Abstract
Colony-forming cyanobacteria of the genus Aphanizomenon form massive blooms in the brackish water of the Baltic Sea during the warmest summer months. There have been recent suggestions claiming that the Baltic Sea Aphanizomenon species may be different from Aphanizomenon flos-aquae found in lakes. In this study, we examined variability in the morphology and 16S-23S rRNA internal transcribed spacer (ITS) sequences of A. flos-aquae populations along a salinity gradient from a string of lakes to a fjord-like extension of the Baltic Sea to the open Baltic Sea. Morphological differences among the populations were negligible. We found that the Baltic Sea was dominated (25 out of 27 sequences) by one ITS1-S (shorter band of ITS 1 [ITS1]) genotype, which also was found in the lakes. The lake populations of A. flos-aquae tended to be genetically more diverse than the Baltic Sea populations. Since the lake ITS1-S genotypes of A. flos-aquae are continuously introduced to the Baltic Sea via inflowing waters, it seems that only one ITS1 genotype is able to persist in the Baltic Sea populations. The results suggest that one of the ITS1-S genotypes found in the lakes is better adapted to the conditions of the Baltic Sea and that natural selection removes most of the lake genotypes from the Baltic Sea A. flos-aquae populations.
Planktonic cyanobacteria of the genus Aphanizomenon (L.) Ralf ex Bornet et Flahault are salt-tolerant organisms that are common in nutrient-rich freshwaters (27). Aphanizomenon spp., together with the cyanobacteria Nodularia spumigena Mertens and Anabaena sp. strain Bory, form heavy blooms in the Baltic Sea during the warmest summer months (13, 15). The Baltic Sea, with a total area of almost 400,000 km2, is a brackish water basin with surface water salinity ranging from 9 practical salinity units (psu) in the southernmost parts of the sea to 1 psu in the eastern and northernmost parts of the sea (32). At least one isolate of Aphanizomenon from a lake near the Baltic Sea has been reported to be neurotoxic (31). However, toxins have not been found in an Aphanizomenon isolate from the Baltic Sea (18). Aphanizomenon spp. are abundant in the Baltic Sea during winter as well (24), and although they occur in all areas of the sea, their abundance decreases toward the more saline southern waters (25). In experiments with a salinity range of 0 to 30 psu, an Aphanizomenon isolate from the Baltic Sea grew best at salinities of 0 to 5 psu (18).
In the Baltic Sea, Aphanizomenon is found mainly in colonies or organized in bundles of parallel trichomes. According to the botanical taxonomy of cyanobacteria, the only species of Aphanizomenon that always occurs in colonies is the type species of the genus, Aphanizomenon flos-aquae (L.) Ralfs ex Bornet et Flahault (14). This species has been divided into two subspecies, A. flos-aquae subsp. flos-aquae and A. flos-aquae subsp. klebahnii, which are discriminated on the basis of the sizes of the different kinds of cells (14). The taxonomic status of colony-forming Aphanizomenon in the Baltic Sea has been questioned because, for example, the ultrastructure of the cells of Aphanizomenon colonies collected from the Baltic Sea was different from that of freshwater A. flos-aquae (12). In addition, a difference between Aphanizomenon collected from the Baltic Sea and an A. flos-aquae isolate from a lake in Northern Ireland was reported based on DNA sequence data from the phycocyanin-encoding operon (PC-IGS) and the 16S rRNA gene (2).
Population genetic studies of cyanobacteria, as well as of other phytoplankton groups, are still rare (e.g., 1, 2, 9, 30). Little is known about the geographical patterns of genetic diversity in populations of cyanobacteria and of the factors that shape the genetic structure of these populations. A study based on the sequence variability of the PC-IGS region of Aphanizomenon from the Baltic Sea and North American lakes reported that the Baltic Sea populations were genetically homogeneous (2). Low genetic diversity was suggested to be caused by selection or by only one PC-IGS genotype initially colonizing the Baltic Sea (2). On the other hand, the populations of N. spumigena in the Baltic Sea were found to be genetically heterogeneous (1, 3, 9). The distribution of the genotypes was nonrandom, but correlations between the distribution of the genotypes and temperature, salinity, or major nutrients were not found (1).
Most molecular biological studies of cyanobacterial populations to date have used cultured organisms. However, culture media are selective; hence, the cultures do not necessarily represent the diversity found in nature. According to one estimate, far less than 5% of cyanobacterial species have been established in cultures, partly because certain types are impossible to grow on artificial media (5). Aphanizomenon spp. from the Baltic Sea have been difficult to culture. In the laboratory of K. Sivonen, only one strain, TR183, has been successfully established in cultures after numerous attempts. To avoid problems related to selective culturing, recent studies of populations of filamentous cyanobacteria in the Baltic Sea have used direct PCR amplification of DNA from single cyanobacterial colonies or filaments picked from water samples (2, 9, 16). In addition, morphological variability, as presented in species descriptions of botanical taxonomy of cyanobacteria, can be most reliably assessed from natural populations, since cultures tend to change morphologically.
In the present study, we examined the morphological and 16S-23S rRNA internal transcribed spacer (ITS) 1 (ITS1) sequence diversity of Aphanizomenon populations along a salinity gradient leading from a string of lakes to a fjord-like extension of the Baltic Sea to the open Baltic Sea. The aim of this study was to observe geographical patterns in genetic diversity and to ascertain whether there are morphological or genetic differences between the lake and the Baltic Sea populations of Aphanizomenon.
MATERIALS AND METHODS
Study areas and sampling.
