tris-HCl, 40 mM EDTA; pH 9.0) and 50 μl of 10% sodium dodecyl sulfate. Three hundred milligrams of glass beads (diameter, 0.1 mm) and 500 μl of buffer-saturated phenol were added to the suspension, and the mixture was vortexed vigorously for 30 s by using a FastPrep FP 120 (Bio 101, Vista, Calif.) at a power level of 5.0. After centrifugation at 14,000 × g for 5 min, the supernatant was collected. Subsequently, phenol-chloroform extractions were performed, and DNA was obtained by isopropanol precipitation. Finally, the DNA was suspended in 1 ml of TE buffer. Routinely, 1 μl of the fecal DNA solution was used for the PCR analysis.
RESULTS
Specificity of primers.
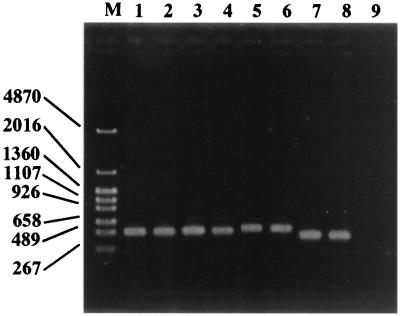
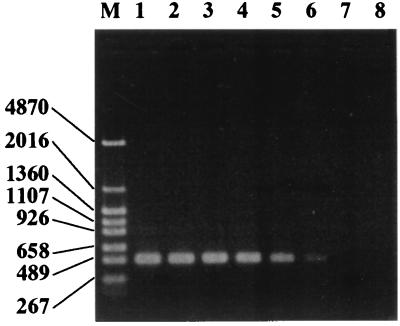
When group-specific amplification was performed with the newly developed primers, PCR products of the expected size were obtained (Fig. 1). The specificity of each primer was experimentally tested by using DNA extracts from strains representing 90 different bacterial species (Table 1). The specific primers gave positive PCR results for the corresponding target bacteria and did not cross-react with any of the nontarget microorganisms. The detection limits for these group-specific PCR techniques were also determined with DNA extracted from pure cultured bacteria. Figure 2 shows that B. fragilis NCTC 9343T was detected with the g-Bfra primers at a concentration of 10 cells per PCR mixture. Similar results were obtained for Bacteroides vulgatus ATCC 8424T, Prevotella melaninogenica JCM 6321, Bifidobacterium adolescentis ATCC 15703T, Bifidobacterium longum ATCC 15707T, Clostridium clostridioforme JCM 1291T, and C. coccoides JCM 1395T with their specific primers (data not shown).
FIG. 1.
PCR products obtained for eight species with group-specific primers. Lane M, DNA size markers (sizes [in bases] are indicated on the left); lane 1, Bacteroides fragilis NCTC 9343T; lane 2, Bacteroides vulgatus ATCC 8424 T; lane 3, Prevotella melaninogenica JCM 6321; lane 4, Prevotella buccae DSM 20615; lane 5, Bifidobacterium bifidum ATCC 29521T; lane 6, Bifidobacterium longum ATCC 157071T; lane 7, Clostridium coccoides JCM 1395T; lane 8, Clostridium clostridioforme JCM 1291T; lane 9, negative control (PCR performed with g-Bfra primers and Escherichia coli ATCC 11775T).
FIG. 2.
Detection limits of the group-specific PCR methods, as determined by using DNA extracted from pure cultured B. fragilis NCTC 9343T. Lane M, DNA size markers (sizes [in bases] are indicated on the left); lane 1, 106 cells per PCR mixture; lane 2, 105 cells per PCR mixture; lane 3, 104 cells per PCR mixture; lane 4, 103 cells per PCR mixture; lane 5, 102 cells per PCR mixture; lane 6, 10 cells per PCR mixture; lane 7, 1 cell per PCR mixture; lane 8, no cells (negative control).
Bacterial counts.
