Abstract
While the genome sequence and gene content are available for an increasing number of organisms, eukaryotic selenoproteins remain poorly characterized. The dual role of the UGA codon confounds the identification of novel selenoprotein genes. Here, we describe a comparative genomics approach that relies on the genome-wide prediction of genes with in-frame TGA codons, and the subsequent comparison of predictions from different genomes, wherein conservation in regions flanking the TGA codon suggests selenocysteine coding function. Application of this method to human and fugu genomes identified a novel selenoprotein family, named SelU, in the puffer fish. The selenocysteine-containing form also occurred in other fish, chicken, sea urchin, green algae and diatoms. In contrast, mammals, worms and land plants contained cysteine homologues. We demonstrated selenium incorporation into chicken SelU and characterized the SelU expression pattern in zebrafish embryos. Our data indicate a scattered evolutionary distribution of selenoproteins in eukaryotes, and suggest that, contrary to the picture emerging from data available so far, other taxa-specific selenoproteins probably exist.
Introduction
Selenium is a micronutrient found in proteins in the eubacterial, archaeal and eukaryotic domains of life. It is present in selenoproteins in the form of selenocysteine (Sec), the 21st amino acid. Sec is inserted co-translationally in response to UGA codons, a stop signal in the canonical genetic code. The alternative decoding of UGA depends on several cis- and trans-acting factors. In eukaryotes, the main cis-factor is an mRNA element, the selenocysteine insertion sequence (SECIS), located in the 3′UTR of selenoprotein genes (Walczak et al, 1998; Grundner-Culemann et al, 1999). About 25 Sec-containing proteins have been identified in eukaryotes (Kryukov et al, 2003), but distribution among taxa varies greatly. For instance, no selenoproteins have been found in yeast and land plants, only one in worms and three in flies. The majority of selenoproteins have homologues in which Sec is replaced by cysteine (Cys), even in genomes lacking the Sec-containing gene.
Because of the dual role of the UGA codon, identification of novel selenoproteins in eukaryotes is very difficult. The more direct approach is to search for occurrences of the SECIS structural pattern. Although this approach has been successfully applied in expressed sequence tag (EST) and other cDNA sequences (Kryukov et al, 1999; Lescure et al, 1999), the low specificity of SECIS searches produces a large number of predictions when applied to eukaryotic genomes. Thus, for the analysis of Drosophila melanogaster (Castellano et al, 2001, Martin-Romero et al, 2001), we devised a strategy that coordinated SECIS identification with prediction of genes with in-frame TGA codons. Again, while this strategy efficiently identified novel selenoproteins in the fly, it resulted in a large number of potential selenoprotein candidates when applied to larger and more complex vertebrate genomes.
Here, we describe a comparative genomics strategy to target bona fide selenoproteins in such complex genomes. Underlying comparative genome methods is the assumption that conservation of function is often reflected in sequence conservation. Indeed, we have already used the fact that SECIS sequences are characteristically conserved between orthologous genes in our recent characterization of human and mouse selenoproteomes (Kryukov et al, 2003). Here, we compare computational predictions of genes with in-frame TGA codons in two different vertebrate genomes, and then search for sequence alignments with conservation around Sec–Sec or Cys–Sec aligned pairs, as suggestive of selenoprotein function. The underlying assumption is that sequence conservation in regions flanking a UGA codon strongly argues for protein coding function across the codon.
We have applied this strategy to human (Homo sapiens) and puffer fish (Takifugu rubripes) genomes. Our method led to the discovery of a novel selenoprotein family (SelU) in puffer fish, whereas its human counterpart contained Cys. In addition, Sec-containing homologues exist in other fish, chicken, sea urchin, green algae and diatoms. The results presented argue for a scattered phylogenetic distribution of selenoprotein genes, suggesting a quite dynamic Sec/Cys evolutionary exchange.
Results
Comparative gene prediction of novel selenoproteins
We used the geneid program (Guigó et al, 1992; Parra et al, 2000) to predict standard and TGA-containing genes. geneid predicted 42,357 and 41,127 standard genes in the human and fugu genomes respectively, and 27,605 and 28,603 TGA-containing genes (see Methods and supplementary information online). In all, 20 out of the 23 human selenoprotein genes and 18 out of the 22 fugu selenoprotein genes that were mapped on these genomes were among the predicted TGA-containing genes.