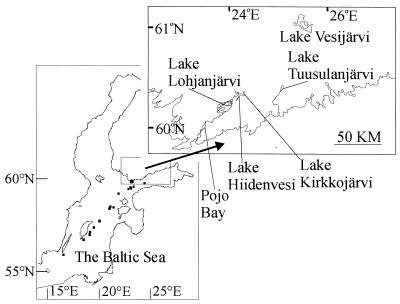
Sampling was carried out in the Baltic Sea between 23 April 1998 and 6 October 1999 (Table 1). The 26 different sampling points covered areas in the open Baltic Sea proper as well as outer and inner archipelago stations off the southwestern coast of Finland (Fig. 1). In addition, Pojo Bay, which is a fjord-like bay ca. 14 km long and 1 to 2 km wide and extending northeast from the entrance area of the Gulf of Finland (24), was sampled. In Pojo Bay, surface water salinity increases from about 0 psu in the north to 3 to 4 psu at the entrance to the bay and then increases further to about 6 psu as one approaches the open sea area (24). Four coastal lakes (Lake Lohjanjärvi, Lake Hiidenvesi, Lake Kirkkojärvi, and Lake Tuusulanjärvi) were each sampled (Table 1 and Fig. 1). Lake Kirkkojärvi is the highly eutrophic northernmost subbasin of Lake Hiidenvesi (28). Lake Hiidenvesi is connected to Lake Lohjanjärvi by the River Vääntee, which flows toward the sea and had a mean discharge rate of 14.4 m3 s−1 in 1998 (29). Lake Lohjanjärvi is the largest lake in southern Finland, and it is connected to Pojo Bay by the River Mustio, which flows toward the sea and had a mean discharge of 20 m3 s−1 in 1998 (10) (Fig. 1). By sampling the ca.70-km passage from Lake Kirkkojärvi through the salinity gradient of Pojo Bay and the archipelago to the open sea, we were able to assess the effects of gradually changing salinity conditions on the morphological and genetic compositions of the Aphanizomenon populations. In addition, samples were taken from Lake Tuusulanjärvi, which is a shallow, highly eutrophic lake connected to the sea by the River Vantaa (Table 1 and Fig. 1).
TABLE 1.
Sampling locations of A. flos-aquae colonies, including dates, positions, and salinity
| Location | Datea | Position (°N, °E) | Salinity (psu) |
A. flos-aquae colonies used forb:
|
||
|---|---|---|---|---|---|---|
| Microscopy (n) | Sequencing | DGGE | ||||
| Baltic Sea | 23 Apr 1998 | 57.16, 19.00 | 7.2 | 11 | BS10 | |
| 25 May 1998 | 58.54, 20.99 | 7.4 | 5 | |||
| 25 May 1998 | 59.59, 23.49 | 6.4 | 4 | |||
| 16 Jun 1998 | 59.59, 23.49 | 6.4 | 17 | |||
| 10 Jul 1998 | 59.51, 22.99 | 6.3 | 10 | BS9, BS13 | ||
| 8 Sep 1998 | 59.47, 23.16 | 6.3 | 30 | |||
| 8 Sep 1998 | 59.40, 23.14 | 6.3 | 40 | |||
| 10 Sep 1998 | 59.51, 23.13 | 6.1 | 10 | BS44, BS56, BS77 | ||
| 10 Sep 1998 | 59.53, 23.18 | 4.7 | 10 | BS1, BS19, BS27, BS33 | ||
| 10 Sep 1998 | 59.55, 23.20 | 4.3 | 10 | |||
| 10 Sep 1998 | 59.56, 23.23 | 3.5 | 10 | BS163 | ||
| 16 Mar 1999 | 59.45, 23.19 | 5.4 | 60 | BS150 | ||
| 17 Mar 1999 | 59.47, 23.19 | 5.6 | 60 | |||
| 17 Mar 1999 | 59.51, 23.13 | 3.3 | 60 | BSW9, BSW14, BSW17, BSW23, BSW27 | ||
| 7 Sep 1999 | 55.97, 16.49 | 6.6 | 3 | |||
| 7 Sep 1999 | 56.84, 18.49 | 6.6 | 1 | |||
| 7 Sep 1999 | 57.18, 18.99 | 6.4 | 1 | |||
| 7 Sep 1999 | 57.53, 19.49 | 6.2 | 3 | |||
| 7 Sep 1999 | 57.86, 20.00 | 6.3 | 2 | |||
| 7 Sep 1999 | 57.88, 20.03 | 6.0 | 5 | BS11 | ||
| 7 Sep 1999 | 58.54, 20.97 | 6.0 | 5 | BS16 | ||
| 7 Sep 1999 | 58.55, 21.00 | 6.1 | 6 | BS22, BS24, BS25, BS26 | ||
| 7 Sep 1999 | 58.59, 21.04 | 6.1 | 4 | BS28, BS29, BS30 | ||
| 5 Oct 1999 | 58.58, 20.50 | 6.3 | 6 | |||
| 5 Oct 1999 | 59.17, 22.00 | 6.4 | 6 | |||
| 6 Oct 1999 | 59.75, 24.50 | 5.8 | 3 | |||
| Pojo Bay | 10 Sep 1998 | 59.58, 23.24 | 2.1 | 10 | PB122, PB127 | |
| 10 Sep 1998 | 59.59, 23.27 | 2.4 | 10 | |||
| 10 Sep 1998 | 60.01, 23.28 | 1.8 | 10 | PB98 | ||
| 10 Sep 1998 | 60.03, 23.31 | 1.2 | 10 | PB91, PB94 | ||
| 10 Sep 1998 | 60.05, 23.33 | 0.1 | 14 | |||
| Lake Lohjanjärvi | 28 Sep 1998 | See Fig. 1 | NDc | 50 | LJ23, LJ28, LJ31, LJ37, LJ38, LJ43 | |
| Lake Hiidenvesi | 2 Sep 1999 | See Fig. 1 | ND | 50 | HV5, HV6, HV7, HV14, HV21, HV25, HV392, HV398 | HV20, HV21,HV25, HV34, HV35, HV387, HV392, HV395, HV398 |
| Lake Kirkkojärvi | 19 Aug 1999 | See Fig. 1 | 0d | 30 | KJ3, KJ5, KJ8, KJ19, KJ21, KJ24, KJ25, KJ26, KJ29 | KJ3, KJ8, KJ9, KJ21, KJ25, KJ29 |
| Lake Tuusulanjärvi | 23 Aug 1999 | See Fig. 1 | 0d | 30 | TJ1, TJ21, TJ26, TJ24, TJ22 | TJ2, TJ8, TJ11, TJ12, TJ13, TJ22, TJ23, TJ30 |
| Total | 596 | 59 | 23 | |||
Apr, April; Jun, June; Jul, July; Sep, September; Mar, March; Oct, October; Aug, August.