According to DAPI staining, there were 2.3 × 1011, 3.8 ×1011, 1.1 × 1011, 2.7 × 1011, 6.3 × 1010, and 4.0 × 1010 cells per g (wet weight) of feces in samples from volunteers A, B, C, D, E, and F, respectively (mean ± standard deviation, 1.8 × 1011 ± 1.3 × 1011 cells per g [wet weight]); the numbers of cultivated bacteria in the anaerobic chamber with Medium 10 were 1.1 × 1011, 9.3 × 1010, 6.7 × 1010, 1.8 × 1011, 4.8 × 1010, and 2.0 × 1010 cells per g, respectively (mean ± standard deviation, 8.6 × 1010 ± 5.6 × 1010 cells per g). These results indicated that organisms which grew anaerobically on a nonselective medium accounted for 48, 24, 61, 67, 76, and 50% of the bacteria counted by DAPI staining, respectively (mean ± standard deviation, 54% ± 18%).
Identification of the isolates.
By using the group-specific primers, 300 isolates from feces of six volunteers were identified as 117 isolates of the B. fragilis group, 65 isolates of the C. coccoides group, 22 isolates of Bifidobacterium, and 17 isolates of Prevotella; 79 isolates remained unidentified. All of the isolates identified as Bacteroides and Prevotella were gram-negative rods, while the Bifidobacterium isolates were gram-positive rods. On the other hand, the Gram staining results and the morphology of the isolates identified as members of the C. coccoides group were diverse, and the organisms ranged from gram-negative rods to gram-positive cocci. The compositions of the bacterial groups in each sample are summarized in Table 4. On average, the B. fragilis group accounted for 39% ± 31% of the total culturable population, while the C. coccoides group accounted for 22% ± 11%. Although Bifidobacterium was not isolated from volunteers E and F, this group of bacteria accounted for 7.3% ± 8.5%. Prevotella was detected only in volunteers C and D and accounted for 5.7% ± 9.3% of the total culturable population.
TABLE 4.
Identification of 300 strains isolated from feces of six volunteers
| Bacterial group | No. of strains obtained from the following volunteers:
|
Total no. of strains | |||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | ||
| Isolates identified with group-specific primers | |||||||
| Bacteroides fragilis group | 27 | 17 | 6 | 1 | 21 | 45 | 117 |
| Bifidobacterium | 2 | 10 | 8 | 2 | 0 | 0 | 22 |
| Clostridium coccoides group | 10 | 8 | 11 | 18 | 16 | 2 | 65 |
| Prevotella | 0 | 0 | 11 | 6 | 0 | 0 | 17 |
| Isolates identified by sequencing of 16S rDNAa | |||||||
| Collinsella | 3 | 6 | 8 | 20 | 3 | 0 | 40 |
| Clostridium leptum subgroup | 4 | 6 | 3 | 2 | 8 | 1 | 24 |
| Disparate cluster | 4 | 3 | 3 | 1 | 2 | 2 | 15 |
The results of a 16S rDNA phylogenetic analysis of isolates that were not identified with the group-specific primers are shown in Table 5.
16S rDNA sequence analysis of unidentified isolates.
The 16S rDNA sequences of the 79 isolates which were not identified with the group-specific primers were determined. These sequences were compared to those available in public databases in order to ascertain their closest relatives (Table 5). Forty isolates were identified as Collinsella aerofaciens. Twenty-four isolates were included in the Clostridium leptum subgroup (18), which is equivalent to Clostridium cluster IV (4). The remaining 15 isolates were members of other phylogenetic groups, such as the Porphylomonas macacae subgroup (isolates A14, A45, B18, B39, B44, and E09), the Rikenella microfusus subgroup (isolates C24 and E44), the Acholeplasma-Anaeroplasma group (isolates C13 and D47), and other groups (18). Collinsella accounted for 13% ± 14% of the total culturable population, while the C. leptum subgroup accounted for 8.0% ± 5.2% (Table 4).
TABLE 5.