Inter- and intragenomic comparisons in search of Sec–Sec- and Sec–Cys-containing conserved alignments reduced the set of TGA-containing predictions to 133 selenoprotein candidates: 49 orthologous human–fugu selenoprotein predictions, including the 17 known selenoproteins that mapped to both genomes; 58 human selenoproteins with standard fugu orthologues; and 26 fugu selenoproteins with standard human orthologues. Here, we rely on the assumption that coding sequence conservation across a UGA codon between two DNA sequences from different species is strongly suggestive of Sec coding function.
To validate the resulting human–fugu pairs, we undertook an exhaustive search against a number of databases of known coding (proteins and ESTs) and genomic sequences (see supplementary information online). These searches narrowed the number of predicted selenoproteins to 19. This set included two novel human–fugu pairs. Both pairs contained a human standard gene and a fugu selenoprotein gene orthologue, and belonged to the same family. A similar secondary structure pattern around the Sec or Cys residue common to the majority of selenoproteins was found (Castellano et al, 2001).
We tested whether newly discovered selenoproteins had SECIS elements in their 3′UTRs. SECIS element prediction was performed in the genomic regions of the two predicted fugu selenoproteins using SECISearch 2.0 (Kryukov et al, 2003) with a loose pattern (see Methods). A type 1 SECIS was found for each gene that fitted the established free-energy criteria.
Further homology searches in the fugu and human genomes expanded the fugu selenoprotein family with a third member having also Sec in fugu and Cys in human. This third SelU fugu gene bears a form 2 SECIS and it was not predicted because it lies in a partial contig, missing the 5′ end of the gene.
SelU in Takifugu rubripes
The Fugu SelU family (Fig 1) is composed of four members: SelUa and SelUb both have five coding exons with the in-frame TGA located in the second exon; SelUc has four coding exons (although the prediction is incomplete because of the lack of upstream genomic sequence) and the in-frame TGA lies in the first exon; and SelUd has Cys and its gene structure is not known.
Figure 1.
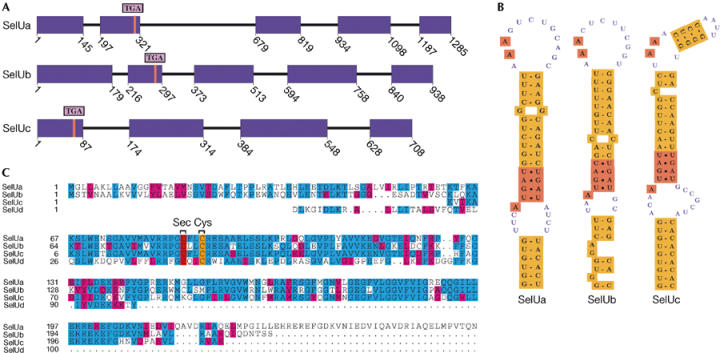
Fugu SelU family. (A) Gene structure (coding exons in purple) plotted using gff2ps (Abril & Guigó, 2000). Red lines mark the TGA triplet. SelUc is a partial gene lacking the upstream region. (B) SECIS structures. SelUa and SelUb bear a type 1 SECIS and SelUc a type 2 SECIS. (C) Alignment of SelU paralogues using CLUSTAL_W (Thompson et al, 1994). U is Sec.
SelU in Homo sapiens
The human SelU family (Fig 2) is composed of three Cys-containing members. They are uncharacterized predictions by the Ensembl system: ENSG00000122378 is a five-exon gene on chromosome 10, ENSG00000158122 is a six-exon gene on chromosome 9, and ENSG00000157870 has seven exons and maps to chromosome 1. Sequence homology does not apparently suffice to establish the unambiguous orthologous genealogy of the fugu and human SelU proteins (human SelUs named 1–3 in Fig 3).
Figure 2.