Number of A. flos-aquae colonies collected for microscopy at each location and codes and total numbers of the colonies used for microscopy that were also used for ITS1-S sequencing or DGGE. Of the colonies used for sequencing, those that yielded ambiguous nucleotides in sequence raw data and that were subsequently omitted from further analyses are underlined. Colonies that were used for sequencing of the DGGE bands are indicated by bold type.
ND, not determined.
The value is an estimate for this freshwater body.
FIG. 1.
Sampling sites in the Baltic Sea (indicated with black squares), Pojo Bay, Lake Lohjanjärvi, Lake Hiidenvesi, Lake Kirkkojärvi, and Lake Tuusulanjärvi. In addition, Lake Vesijärvi, the site of isolation of A. flos-aquae strain 202, is also shown.
In the Baltic Sea coastal sampling points, Pojo Bay, and the lakes, sampling was carried out with a 50- or 100-μm-mesh-size plankton net in the 0- to 2-m surface layer. In March 1999, sampling was done by a scuba diver dragging the plankton net under 40- to 50-cm-thick ice. In the coastal stations as well as Pojo Bay, salinities were determined with a sonde that measures connectivity, temperature, and density. The net samples were stored in cool and dark conditions until their examination the following day. The Aphanizomenon samples and salinity data for the open Baltic Sea sampling points were obtained from unattended data collection systems on merchant ships (19).
Colonies were assigned numbers, and the different source populations are indicated by the following abbreviations: BS, Baltic Sea; PB, Pojo Bay; LJ, Lake Lohjanjärvi; HV, Lake Hiidenvesi; KJ, Lake Kirkkojärvi; and TJ, Lake Tuusulanjärvi.
Processing of the colonies, PCR, and sequencing
In the laboratory, Aphanizomenon colonies were picked from water samples under a dissecting microscope by using a stretched Pasteur pipette. The colonies were transferred to slides with a drop of GF/F-filtered seawater or, for freshwater samples, transferred to slides with a drop of sterile water; each slide was then covered with a cover glass. With a phase-contrast light microscope, the dimensions of the different types of cells (intercalary vegetative cells, terminal cells, heterocytes, and akinetes) were measured. The cover glass was removed carefully, and the colony was transferred to a 200-μl PCR tube containing 25 μl of 10× DyNAzyme polymerase buffer (Finnzymes, Espoo, Finland). The samples were kept frozen at −20°C until further treatment. The colonies from which the 16S-23S rRNA ITS1 region was sequenced (n = 59) were chosen randomly from among those analyzed microscopically (Table 1).
The 16S-23S rRNA ITS1 is considered a highly variable and appropriate region for investigations of intraspecific variability in cyanobacteria (e.g., 23). ITS1 with 150 bp from the flanking 16S rRNA region and 46 bp from the 23S rRNA region was amplified with a 250 nM concentration of cyanobacterium-specific primers 16CITS and 23CITS (22). The reaction mixture (25 μl) contained 200 μM each deoxynucleoside triphosphate (Finnzymes), 700 μM MgCl2 solution, 1 U of DyNAzyme DNA polymerase (Finnzymes), and 4 μl of Aphanizomenon sample as a template. The PCR protocol consisted of an initial denaturation step at 96°C for 3 min; 35 cycles of 94°C for 20s, 50°C for 30s, and 72°C for 1 min; and an extension step at 72°C for 5 min. After the PCR, the whole reaction was electrophoresed through 1% agarose gels (85 V, 40 min), stained with ethidium bromide, and photographed under UV light. Of the two ITS1 products that were observed in the agarose gels, the shorter band of ca. 470 bp (ITS1-S) was chosen for sequencing because that band was usually heavier and more sequence data exist for ITS1 of that length in GenBank. The larger band was thought to represent a longer copy of ITS1 (ITS1-L).
The ITS1-S band was excised from the gels and purified for sequencing by using Ultrafree-DA spin columns (Millipore, Bedford, Mass.). To avoid errors created by PCR, for each colony Ultrafree-DA-cleaned ITS1-S bands originating from three separate PCRs were pooled for sequencing. Sequencing was carried out with an ABI Prism 310 genetic analyzer and an ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction kit by following the manufacturer's instructions (PE Applied Biosystems, Foster City, Calif.). The sequencing primers were the same as for the initial amplifications. The PCR products were sequenced on both strands at least twice. In addition to the colonies of A. flos-aquae from the natural populations, ITS1-S of A. flos-aquae strain 313, isolated from Lake Tuusulanjärvi in 1997 and maintained in the laboratory of K. Sivonen, was also sequenced. The initial ITS PCR amplification was done with 2 μl of culture as a template; otherwise, the PCR conditions used were similar to those for the colonies of A. flos-aquae obtained in the field.
DGGE.
As it was noted that some of the ITS1-S sequences from the lakes and Pojo Bay had ambiguous nucleotides, possibly indicating the presence of more than one ITS1-S with similar lengths and various nucleotide sequences, denaturing gradient gel electrophoresis (DGGE) was used to enable detection of the number of different copies and separation of the copies for sequencing. In addition, DGGE enabled screening for ITS1 sequence variability within and between the lake populations. Altogether, 23 colonies from Lake Hiidenvesi, Lake Kirkkojärvi, and Lake Tuusulanjärvi were used for DGGE analyses (Table 1).
The ITS1 PCR products used for DGGE analyses were amplified with primers 16CITS and 23CITS, with a 40-nucleotide GC-rich sequence added to the 5′ end of the forward 16CITS primer. The PCR products were loaded on 6% acrylamide-bisacrylamide (37.5:1) gels with a vertical denaturing gradient from 35 to 44% (100% denaturant consisting of 7 M urea and 40% formamide) and electrophoresed at 150 V for 4.5 h at 60°C in 20 mM Tris-acetate- 0.5 mM EDTA (pH 8) buffer. The gels were stained with a nucleic acid gel stain (GelStar, Rockland, Maine) for 40 min and photographed under UV light. The two ITS1-S bands of colonies HV21, HV25, KJ21, and KJ25 were excised from the gels, dissolved in 30 μl of sterile water, and stored at −20°C (Table 1). Five microliters of dissolved material from the excised DGGE bands was used as a template, and PCR was run as for the initial PCR of the colonies. The PCR products were purified with Ultrafree-DA and sequenced as described above.
Sequence analyses and morphological data.