16S rDNA phylogenetic analysis of 79 isolates that were not identified with the newly developed group-specific primers
| Isolate(s) | Microorganism with most similar sequence in database (accession no.) | % Simi- larity |
|---|---|---|
| Collinsella | ||
| A15, A46, A46, B02, B03, B08, B28, B49, C08, C25, C28, C35, D05, D06, D09, D10, D14, D15, D17, D18, D19, D21, D22, D26, D30, D33, D34, D41, D44, D49, E13, E19, E23 | Collinsella aerofaciens (AB011816) | 100 |
| C11, C15, C18, C41 | Collinsella aerofaciens (AB011816) | 99 |
| B17, D04, D23 | Collinsella aerofaciens (AB011816) | 98 |
| Clostridium leptum subgroup | ||
| A05, E10, E33 | Fusobacterium prausnitzii (X85022) | 99 |
| B26 | Fusobacterium prausnitzii (X85022) | 98 |
| A42, D50, E32, E34 | Fusobacterium prausnitzii (X85022) | 97 |
| C29, C44, D45 | Fusobacterium prausnitzii (X85022) | 96 |
| E47 | Fusobacterium prausnitzii (X85022) | 95 |
| C45 | Fusobacterium prausnitzii (X85022) | 94 |
| B20, B50, E07, E49 | Ruminococcus albus (L76598) | 95 |
| B01, B29 | Ruminococcus albus (L76598) | 94 |
| A02, F30 | Eubacterium desmolans (L34618) | 98 |
| E25 | Ruminococcus bromii (L76600) | 100 |
| B34 | Ruminococcus bromii (L76600) | 94 |
| A50 | Clostridium orbiscindens (Y18187) | 98 |
| Disparate cluster | ||
| B44, E09 | Bacteroides distasonis (M86695) | 100 |
| A45 | Bacteroides distasonis (M86695) | 97 |
| A14 | Bacteroides distasonis (M86695) | 94 |
| B18, B39 | Bacteroides distasonis (M86695) | 93 |
| C24 | Bacteroides putredenis (L16497) | 97 |
| E44 | Bacteroides putredenis (L16497) | 96 |
| A26 | Clostridium propionicum (X77841) | 96 |
| A38 | Clostridium aldrichii (X71846) | 92 |
| C13, D47 | Clostridium ramosum (M23731) | 93 |
| C46 | Megasphaera elsdenii (U95029) | 100 |
| F11 | Clostridium hiranonis (AB023971) | 100 |
| F22 | Anaerovibria glycerini (AJ010960) | 91 |
Group-specific PCR detection.
Group-specific PCR assays were applied to DNA extracted from fecal samples from the six volunteers. The B. fragilis group, Bifidobacterium, and the C. coccoides group were detected in all the samples, whereas Prevotella was detected only in samples from volunteers C and D.
DISCUSSION
To investigate the population structure of the human fecal microflora, new oligonucleotide primers for the B. fragilis group, Bifidobacterium, the C. coccoides group, and Prevotella were designed, validated, and used for detection and identification of the predominant bacteria in human feces.
The group-specific g-Bfra primers were developed to detect the B. fragilis group (18). Species of this cluster are isolated primarily from human feces. Although Bacteroides distasonis and Bacteroides putredenis are isolated from human feces, these two species are not members of the B. fragilis group (18). Therefore, the specificity of these primers is consistent with the phylogenetic relationships based on the 16S rDNA sequence. The g-Prevo primers are designed for specific detection of Prevotella. Although group-specific primers for Bifidobacterium were prepared by Kok et al. (16) and Kauffmann et al. (15), we found other specific sequences which are highly conserved in the genus Bifidobacterium. The g-Bifid primers gave positive PCR results with Gardnerella vaginalis as well. Although G. vaginalis is not a member of the genus Bifidobacterium, it is difficult to distinguish between these two genera on the basis of 16S rDNA sequences (18, 22). As G. vaginalis has not been isolated from human feces, the g-Bifid primers would be useful for analysis of the fecal flora. The members of the genus Clostridium do not form a monophyletic cluster on the basis of 16S rRNA sequences (4). Therefore, primers for phylogenetic groups or clusters had to be considered. Members of the C. coccoides group, which corresponded to Clostridium cluster XIVa (4), have been reported to be major components of the human fecal flora (7, 32). Although this group contains members of the genera Clostridium, Coprococcus, Eubacterium, Lachnospira, and Ruminococcus, the organisms falling into this branch are phylogenetically very similar to one another.