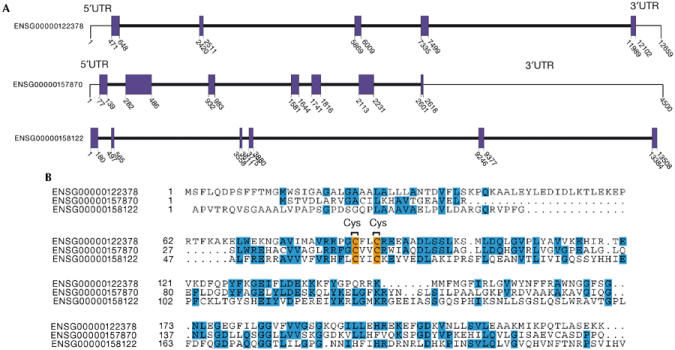
Ensembl human SelU family. (A) Gene structure (coding exons) for ENSG00000122378, ENSG00000157870 and ENSG00000158122 genes. (B) Alignment of SelU paralogues.
Figure 3.
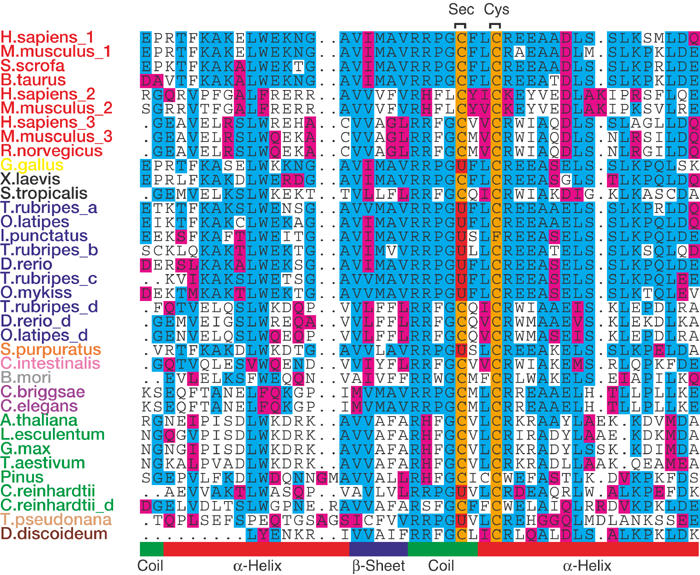
Multiple alignment of SelU proteins across the eukaryotic lineage (the sequence around the Sec (U) amino acid in red and Cys (C) in orange is shown). The sequences are clustered phylogenetically and by sequence similarity. The predicted protein secondary structure is shown at the bottom (also see supplementary information online). Species colours: mammals, red; birds, yellow; amphibians, black; fish, blue; echinoderms, orange; tunicates, pink; arthropods, grey; worms, violet; plants, green; diatoms, light orange; slime molds, brown.
SelU distribution in eukaryotes
The SelU family is widely distributed across the eukaryotic domain with either Cys- or Sec-containing proteins (Fig 3). Available sequences show that mammals, land plants, arthropods, worms, amphibians, tunicates and slime molds have Cys-containing SelUs, whereas fish, birds, echinoderms, green algae and diatoms carry Sec-containing proteins, although fish and possibly other genomes also have Cys paralogues. Apparently, yeast and flies (among arthropods) lack proteins of this family. Sec is located in SelU proteins close to a conserved Cys such that the two residues form a motif that resembles the CxxC motif that is present in various thiol-dependent redox proteins. Similar motifs are present in a number of eukaryotic selenoproteins, including SelP, SelW, SelV, SelT, SelM and SelH. Conversely, no SelU homologue is present in prokaryotes (see supplementary information online).
Metabolic labelling of SelU with 75Se
To determine whether the SelU family indeed contains Sec (Fig 4), we developed a construct containing the green fluorescent protein (GFP), fused to the carboxy (C)-terminal region of chicken SelU, and the entire 3′UTR (including the predicted SECIS element). The fusion protein was designed such that its size would be different from those of endogenous mammalian selenoproteins. Monkey CV-1 cells transfected with the construct were metabolically labelled with 75Se, and 75Se-containing selenoproteins were analysed by SDS–polyacrylamide gel electophoresis (SDS–PAGE) and a PhosphorImager analysis. This experiment revealed the presence of a 75Se-labelled band corresponding in size to the GFP–SelU fusion protein, if TGA encoded Sec. Thus, SelU is a true selenoprotein.