The ITS1-S sequences for each Aphanizomenon colony and strain 313 were checked by alignment with PILEUP from the GCG package, version 10.1 (Genetics Computer Group, Madison, Wis.), and manually edited with the GeneDoc multiple-sequence-alignment editor. Using ClustalW and manual editing, the sequences for each Aphanizomenon colony and strain 313 were aligned with Nostoc sp. strain PCC7120 (GenBank accession no. AF180969) and reference Aphanizomenon ITS1-S sequences. The reference Aphanizomenon sequences were from strain 202, an isolate from Lake Vesijärvi (Fig. 1) (GenBank accession no. AJ293198) (7), strain TR183, an isolate from the Baltic Sea (GenBank accession no. AJ293212) (7), and strain 326, an isolate from Lake Lohjanjärvi (GenBank accession no. AJ293199 and AJ293200) (7). The latter strain had two equal-length copies of ITS1-S (sequences 326a and 326b) which had been sequenced from cloned PCR fragments (7). To improve the quality of the alignment, the cyanobacterial conserved domains of the ITS1 region (11) were used to aid the alignment. Maximum-parsimony and neighbor-joining trees with Jukes-Cantor-corrected distances were created by using the PHYLIP, version 3.6 (alpha2), package (6). The trees were constructed from alignments that included insertions and deletions, and they were statistically evaluated by 100 bootstrap resamplings.
Data on cell dimensions were compared with botanical descriptions of Aphanizomenon taxa (14). Furthermore, comparisons of cell sizes among the different populations were made.
Nucleotide sequence accession numbers.
All sequences produced in this study have been deposited in GenBank under accession numbers AF431745 to AF431755.
RESULTS
Morphology and environmental conditions.
All Aphanizomenon populations formed colonies that were spindle- or flake-like and thus were identified as A. flos-aquae. Within populations, the sizes of the cells varied (data not shown), and both botanically described subspecies of A. flos-aquae, A. flos-aquae subsp. flos-aquae and A. flos-aquae subsp. klebahnii, could be identified in the Baltic Sea population. No significant differences were apparent in cell dimensions among the populations, except for the terminal cells of the Lake Lohjanjärvi population, which were generally slightly longer than those of the other populations.
In the Baltic Sea and Pojo Bay, the surface water salinity varied from 7.4 psu in the Baltic Sea proper to 0.1 psu in the northernmost end of Pojo Bay (Table 1).
ITS1 PCR, DGGE, and sequencing.
ITS1 PCR of the A. flos-aquae colonies and strain 313 yielded two PCR products of different lengths. The longer product, ITS1-L, was slightly over 700 bp, while the shorter product, ITS1-S, was approximately 470 bp.
The ITS1-S PCR products from 26 A. flos-aquae colonies from the Baltic Sea were sequenced, including 5 colonies obtained from under-ice populations in March 1999 (Table 1). The sequences of the colonies from the Baltic Sea were all unambiguous. Sequencing was initially attempted for 33 colonies from the lakes and Pojo Bay, but 14 of the colonies yielded ITS1-S sequences that contained ambiguous nucleotides (double peaks in the raw sequencing data); thus, those sequences were excluded from further analyses (Table 1). Altogether, 19 unambiguous ITS1-S sequences were obtained from the colonies that originated from the lake populations and Pojo Bay (Table 1), in addition to the sequence of strain 313 from Lake Tuusulanjärvi.
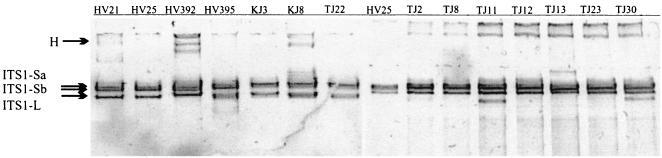
In the DGGE analysis of the ITS1 regions amplified from colonies from the lakes, a separate single band and a tight cluster of two bands were observed (Fig. 2). In addition, a faint band observed near the wells was considered to be a heteroduplex. PCR of the excised DGGE bands indicated that the lowest, separate band was ITS1-L of about 700 bp and that the two bands in the tight cluster both had lengths of about 470 bp, corresponding to the length of ITS1-S. Only slight variations in the position of the ITS1-S bands in the DGGE analysis were observed between and within populations, indicating only slight sequence variability (Fig. 2). To study the sequence differences between the two similarly sized copies of ITS1-S, hereafter referred to as ITS1-Sa and ITS1-Sb, the two ITS1-S bands of two A. flos-aquae colonies from Lake Hiidenvesi (HV21 and HV25) and Lake Kirkkojärvi (KJ21 and KJ25) were excised from the gels and sequenced. With those sequences, the number of ITS1-S sequences obtained from A. flos-aquae colonies and strain 313 totaled 54.
FIG. 2.
Composite of two DGGE runs with ITS1 amplification products from 14 different A. flos-aquae colonies. H, presumed heteroduplex. The ITS1-Sa and ITS1-Sb bands of colonies HV21 and HV25 were excised from the gel for sequencing. For details on DGGE, see Materials and Methods.
All of the Aphanizomenon sequences obtained during this study had identical nucleotides in both flanking 16S rRNA and 23S rRNA regions, and these regions were omitted from phylogenetic analyses. The lengths of the Aphanizomenon ITS1-S sequences (including the four reference sequences from GenBank) varied from 266 to 288 bp. The alignment of all 58 Aphanizomenon sequences with insertions and deletions included revealed 96 variable positions (33%); the same alignment with insertions and deletions excluded revealed 70 variable positions (27%). The alignment of Aphanizomenon sequences with the ITS1-S sequence of outgroup Nostoc sp. strain PCC7120 contained 312 positions, of which 160 positions (51%) had a variable nucleotide or an insertion or deletion. The latter alignment was used for the construction of the final trees and is available at GenBank.
Phylogenetic analyses of ITS1-S sequences.