Extensive efforts have been made in the past to cultivate the bacteria found in human feces, with the result that the human intestinal flora is one of the most successfully studied natural communities of bacteria (5, 6, 23, 24). The total bacterial counts as shown by DAPI staining were in general agreement with the values obtained by other investigators (13, 17). On the other hand, considerable variation has been reported for culturable cell counts. According to some investigators, the majority of the fecal flora is culturable (6, 23), whereas other researchers have reported that the plate counts of total anaerobes were 5- or 10-fold lower than the total cell counts (9, 17, 35). The difference may be explained by the different culture methods and media used. In our hands, the culturable fraction was 54% of the total DAPI counts. The percentage in our study was in good agreement with the results obtained by Wilson and Blitchington (41), who used the same nonselective agar, Medium 10.
When a panel of four pairs of primers was used, 74% of the isolates were identified in the present study (Table 4). The results of identification with specific primers are consistent with the Gram staining results and the morphology of the isolates, although the C. coccoides group showed considerable variation. The proportion of the B. fragilis group and Bifidobacterium enumerated was consistent with current knowledge obtained by both culture-based and molecule-based methods (7, 23, 41). The proportions of the C. coccoides group and the C. leptum subgroup were comparable to the results obtained by other investigators (7, 32). C. aerofaciens is also well recognized as the predominant bacterium in the human fecal flora (10, 14, 23, 24). Although Prevotella is a genus found in both the oral microflora and the rumen microflora (27), this genus has been detected in the adult fecal flora by direct 16S rDNA analysis (35). The results of this study show that 95% of the cultivated bacteria could be assigned to six major phylogenetic lineages (the B. fragilis group, the C. coccoides group, Bifidobacterium, Prevotella, Collinsella, and the C. leptum subgroup). Therefore, group-specific primers for Collinsella and the C. leptum subgroup should be prepared.
By using DNAs extracted from fecal samples, qualitative PCR detection of the B. fragilis group, Bifidobacterium, the C. coccoides group, and Prevotella was accomplished. Targeted bacteria were detected when they were present at a concentration of at least 10 cells per PCR mixture, indicating that the detection limit for the procedures described here was 106 cells per g of feces. In contrast, the detection limit of the culture method for minor species was 2% of the total bacterial counts in this study (for example, 9.6 × 108 and 4.0 × 108 cells per g in samples from volunteers E and F, respectively). This accounts for the fact that Bifidobacterium was detected in volunteers E and F by the specific PCR technique but was not detected by the culture method.
Establishing a procedure for quantitative detection of these bacterial groups is a task for the future, and research into real-time quantitative detection is proceeding (28). The primers described here should also be used for group-specific PCR and denaturing gradient gel electrophoresis to monitor the diversity of the target bacterial groups in human feces (30) and for identification of the cloned 16S rRNA genes that were directly amplified from fecal DNA (35). Therefore, the techniques for detection and identification of predominant bacteria with the group-specific primers described here should create new opportunities for noncultivation studies of the human intestinal microflora.
REFERENCES
- 1.Alm, E. W., D. B. Oerther, N. Larsen, D. A. Stahl, and L. Raskin. 1996. The oligonucleotide probe database. Appl. Environ. Microbiol. 62:3557-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Amann, R. I., W. Ludwig, and K. H. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins, M. D., P. A. Lawson, A. Willems, J. J. Cordoba, J. Fernandez-Garayzabal, P. Garcia, J. Cai, H. Hippe, and J. A. Farrow. 1994. The phylogeny of the genus Clostridium: proposal of five new genera and eleven new species combinations. Int. J. Syst. Bacteriol. 44:812-826. [DOI] [PubMed] [Google Scholar]
- 5.Finegold, S. M., H. R. Attebery, and V. L. Sutter. 1974. Effect of diet on human fecal flora: comparison of Japanese and American diets. Am. J. Clin. Nutr. 27:1456-1469. [DOI] [PubMed] [Google Scholar]
- 6.Finegold, S. M., V. S. Sutter, and G. E. Mathisen. 1983. Normal indigenous intestinal flora, p. 3-31. In D. J. Hentges (ed.), Human intestinal microflora in health and disease. Academic Press, New York, N.Y.