Figure 4.
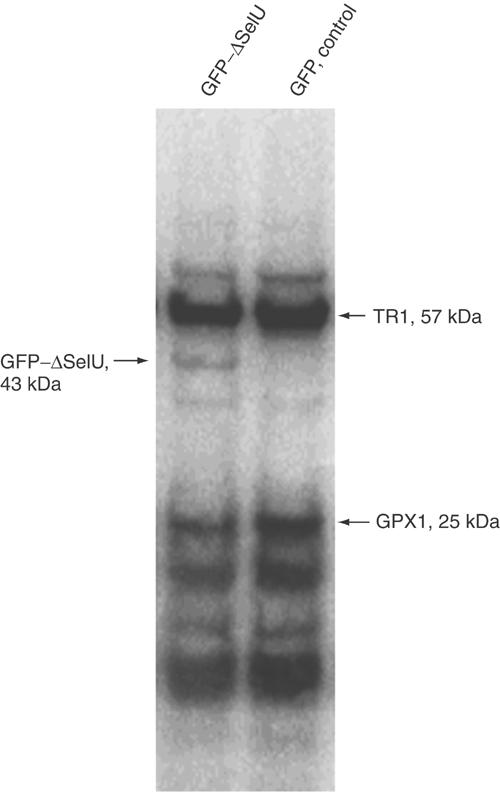
Detection of 75Se-labelled SelU. CV-1 cells were transfected with either GFP–ΔSelU fusion construct (left line) or GFP vector as a control (right line), and grown in the presence of 75Se[selenite] for 24 h. Cell extracts containing 75Se-labelled selenoproteins were resolved by SDS–polyacrylamide gel electrophoresis and visualized with a PhosphorImager System. Locations of major endogenous selenoproteins TR1 (57 kDa) and GPX1 (25 kDa) are shown on the right, and the GFP–ΔSelU fusion protein on the left.
Expression of SelU during zebrafish embryogenesis
Tissue and temporal expression of the SelU gene during embryogenesis was addressed in the zebrafish model. A probe complementary to the zebrafish SelU cDNA (EST fz58h06.y2, homologue to fugu SelUa) was designed, and in situ hybridization was performed on whole zebrafish embryos from different developmental stages. The hybridization sites were revealed by a chromogenic reaction and the expression patterns were analysed. The SelU gene was widely expressed in all embryonic tissues from all stages (Fig 5). Expression was already detectable at the early stages from gastrula and somitogenesis (Fig 5A–C), but within the embryonic tissues only; there was no expression within the nutrient cells of the yolk syncytial layer. Later in development, expression remained high and nonrestricted (Fig 5D–F), demonstrating ubiquitous expression of the SelU gene.
Figure 5.
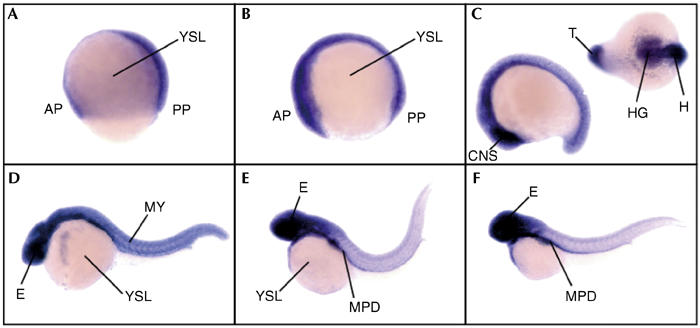
Expression pattern of the SelU gene during development in zebrafish embryos. Developmental stages are (A) gastrula, (B) early somitogenesis, (C) late somitogenesis, (D) 24 h postfertilization, (E) 36 h postfertilization and (F) 48 h postfertilization. All views are lateral except the one in the upper right corner in (C) which is dorsoventral. AP, anterior pole; CNS, central nervous system; E, eye; H, head; HG, hatching gland; MPD, medial part of the pronephric duct; MY, myotomes; PP, posterior pole; T, tail; YSL, yolk syncytial layer.
