Abstract
We identify and characterize MlaA, a novel protein, which is found in a conserved operon with Mre11 and Rad50 in archaeal genomes. MlaA is fused with Mre11 in Methanobacter thermoautotrophicus, suggesting the MlaA is functionally linked to the Mre11 complex. MlaA preferentially and cooperatively binds double-stranded and secondary structure containing DNA and has double-stranded but not single-stranded DNA-stimulated ATPase activity. Electron microscopy reveals that MlaA forms a 360-kDa hexameric ring structure with a central hole. Our data suggest that the archaeal Mre11 complex is associated with a novel hexameric ATPase that could be required for the processing of DNA double-stranded breaks and recombination intermediates.
Introduction
Homologous recombination of DNA is an evolutionarily conserved process that is implicated in DNA double-stranded break (DSB) repair, rescue of stalled replication forks and meiosis (Haber, 2000). A key player in homologous recombination is the Mre11 complex, which contains the nuclease Mre11 and the ATPase Rad50 (Connelly & Leach, 2002; D'Amours & Jackson, 2002). The Mre11 complex degrades DNA ends and hairpins in an ATP-dependent manner (Connelly et al, 1998; Furuse et al, 1998; Paull & Gellert, 2000; Trujillo & Sung, 2001) and is suggested to bridge sister chromatids and/or DNA ends in recombination and repair (de Jager et al, 2001; Hopfner et al, 2002).
In eukaryotes, the Mre11 complex contains a third component, Nbs1/Xrs2, which is implicated in checkpoint regulation and association of the Mre11 complex with chromatin (Carney et al, 1998; D'Amours & Jackson, 2001; Tauchi et al, 2002). In addition, interactions with the breast cancer-associated protein 1 (Brca1), Rad50-interacting protein 1 and the mediator of damage checkpoint Mdc1 (Zhong et al, 1999; Wang et al, 2000; Xiao et al, 2001; Goldberg et al, 2003; Stewart et al, 2003) suggest that the Mre11 complex tightly cooperates with other proteins in conducting damage recognition and repair.
The Mre11 complex is highly conserved not only in eukaryotes but also in archaea, with similar structural and functional features (Hopfner et al, 2000). Archaea can be resistant to ionizing radiation, indicating that they have a very efficient recombination machinery (Gerard et al, 2001). Like eukaryotes, archaea do not possess the eubacterial recombination factors RecBCD and RecFOR, suggesting that in archaea and eukaryotes the Mre11 complex has similar key roles in recombination. Little is known yet about interacting partners of archaeal Mre11 complex homologues.
In this report, we identify and characterize MlaA, a 60-kDa component of the conserved Mre11 and Rad50-containing operon in archaeal genomes. We show that MlaA from Pyrococcus abyssii is an ATPase and preferentially binds double-stranded (ds)DNA with secondary structure. Moreover, ATPase activity is stimulated by dsDNA but not single-stranded (ss)DNA binding. In the genome of Methanobacter thermoautotrophicus, MlaA is evidently fused to Mre11, indicating that it processes DNA in concert with the Mre11/Rad50 complex. Electron microscopy reveals a hexameric ring, indicating that MlaA is a member of the hexameric AAA family ATPases. This novel DNA-binding ATPase could be involved in the processing of stalled replication/recombination intermediates in DSB repair in conjunction with the Mre11 complex, and suggests that similar activities exist in eukaryotes.
Results
The Mre11/Rad50 operon in archaeal genomes
Mre11 and Rad50 open reading frames are found in operon-like structures in archaeal genomes. This operon contains two additional conserved open reading frames (Fig 1). The first open reading frame has recently been identified to code for a 5′–3′ nuclease (NurA), suggesting that the operon is linked to DSB repair and recombination in archaea (Constantinesco et al, 2002).The second open reading frame codes for a novel 60-kDa protein (termed MlaA in the following). The conservation of this archaeal operon suggests that these four proteins could participate in the same repair/recombination pathway.
Figure 1.