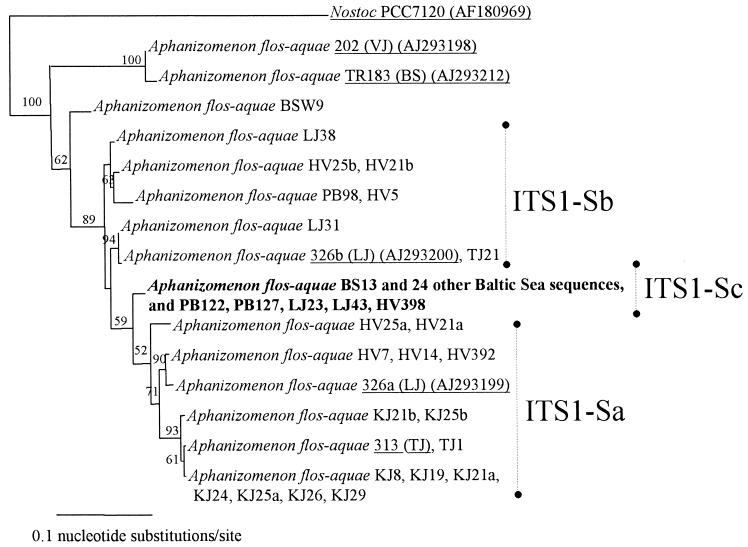
Essentially similar groupings emerged in the neighbor-joining and parsimony trees. Thus, only the neighbor-joining tree is shown (Fig. 3).
FIG. 3.
Neighbor-joining tree of the 16S-23S rRNA ITS1 sequences (length, 266 to 288 nucleotides) of A. flos-aquae colonies from the Baltic Sea (BS), Pojo Bay (PB), Lake Lohjanjärvi (LJ), Lake Hiidenvesi (HV), Lake Kirkkojärvi (KJ), and Lake Tuusulanjärvi (TJ) together with sequences (underlined) of strain 313 from Lake Tuusulanjärvi and reference A. flos-aquae sequences (underlined) (GenBank accession numbers) of strains 202 (AJ293198) from Lake Vesijärvi (VJ), TR183 (AJ293212) from the Baltic Sea, and 326b (AJ293200) and 326a (AJ293199) from Lake Lohjanjärvi. The dominating ITS1-S allele in the Baltic Sea is shown in bold type. Three subgroups, ITS1-Sb, ITS1-Sc, and ITS1-Sa, are indicated (for details, see the text). The scale bar indicates branch length, showing 0.1 nucleotide substitution per site. Only bootstrap values of >50% are shown.
The reference ITS1-S sequence of A. flos-aquae strain 202 from Lake Vesijärvi and the ITS1-S sequence of A. flos-aquae strain TR183 from the Baltic Sea grouped separately from the other A. flos-aquae sequences with 100% bootstrap support (Fig. 3). The sequence for A. flos-aquae colony BSW9, obtained from under the ice in the Baltic Sea archipelago, also clustered slightly separately from the other sequences. The rest of the sequences formed one large cluster that was supported by 89% of the bootstrap resamplings (Fig. 3). The large cluster consisted of 12 different clades. The outgroup Nostoc sp. strain PCC7120 ITS1-S sequence had a similarity of 57 to 62% with the Aphanizomenon ITS1-S sequences. Among all Aphanizomenon ITS1-S sequences, similarities ranged from 68 to 100%, and within the large cluster, they ranged from 87 to 100%.
The ITS1-S sequences within the large cluster were separated into two poorly resolved subgroups, ITS1-Sb and ITS1-Sa, and a separate clade, ITS1-Sc (Fig. 3). The two reference sequences, 326a and 326b, which represented ITS1-S copies from Lake Lohjanjärvi isolate 326 with equal lengths but different sequences and which had been resolved by cloning, were positioned in the separate subgroups ITS1-Sb and ITS1-Sa (Fig. 3). Two sequenced DGGE bands, HV21b and HV25b (Fig. 2), clustered near 326b, and two other bands from the same colonies, HV21a and HV25a, clustered closer to 326a (Fig. 3). The sequences in subgroup ITS1-Sb had a three-base fragment, CAA, at positions 146 to 148 of the alignment that was missing from the sequences in subgroup ITS1-Sa. The sequences in subgroup ITS1-Sa had a C at position 129 that was missing from the sequences in subgroup ITS1-Sb. In addition, ITS1-Sa and ITS1-Sb had different nucleotides at 17 other positions. Sequences within subgroup ITS1-Sa were considered to represent the ITS1-Sa copy, and those in subgroup ITS1-Sb were considered to represent the ITS1-Sb copy. Hence, the ITS1-S sequences that had not been separated by DGGE but that had been sequenced from agarose gels and that had been found unambiguous also were thought to represent either ITS1-Sa or ITS1-Sb.
The ITS1-S sequence representing colony BS13 and 24 other sequences from the Baltic Sea were intermediate relative to copies ITS1-Sa and ITS1-Sb. The middle part of the sequence (from positions 183 to 219 of the alignment) resembled the ITS1-Sa copy, whereas the rest of the sequence was more similar to the ITS1-Sb copy. This sequence type is hereafter referred to as ITS1-Sc (Fig. 3). Even though the ITS1-S sequences from the DGGE bands of colonies HV21 and HV25 appeared to be highly similar to either of the reference sequences 326a and 326b, both ITS1-S sequences from the DGGE bands of Lake Kirkkojärvi colonies KJ21 and KJ25 clustered with 326a in the ITS1-Sa subgroup and had a difference of only 2 nucleotides between them. The sequences of strains 202 and TR183 and colony BSW9 could not be assigned to any of the three ITS1-S subgroups, as they were highly different from the sequences of the large cluster.
The same ITS1-S types (e.g., ITS1-Sb) from different populations were more similar among them (for example, sequences within the subgroup ITS1-Sb were from 94 to 99% similar among them) than the different ITS1-S types from the same colonies (e.g., ITS1-Sa and ITS1-Sb sequences of colony HV21 were 88% similar). Consequently, the ITS1-S sequence diversities of the different populations are considered separately for each ITS1-S type.
ITS1-S sequence diversity of A. flos-aquae populations.