Discussion
A growing body of evidence relates selenium to cancer prevention, immune system function, male fertility, cardiovascular and muscle disorders and prevention and control of the ageing process (Hatfield, 2001). Selenoproteins are thought to be responsible for a majority of these biomedical effects of selenium. To understand the role of selenium in health, the identification and characterization of eukaryotic selenoproteins is thus essential. Despite the increasing availability of eukaryotic genome sequences, the dual role of the UGA codon limits our ability to identify novel selenoproteins. The discovery here of the SelU family shows that comparative genomics could play an important role in overcoming this limitation.
While our comparative method aims at the exhaustive characterization of selenoproteomes, it is certainly unclear how complete is our set of fugu selenoproteins. However, recognition of the majority of known selenoproteins in this organism by this method argues for the identification of all or almost all fugu selenoproteins. In addition, because it assumes no restriction in the SECIS structure, our approach can identify genes with noncanonical SECIS. Although no such elements were found here, they may exist in more divergent lower eukaryotic genomes.
At present, neither sequence database searches nor more specialized motif searches identify similar proteins of known function (data not shown). However, in situ hybridization shows ubiquitous expression of SelU in fish embryos (Fig 5), and EST searches also suggest a widespread expression of SelU in human adult tissues (data not shown) pointing to a basic function in the cell.
The SelU family is widely distributed across the eukaryotic lineage, either as Sec- or Cys-containing proteins (Fig 3), but lacks the counterpart in prokaryotes. The scattered and taxa-specific distribution of Sec and Cys forms of a SelU, although common in prokaryotic selenoprotein families, is unexpected in eukaryotes. Besides SelU, other eukaryotic families show an unbalanced distribution, but are constantly present in mammals as true selenoproteins. Therefore, it has been implicitly assumed that mammalian selenoproteins recapitulate the eukaryotic selenoproteome. Our finding challenges this statement and suggests a more discrete distribution of Sec-containing proteins. This hypothesis is reinforced by the recent discovery that methionine-S-sulphoxide reductase (MsrA) occurs as a selenoprotein in Chlamydomonas reinhardtii, a green algae, but has Cys in vertebrates (including mammals) and other invertebrates (Fu et al, 2002; Novoselov et al, 2002). Furthermore, a glutathione peroxidase homologue (GPX6) was recently reported to have Sec in humans and pigs, but Cys in rodents (Kryukov et al, 2003).
The fact that selenoproteins are distributed discretely at very different taxonomic levels raises the question of whether Sec loss or Sec gain is favoured by evolution. Arguments exist in favour of both possibilities. Replacement of Sec by Cys is plausible because it yields a protein with diminished, but still functional, catalytic activity (Axley et al, 1991; Berry et al, 1992), and allows an organism to be independent of the supply of the trace element selenium. The fact that a ‘fossil' SECIS has been identified in the Cys-containing GPX6 in rodents (Kryukov et al, 2003) and in human GPX5 (data not shown) suggests that this event has indeed occurred during evolutionary time. In this regard, we searched for vestigial SECIS in human, rodent, amphibian and fish (Cys paralogues) SelU UTRs (see supplementary information online) with inconclusive results. The conversion in the other direction, a Cys to Sec mutation, is apparently more difficult, since the introduction of an in-frame stop codon must be compensated by the simultaneous emergence of a functional SECIS element in the 3′UTR of the gene. However, gene duplications, the pre-existence of SECIS-like signals, mobile genomic elements, horizontal transfer and the superior catalytic efficiency of Sec could make this process feasible. In any case, it remains to be settled why some organisms prefer Sec, while others prefer Cys-containing forms of orthologous proteins. The presence of SelU Sec and Cys paralogues in fish genomes, however, is suggestive of a particular history for each family and taxa, mediated by an ongoing evolutionary process of Sec/Cys interconversion, in which contingent events could play a role as important as functional constraints.