- 7.Franks, A. H., H. J. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuller, R. 1989. Probiotics in man and animals. J. Appl. Bacteriol. 66:365-378. [PubMed] [Google Scholar]
- 9.Harmsen, H. J., G. R. Gibson, P. Elfferich, G. C. Raangs, A. C. Wildeboer-Veloo, A. Argaiz, M. B. Roberfroid, and G. W. Welling. 2000. Comparison of viable cell counts and fluorescence in situ hybridization using specific rRNA-based probes for the quantification of human fecal bacteria. FEMS Microbiol. Lett. 183:125-129. [DOI] [PubMed] [Google Scholar]
- 10.Harmsen, H. J., A. C. Wildeboer-Veloo, J. Grijpstra, J. Knol, J. E. Degener, and G. W. Welling. 2000. Development of 16S rRNA-based probes for the Coriobacterium group and the Atopobium cluster and their application for enumeration of Coriobacteriaceae in human feces from volunteers of different age groups. Appl. Environ. Microbiol. 66:4523-4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harmsen, H. J. M., P. Elfferich, F. Schut, and G. W. Welling. 1999. A 16S rRNA-targeted probe for detection of lactobacilli and enterococci in faecal samples by fluorescent in situ hybridization. Microb. Ecol. Health Dis. 11:3-12. [Google Scholar]
- 12.Holdman, L. V., E. P. Cato, and W. E. C. Moore. 1977. Anaerobe laboratory manual, 4th ed. Southern Printing Co., Blacksburg, Va.
- 13.Jansen, G. J., A. C. Wildeboer-Veloo, R. H. Tonk, A. H. Franks, and G. W. Welling. 1999. Development and validation of an automated, microscopy-based method for enumeration of groups of intestinal bacteria. J. Microbiol. Methods 37:215-221. [DOI] [PubMed] [Google Scholar]
- 14.Kageyama, A., M. Sakamoto, and Y. Benno. 2000. Rapid identification and quantification of Collinsella aerofaciens using PCR. FEMS Microbiol. Lett. 183:43-47. [DOI] [PubMed] [Google Scholar]
- 15.Kaufmann, P., A. Pfefferkorn, M. Teuber, and L. Meile. 1997. Identification and quantification of Bifidobacterium species isolated from food with genus-specific 16S rRNA-targeted probes by colony hybridization and PCR. Appl. Environ. Microbiol. 63:1268-1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kok, R. G., A. de Waal, F. Schut, G. W. Welling, G. Weenk, and K. J. Hellingwerf. 1996. Specific detection and analysis of a probiotic Bifidobacterium strain in infant feces. Appl. Environ. Microbiol. 62:3668-3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genus-specific 16S rRNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matsuki, T., K. Watanabe, R. Tanaka, and H. Oyaizu. 1998. Rapid identification of human intestinal bifidobacteria by 16S rRNA-targeted species- and group-specific primers. FEMS Microbiol. Lett. 167:113-121. [DOI] [PubMed] [Google Scholar]
- 21.Mitsuoka, T., K. Hayakawa, and N. Kimura. 1974. Die Faekalflora bei Menschen. II. Mitteilung: Die Zusammensetzung der Bifidobakerien flora der verschiedenen Altersgruppen. Zentbl. Bakteriol. Parasitenkd. Infektionskr. Abt. 1 Orig. Reihe A 226:469-478. [PubMed] [Google Scholar]
- 22.Miyake, T., K. Watanabe, T. Watanabe, and H. Oyaizu. 1998. Phylogenetic analysis of the genus Bifidobacterium and related genera based on 16S rDNA sequences. Microbiol. Immunol. 42:661-667. [DOI] [PubMed] [Google Scholar]
- 23.Moore, W. E. C., and L. V. Holdeman. 1974. Human fecal flora: the normal flora of 20 Japanese-Hawaiians. Appl. Microbiol. 27:961-979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moore, W. E. C., and L. H. Moore. 1995. Intestinal floras of populations that have a high risk of colon cancer. Appl. Environ. Microbiol. 61:3202-3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mutai, M., and R. Tanaka. 1987. Ecology of Bifidobacterium in the human intestinal flora. Bifidobacteria Microflora 6:33-41. [Google Scholar]