In the Baltic Sea, genetic diversity within the population was low, as 25 out of the 27 sequences (93%) from the Baltic Sea were completely identical (Table 2 and Fig. 3). The sequence of strain TR183 was 73% similar and the sequence of colony BSW9 was 84% similar to the dominant ITS1-S sequence found in the Baltic Sea. In fact, the sequences of TR183 and BSW9 were most similar to the sequences of samples from the lakes (Fig. 3). The populations in Lake Lohjanjärvi and Lake Hiidenvesi were more diverse than those in the Baltic Sea. In Lake Lohjanjärvi, three different alleles of ITS1-Sb with similarities of 97 to 99% were found among three sequences (Table 2). In Lake Hiidenvesi, two different ITS1-Sb alleles with a similarity of 97% among three sequences and two different ITS1-Sa alleles with a similarity of 98% among five sequences were found (Table 2). In addition, ITS1-Sc, the allele that was dominant in the Baltic Sea, was also found in Lake Lohjanjärvi and Lake Hiidenvesi populations (Table 2 and Fig. 3).
TABLE 2.
ITS1-S sequence diversity within A. flos-aquae populations of the Baltic Sea, Pojo Bay, and various lakesa
| Location | ITS1-Sa
|
ITS1-Sb
|
ITS1-Sc
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| No. of:
|
% Similarity | No. of:
|
% Similarity | No. of:
|
% Similarity | ||||
| Sequences | Different alleles | Sequences | Different alleles | Sequences | Different alleles | ||||
| Baltic Sea | 25 | 1 | 100 | ||||||
| Pojo Bay | 1 | 1 | 2 | 1 | 100 | ||||
| Lake Lohjanjärvi | 1 | 1 | 3 | 3 | 97-99 | 2 | 1 | 100 | |
| Lake Hiidenvesi | 5 | 2 | 98 | 3 | 2 | 97 | 1 | 1 | |
| Lake Kirkkojärvi | 9 | 2 | 99 | ||||||
| Lake Tuusulanjärvi | 2 | 1 | 100 | 1 | 1 | ||||
See the text for details on the determination of the different types. Similarity is the percent sequence similarities among the different alleles.
In Lake Kirkkojärvi, Lake Tuusulanjärvi, and Pojo Bay, A. flos-aquae populations seemed less diverse. In Lake Kirkkojärvi, two different ITS1-Sa alleles that were 99% similar were found among nine sequences (Table 2). In Lake Tuusulanjärvi, one allele of ITS1-Sb and one allele of ITS1-Sa were found. In Pojo Bay, one allele of ITS1-Sb and one allele of ITS1-Sc were found.
DISCUSSION
The Baltic Sea population of A. flos-aquae was morphologically diverse, and colonies resembling both botanically described subspecies of A. flos-aquae, A. flos-aquae subsp. flos-aquae and A. flos-aquae subsp. klebahnii, were found. Nevertheless, the population was dominated by only one ITS1-S genotype. This finding does not genetically support the division of the species A. flos-aquae into the two subspecies listed above. ITS1-S-based phylogenies have been approximately similar to 16S rRNA-based phylogenies in the genera Aphanizomenon and Anabaena (7) as well as in Nodularia (16, 17). Therefore, identical ITS1-S sequences are likely to represent one subspecies rather than two different taxa. In N. spumigena, cultures with identical PC-IGS and ITS1-S genotypes were shown to be morphologically highly variable (16), and in the cyanobacterial genus Merismopedia, even a high level of morphological diversity within one 16S rRNA genotype is possible (26).
There were no consistent morphological differences between the Baltic Sea and lake populations of A. flos-aquae in this study. Furthermore, the ITS1-S genotype that was dominant in the Baltic Sea was found in Lake Lohjanjärvi as well. Hence, our data do not support the suggestion of a separate Baltic Sea species of Aphanizomenon and freshwater A. flos-aquae, as suggested by ultrastructures (12) and PC-IGS and 16S rRNA sequences (2). Instead, Aphanizomenon found in the Baltic Sea fits the morphological description of A. flos-aquae (14) and is not an exclusive Baltic Sea species.
Our data on ITS1-S sequence diversity indicated a higher level of genetic diversity in the lakes than in the Baltic Sea. These results are in line with those of a recent study on genetic diversity within populations of colony-forming Aphanizomenon in the Baltic Sea and two North American lakes (2). In that study, only one PC-IGS genotype of Aphanizomenon was found in the Baltic Sea, whereas two PC-IGS genotypes were found in Little Crooked Lake (northern Indiana); on that basis it was concluded that the North American lakes supported a genetically more diverse population (2). The lower level of genetic diversity in the Baltic Sea was suggested to have arisen from only one genotype of Aphanizomenon initially colonizing the Baltic Sea, from natural selection, or from a population bottleneck that would have reduced the diversity to only one fixed PC-IGS allele (2). In the present study, we showed that the lake waters ultimately flowing into the Baltic Sea harbored genetically more diverse A. flos-aquae populations than the Baltic Sea. This finding implies that the lake ITS1-S genotypes are continuously introduced into the Baltic Sea, and yet only one ITS1-S genotype dominates the population.
The dominant ITS1-S genotype of the Baltic Sea was found in the lakes as well, suggesting that it represents a phenotype that grows both in the lakes and in the Baltic Sea. It is possible that the dominant Baltic Sea ITS1-S genotype is more tolerant of the Baltic Sea conditions than the other ITS1-S genotypes found in the lakes, because the lake ITS1-S genotypes were rare in the Baltic Sea. Hence, the results may imply natural selection, with removal of the less fit lake A. flos-aquae genotypes from the Baltic Sea population. Since A. flos-aquae is well known as a freshwater species, the most obvious selecting factor would seem to be salinity. However, a range of other factors, such as different mixing depths in the lakes and the sea, light or nutrient conditions, chemical environment in general, or accompanying biota, may also affect the fitness of the different ITS1-S genotypes in the Baltic Sea.
According to the central-marginal model in evolutionary biology, populations in the margins of the species range are often genetically more homogeneous than populations in the center of the range (e.g., reference 4 and references therein). Even though the model has been mostly applied to sexually reproducing eukaryotes, its extension to the studied A. flos-aquae populations would imply that the lakes are the center of the range of this species and the Baltic Sea is the margin of the range. This view is coherent with the perception that A. flos-aquae is a freshwater species. On the other hand, populations of N. spumigena are genetically diverse in the Baltic Sea (1, 3, 9), supporting the idea of a true brackish water species.