In any case, if the results obtained here through the analysis of the fugu genome are representative of more divergent eukaryotic genomes, the certain conclusion is that we comprehend today only a fraction of the selenium-dependent world.
Methods
Prediction of selenoproteins in nucleotide sequences. A general scheme is shown in Fig 6. Briefly, for each genome, we predict independently standard and selenoprotein genes, using the standard geneid and a modification that allows the prediction of genes interrupted by in-frame TGA (Castellano et al, 2001) (see supplementary information online).
Figure 6.
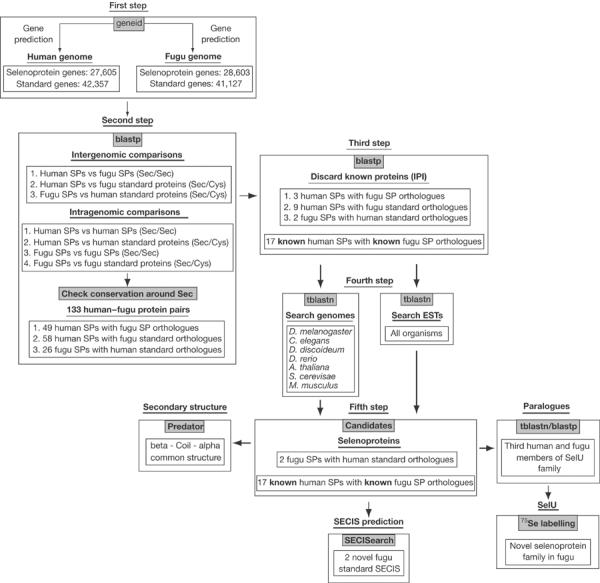
General schema for selenoprotein identification.
Protein sequence comparisons: identification of Sec–Sec and Sec–Cys conserved pairs. Proteins predicted in fugu and human are compared using blastp (Altschul et al, 1997). Conserved protein sequence alignments with conservation in regions flanking Sec–Sec or Sec–Cys aligned pairs are selected as potential selenoproteins (see supplementary information online).
Prediction of SECIS in nucleotide sequences. SECIS elements are predicted in selected selenoprotein genes with the SECISearch program (Kryukov et al, 2003) (see supplementary information online).
Metabolic labelling of SelU with 75Se. A 760 bp fragment of chicken SelU cDNA coding for a 16 kDa C-terminal portion and 3′UTR (including the SECIS element) was amplified with AGTGCTCGAGGTGATCATGGCTGTGCGAAGAC and TTATGGATCCGGTTTTGCTCCCCTGGGTAGAC primers and cloned into the XhoI/BamHI sites of pEGFP-C3 vector (Clontech). CV-1 cells were transfected with either the resulting construct or corresponding vector as a control. In all, 5 μg of DNA and 20 μl of lipofectamine (Invitrogen) were used for transfection of each 60-mm-diameter plate, followed by incubation of cells with 25 μCi 75Se[selenite] (University of Missouri Research Reactor). Samples were analysed on sodium dodecyl sulphate (SDS)–10% NuPAGE gels (Invitrogen). 75Se-labelled proteins were visualized with a Storm PhosphorImager system (Molecular Dynamics). Transfection efficiency was followed by a parallel transfection of cells with a GFP construct. In addition, CV-1 cells were separately transfected with a human SelM construct and labelled with 75Se, which provided a positive control.
In situ hybridization. Eight different zebrafish ESTs, encoding a protein homologous to the fugu SelU protein, were compiled. These EST sequences generated a 1,292 bp contiguous nucleotide sequence encompassing the entire open reading frame and the 3′UTR containing the SECIS motif. A DNA probe complementary to the entire zebrafish SelU cDNA was PCR amplified from an oligo-dT cDNA library (a gift from C. Thisse and B. Thisse) and cloned with compatible restriction sites into pSK(−). Antisense probe synthesis and whole-mount in situ hybridization were performed according to Thisse et al (1993). The fully detailed protocol is accessible at http://zfin.org/zf_info/zfbook/chapt9/9.82.html. Specificity was assessed using antisense and other irrelevant probes (data not shown).