- 26.Olsen, G. J., C. R. Woese, and R. Overbeek. 1994. The winds of (evolutionary) change: breathing new life into microbiology. J. Bacteriol. 176:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paster, B. J., F. E. Dewhirst, I. Olsen, and G. J. Fraser. 1994. Phylogeny of Bacteroides, Prevotella, and Porphyromonas spp. and related bacteria. J. Bacteriol. 176:725-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Requena, T., J. Burton, T. Matsuki, K. Munro, M. A. Simon, R. Tanaka, K. Watanabe, and G. W. Tannock. 2002. Identification, detection, and enumeration of Bifidobacterium species by PCR targeting the transaldolase gene. Appl. Environ. Microbiol. 68:2420-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Satake, S., N. Clark, D. Rimland, F. S. Nolte, and F. C. Tenover. 1997. Detection of vancomycin-resistant enterococci in fecal samples by PCR. J. Clin. Microbiol. 35:2325-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Satokari, R. M., E. E. Vaughan, A. D. Akkermans, M. Saarela, and W. W. de Vos. 2001. Bifidobacterial diversity in human feces detected by genus-specific PCR and denaturing gradient gel electrophoresis. Appl. Environ. Microbiol. 67:504-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwiertz, A., G. Le Blay, and M. Blaut. 2000. Quantification of different Eubacterium spp. in human fecal samples with species-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 66:375-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sghir, A., G. Gramet, A. Suau, V. Rochet, P. Pochart, and J. Dore. 2000. Quantification of bacterial groups within human fecal flora by oligonucleotide probe hybridization. Appl. Environ. Microbiol. 66:2263-2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Simpson, J. M., V. J. McCracken, H. R. Gaskins, and R. I. Mackie. 2000. Denaturing gradient gel electrophoresis analysis of 16S ribosomal DNA amplicons to monitor changes in fecal bacterial populations of weaning pigs after introduction of Lactobacillus reuteri strain MM53. Appl. Environ. Microbiol. 66:4705-4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Song, Y., N. Kato, C. Liu, Y. Matsumiya, H. Kato, and K. Watanabe. 2000. Rapid identification of 11 human intestinal Lactobacillus species by multiplex PCR assays using group- and species-specific primers derived from the 16S-23S rRNA intergenic spacer region and its flanking 23S rRNA. FEMS Microbiol. Lett. 187:167-173. [DOI] [PubMed] [Google Scholar]
- 35.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Dore. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tannock, G. W. 1995. Normal microflora: an introduction to microbes inhabiting the human body. Chapman & Hall, London, United Kingdom.
- 37.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Walter, J., G. W. Tannock, A. Tilsala-Timisjarvi, S. Rodtong, D. M. Loach, K. Munro, and T. Alatossava. 2000. Detection and identification of gastrointestinal Lactobacillus species by using denaturing gradient gel electrophoresis and species-specific PCR primers. Appl. Environ. Microbiol. 66:297-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, R. F., W. W. Cao, and C. E. Cerniglia. 1996. PCR detection and quantitation of predominant anaerobic bacteria in human and animal fecal samples. Appl. Environ. Microbiol. 62:1242-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, R. F., W. W. Cao, and C. E. Cerniglia. 1997. PCR detection of Ruminococcus spp. in human and animal faecal samples. Mol. Cell. Probes 11:259-265. [DOI] [PubMed] [Google Scholar]
- 41.Wilson, K. H., and R. B. Blitchington. 1996. Human colonic biota studied by ribosomal DNA sequence analysis. Appl. Environ. Microbiol. 62:2273-2278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Woese, C. R. 1987. Bacterial evolution. Microbiol. Rev. 51:221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zoetendal, E. G., A. D. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]