The only ITS1-S sequences from the Baltic Sea that were different from the dominant Baltic Sea ITS1-S allele were those of strain TR183 and of colony BSW9. Both of those sequences were most similar to sequences from the lakes. In fact, colony BSW9 may well have originated from a lake, as it was obtained from a winter population in the inner archipelago at an area that is prone to inflows of less saline water from Pojo Bay, especially during the winter months, as demonstrated by the low salinity (3.3 psu) on 17 March 1999.
The only Aphanizomenon isolate in our laboratory from the Baltic Sea, strain TR183, was genetically most similar to strain 202, an isolate from Lake Vesijärvi. Being different from the dominant ITS1-S allele of the Baltic Sea, strain TR183 is not a good representative of the major populations of the Baltic Sea. It is also possible that this strain has its origins in a lake that is connected to the sea. In general, it has been easier to establish cultures of A. flos-aquae from lakes than from the Baltic Sea, suggesting that there are phenotypic differences between lake A. flos-aquae and Baltic Sea A. flos-aquae.
The ITS1-S region is not linked to any known phenotypic feature. Nevertheless, a number of recent studies have demonstrated that differences in different regions of the genome can be associated with physiological differences among strains of cyanobacteria. In North Atlantic populations of Prochlorococcus, coexisting high-light-adapted strains differed from low-light-adapted strains by a 3% difference in the 16S rRNA gene sequences (21). Similarly, differences in 16S rRNA genes were found between high-temperature-adapted and other populations of Synechococcus in Oregon hot springs (20). In Nodularia, the ability to produce nodularin was reflected in the 16S rRNA genes (17) as well as in the PC-IGS and ITS1-S regions (16). In the diatom Ditylum brightwellii, genetically distinct (DNA fingerprinting) isolates differed significantly in their physiological properties, suggesting that in a single geographical locale, extensive genetic and physiological differentiation within a population is possible (30). Similarly, the different ITS1-S genotypes of A. flos-aquae that we observed in this study may represent different phenotypes that have different levels of fitness under the Baltic Sea conditions.
Although the ITS1-S region contained the variability needed to study the relationships of close relatives, the different copies of ITS1 with similar lengths called for cautious interpretation of the results. The 16S-23S rRNA region of bacteria is prone to intragenomic rearrangements as well as lateral transfer between organisms (8). In fact, the ITS1-Sc copy that was dominant in the Baltic Sea had elements from both ITS1-Sa and ITS1-Sb. It cannot be completely ruled out that ITS1-Sc is an artifact produced by PCR-mediated recombination between ITS1-Sa and ITS1-Sb carried on the same genome, but it seems very unlikely, given the high frequency of completely identical sequences that were obtained. ITS1-Sc may have been produced either by intragenomic rearrangements between the rRNA operons or by genetic exchange. Nevertheless, if the Baltic Sea population were more variable and genetic exchange did occur, it is unlikely that our random sampling would have yielded ITS1-S sequences among which 25 out of 27 sequences (93%) (or, if strain TR183 were excluded, 96%) were completely identical. A recent study demonstrated that among strains of N. spumigena, genetic exchange most likely occurs in the Baltic Sea (1). Those results were obtained by allele-specific PCR of a noncoding spacer next to the gas vacuole protein A-encoding gene and the ITS1 and PC-IGS regions (1). In N. spumigena, different alleles of each of those regions were found. In light of those findings, genetic homogeneity of A. flos-aquae populations in the Baltic Sea, as observed on the basis of ITS1-S in the present study and PC-IGS in the study of Barker et al. (2), seems valid, and genetic exchange is more likely to occur in the genetically more diverse lake populations of A. flos-aquae.
Acknowledgments
Funding for the present study was provided by two grants from the Academy of Finland: 46812 and FIBRE 48008.
We thank the Tvärminne Zoological Station of the University of Helsinki for field assistance and permission to use boats and sampling equipment. We thank Algaline, the system of continuous sampling on ferry boats of the Finnish Institute of Marine Research, for samples from the Baltic Sea proper and the open Gulf of Finland. We thank Juha Flinkman for kindly providing us with samples beneath the ice cover by scuba diving. Comments by anonymous reviewers, David Fewer, Muriel Gugger, Jaana Lehtimäki, Christina Lyra, and Sari Repka helped to improve the article.
REFERENCES
- 1.Barker, G. L., B. A. Handley, P. Vacharapiyasophon, J. R. Stevens, and P. K. Hayes. 2000. Allele-specific PCR shows that genetic exchange occurs among genetically diverse Nodularia (Cyanobacteria) filaments in the Baltic Sea. Microbiology 146:2865-2875. [DOI] [PubMed] [Google Scholar]
- 2.Barker, G. L., A. Konopka, B. A. Handley, and P. K. Hayes. 2000. Genetic variation in Aphanizomenon (Cyanobacteria) colonies from the Baltic Sea and North America. J. Phycol. 36:947-950. [Google Scholar]
- 3.Barker, G. L. A., P. K. Hayes, S. L. O' Mahony, P. Vacharapiyasophon, and A. E. Walsby. 1999. A molecular and phenotypic analysis of Nodularia (Cyanobacteria) from the Baltic Sea. J. Phycol. 35:931-937. [Google Scholar]
- 4.Brussard, P. F. 1984. Geographic patterns and environmental gradients: the central-marginal model in Drosophila revisited. Annu. Rev. Ecol. Syst. 15:25-64. [Google Scholar]
- 5.Castenholz, R. 1992. Species usage, concept, and evolution in the cyanobacteria (blue-green algae). J. Phycol. 28:737-745. [Google Scholar]
- 6.Felsenstein, J. 2001. PHYLIP (phylogeny inference package), version 3.6 (alpha2). Department of Genetics, University of Washington, Seattle.
- 7.Gugger, M., C. Lyra, P. Henriksen, A. Couté, J. F. Humbert, and K. Sivonen. 2002. Phylogenetic comparison of the cyanobacterial genera Anabaena and Aphanizomenon. Int. J. Syst. Evol. Microbiol. 52:1867-1880. [DOI] [PubMed]
- 8.Gürtler, V. 1999. The role of recombination and mutation in 16S-23S rDNA spacer rearrangements. Gene 238:241-252. [DOI] [PubMed] [Google Scholar]
- 9.Hayes, P. K., and G. L. A. Barker. 1997. Genetic diversity within Baltic Sea populations of Nodularia (Cyanobacteria). J. Phycol. 33:919-923. [Google Scholar]
- 10.Holmberg, R., and O. Jokinen. 2000. Mustionjoen, Fiskarsinjoen, Pohjanpitäjänlahden ja Tammisaaren merialueen yhteistarkkailun yhteenveto vuodelta 1998. Länsi-Uudenmaan vesi ja ympäristö ry, Report no. 102.