Data and software availability. Sequence data and software can be found at http://genome.imim.es/databases/spfugu2004
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/7400036s1.pdf).
Supplementary Material
supplementary information
Acknowledgments
We thank the referees for helpful suggestions and J.F. Abril for technical assistance. S. Obrecht-Pflumio, C. Thisse and B. Thisse are gratefully thanked for technical expertise with in situ hybridization. S.C. is the recipient of a predoctoral fellowship from Generalitat de Catalunya. This work was supported by grant BIO2000-1358-C02-02 from Ministerio de Educación y Ciencia (Spain) to R.G. and by NIH grant GM061603 to V.N.G.
References
- Abril JF, Guigó R (2000) gff2ps: visualizing genomic annotations. Bioinformatics 16: 743–744 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axley MJ, Böck A, Stadman TC (1991) Catalytic properties of an Escherichia coli formate dehydrogenase mutant in which sulfur replaces selenium. Proc Natl Acad Sci USA 88: 8450–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MJ, Mai AL, Kieffer J, Harney JW, Larsen P (1992) Substitution of cysteine for selenocysteine in type i iodothyronine deiodinase reduces the catalytic efficiency of the protein but enhances its translation. Endocrinology 131: 1848–1852 [DOI] [PubMed] [Google Scholar]
- Castellano S, Morozova N, Morey M, Berry MJ, Serras F, Corominas M, Guigó R (2001) In silico identification of novel selenoproteins in the Drosophila melanogaster genome. EMBO Rep 2: 697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu LH, Wang XF, Eyal Y, She YM, Donald LJ, Standing KG, Ben-Hayyim G (2002) A selenoprotein in the plant kingdom. Mass spectrometry confirms that an opal codon (UGA) encodes selenocysteine in Chlamydomonas reinhardtii glutathione peroxidase. J Biol Chem 277: 25983–25991 [DOI] [PubMed] [Google Scholar]
- Grundner-Culemann E, Martin GW III, Harney JW, Berry MJ (1999) Two distinct SECIS structures capable of directing selenocysteine incorporation in eukaryotes. RNA 5: 625–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guigó R, Knudsen S, Drake N, Smith TF (1992) Prediction of gene structure. J Mol Biol 226: 141–157 [DOI] [PubMed] [Google Scholar]
- Hatfield DL (ed.) (2001) Selenium: Its Molecular Biology and Role in Human Health. Dordrecht: Kluwer Academic Publishers [Google Scholar]
- Kryukov GV, Kryukov VM, Gladyshev VN (1999) New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J Biol Chem 274: 33888–33897 [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigó R, Gladyshev VN (2003) Characterization of mammalian selenoproteomes. Science 300: 1439–1443 [DOI] [PubMed] [Google Scholar]
- Lescure A, Gautheret D, Carbon P, Krol A (1999) Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J Biol Chem 274: 38147–38154 [DOI] [PubMed] [Google Scholar]
- Martin-Romero FJ, Kryukov GV, Lobanov AV, Carlson BA, Lee BJ, Gladyshev VN, Hatfield DL (2001) Selenium metabolism in Drosophila: selenoproteins, selenoprotein mRNA expression, fertility, and mortality. J Biol Chem 276: 29798–29804 [DOI] [PubMed] [Google Scholar]
- Novoselov SV, Rao M, Onoshko NV, Zhi H, Kryukov GV, Xiang Y, Weeks DP, Hatfield DL, Gladyshev VN (2002) Selenoproteins and selenocysteine insertion system in the model plant cell system, Chlamydomonas reinhardtii. EMBO J 21: 3681–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Blanco E, Guigó R (2000) Geneid in Drosophila. Genome Res 10: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thisse C, Thisse B, Schilling TF, Postlethwait JH (1993) Structure of the zebrafish snail1 gene and its expression in wild-type, spadetail and no tail mutant embryos. Development 119: 1203–1215 [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL_W: improving the sensitivity of progressive sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Res 22: 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walczak R, Carbon P, Krol A (1998) An essential non-Watson–Crick base pair motif in 3′UTR to mediate selenoprotein translation. RNA 4: 74–84 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
supplementary information