- 11.Iteman, I., R. Rippka, N. T. De Marsac, and M. Herdman. 2000. Comparison of conserved structural and regulatory domains within divergent 16S rRNA-23S rRNA spacer sequences of cyanobacteria. Microbiology 146:1275-1286. [DOI] [PubMed] [Google Scholar]
- 12.Janson, S., E. J. Carpenter, and B. Bergman. 1994. Fine structure and immunolocalization of proteins in Aphanizomenon sp. from the Baltic Sea. Eur. J. Phycol. 29:203-211. [Google Scholar]
- 13.Kahru, M., U. Horstmann, and O. Rud. 1994. Satellite detection of increased cyanobacteria blooms in the Baltic Sea: natural fluctuation or ecosystem change? Ambio 23:469-472. [Google Scholar]
- 14.Komárek, J., and L. Kovácik. 1989. Trichome structure of four Aphanizomenon taxa (Cyanophyceae) from Czechoslovakia, with notes on the taxonomy and delimitation of the genus. Plant Syst. Evol. 164:47-64. [Google Scholar]
- 15.Kononen, K., J. Kuparinen, K. Mäkelä, J. Laanemets, J. Pavelson, and S. Nommann. 1996. Initiation of cyanobacterial blooms in a frontal region at the entrance to the Gulf of Finland, Baltic Sea. Limnol. Oceanogr. 41:98-112. [Google Scholar]
- 16.Laamanen, M. J., M. F. Gugger, J. M. Lehtimäki, K. Haukka, and K. Sivonen. 2001. Diversity of toxic and nontoxic Nodularia isolates (cyanobacteria) and filaments from the Baltic Sea. Appl. Environ. Microbiol. 67:4638-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehtimäki, J., C. Lyra, S. Suomalainen, P. Sundman, L. Rouhiainen, L. Paulin, M. Salkinoja-Salonen, and K. Sivonen. 2000. Characterization of Nodularia strains, cyanobacteria from brackish waters, by genotypic and phenotypic methods. Int. J. Syst. E vol. Microbiol. 50:1043-1053. [DOI] [PubMed] [Google Scholar]
- 18.Lehtimäki, J., P. Moisander, K. Sivonen, and K. Kononen. 1997. Growth, nitrogen fixation, and nodularin production by two Baltic Sea cyanobacteria. Appl. Environ. Microbiol. 63:1647-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leppänen, J.-M., E. Rantajärvi, S. Hällfors, M. Kruskopf, and V. Laine. 1995. Unattended monitoring of potentially toxic phytoplankton species in the Baltic Sea in 1993. J. Plankton Res. 17:891-902. [Google Scholar]
- 20.Miller, S. R., and R. W. Castenholz. 2000. Evolution of thermotolerance in hot spring cyanobacteria of the genus Synechococcus. Appl. Environ. Microbiol. 66:4222-4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moore, L. R., G. Rocap, and S. W. Chisholm. 1998. Physiology and molecular phylogeny of coexisting Prochlorococcus ecotypes. Nature 393:464-467. [DOI] [PubMed] [Google Scholar]
- 22.Neilan, B. A., J. L. Stuart, A. E. Goodman, P. T. Cox, and P. R. Hawkins. 1997. Specific amplification and restriction polymorphisms of the cyanobacterial rRNA operon spacer region. Syst. Appl. Microbiol. 20:612-621. [Google Scholar]
- 23.Nelissen, B., A. Wilmotte, J. M. Neefs, and R. De Wachter. 1994. Phylogenetic relationships among filamentous helical cyanobacteria investigated on the basis of 16S ribosomal RNA gene sequence analysis. Syst. Appl. Microbiol. 17:206-210. [Google Scholar]
- 24.Niemi, Å. 1973. Ecology of phytoplankton in the Tvärminne area, SW coast of Finland. I. Dynamics of hydrography, nutrients, chlorophyll a and phytoplankton. Acta Bot. Fenn. 100:1-68. [Google Scholar]
- 25.Niemistö, L., I. Rinne, T. Melvasalo, and Å. Niemi. 1989. Blue-green algae and their nitrogen fixation in the Baltic Sea in 1980, 1982 and 1984. Meri 17:1-59. [Google Scholar]
- 26.Palinska, K. A., W. Liesack, E. Rhie, and W. E. Krumbein. 1996. Phenotype variability of identical genotypes: the need for a combined approach in cyanobacterial taxonomy demonstrated on Merismopedia-like isolates. Arch. Microbiol. 166:224-233. [DOI] [PubMed] [Google Scholar]
- 27.Pankow, H. 1976. Algenflora der Ostsee II. Plankton. Gustav Fischer Verlag, Jena, Germany.
- 28.Ranta, E., and O. Jokinen. 2000. Hiidenveden ja eräiden siihen laskevien vesistönosien yhteistarkkailun yhteenveto vuodelta 1999. Länsi-Uudenmaan vesi ja ympäristö ry, Report no. 104.
- 29.Ranta, E., and O. Jokinen. 1999. Lohjanjärven yhteistarkkailun yhteenveto vuodelta 1998. Länsi-Uudenmaan vesi ja ympäristö ry, Report no. 94.
- 30.Rynearson, T. A., and E. V. Armbrust. 2000. DNA fingerprinting reveals extensive genetic diversity in a field population of the centric diatom Ditylum brightwellii. Limnol. Oceanogr. 45:1329-1340. [Google Scholar]
- 31.Sivonen, K., S. Niemelä, R. M. Niemi, L. Lepistö, T. H. Luoma, and L. A. Räsänen. 1990. Toxic cyanobacteria (blue-green algae) in Finnish fresh and coastal waters. Hydrobiologia 190:267-275. [Google Scholar]
- 32.Voipio, A. 1981. The Baltic Sea. Elsevier Scientific Publishing Co., Amsterdam, The Netherlands.