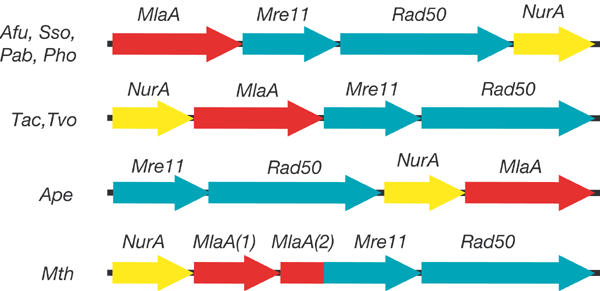
Mre11-containing operon in archaea. Four genes are consistently found in Mre11 and Rad50-containing archaeal operon-like structures: Mre11 and Rad50 (green), the NurA nuclease (yellow) and the uncharacterized MlaA (red). The arrangement of genes varies in different archaea, but the directionality of the open reading frames and the contents of the operon are highly conserved, suggesting that these four proteins are functionally linked. In Mth, the MlaA gene is split into two segments. The C-terminal segment is fused with the Mre11 gene into a single open reading frame, suggesting tight functional coupling of Mre11 and MlaA in archaea (Afu, Archaeglobus fulgidus; Ape, Aeropyrum pernix; Mth, Methanobacter thermoautotrophicus; Pab, Pyrococcus abyssii; Pho, Pyrococcus horikoshii; Sso, Sulfolobus solfataricus; Tac, Thermoplasma acidophilum; Tvo, Thermoplasma volcanium).
In the genome of M. thermoautotrophicus, MlaA is split into two segments (Figs 1, 2). Remarkably, the carboxy (C)-terminal segment of MlaA is fused with Mre11 into a single open reading frame, further suggesting that Mre11 and MlaA interact functionally.
Figure 2.

Sequence alignment and functional motifs of MlaA. Alignment of different archaeal MlaA sequences (for abbreviations see Fig 1) reveal highly conserved motifs (I–IV). MlaA contains putative Walker A (I) and Walker B (III) motifs, suggesting that MlaA is an ATPase. Two other highly conserved motifs (II and IV) are unique to MlaA. Two regions display homology to E. coli FtsK (FtsK-I and FtsK-II). The C-terminal region of MlaA is highly acidic (underlined residues). Mth MlaA is split into two segments (corresponding to residues 1–348 and 349–536). The C-terminal segment is fused with Mre11 (only partially shown) into a single open reading frame. This gene fusion suggests a tight functional linkage between MlaA and Mre11.
MlaA has sequence similarity with AAA family ATPases
To identify conserved sequence motifs of MlaA, we aligned different archaeal sequences (Fig 2). We find conserved putative Walker-A (motif I) and Walker B (motif III) motifs, suggesting that MlaA is an ATPase. We would therefore like to suggest the name MlaA for Mre11 complex-linked ATPase in archaea. Weak homology to FtsK, an ATPase involved in eubacterial DNA dimer resolution (Aussel et al, 2002), indicates that MlaA is a DNA-associated AAA ATPase. Since MlaA does not have the FtsK characteristic transmembrane domains and some organisms have genes for both MlaA and FtsK, MlaA is probably not a functional homologue of the segregation ATPase FtsK in archaea.
MlaA has ATPase activity
To identify biochemical features of MlaA, we expressed MlaA from P. abyssii in Escherichia coli and purified the protein to homogeneity (Fig 3A). Using thin layer chromatography (TLC), we find that MlaA has a strongly temperature-dependent ATPase activity (Fig 3B), consistent with its hyperthermophilic source organism P. abyssii (optimal growth >90°C). To test the sequence-predicted putative ATPase motifs, we introduced Lys169Leu (Walker A) and Glu408Gln (Walker B) point mutations. Both mutations severely impaired ATP hydrolysis, verifying the putative Walker A and B motifs (Fig 3B).
Figure 3.
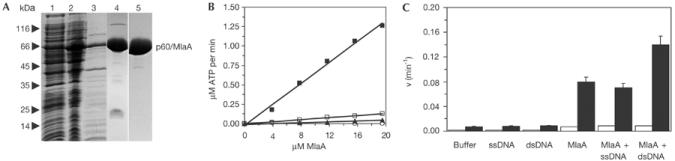
Purification and biochemical characterization of MlaA. (A) SDS–PAGE analysis (Coomassie stained) of P. abyssii MlaA expression and purification. Lane 1: insoluble E. coli fraction; lane 2: soluble E. coli fraction; lane 3: soluble fraction after heat denaturation; lane 4: MlaA after Source Q ion exchange chromatography; lane 5: MlaA after gel filtration chromatography. (B) ATPase activity assays of MlaA using TLC. The ATPase activity is greatly enhanced at 60°C (filled squares) compared to 30°C (open squares) consistent with the hyperthermophilic source organism (P. abyssii). Lys169Leu (open circles) and Glu408Gln (filled triangles) mutations dramatically reduce ATPase activity and verify MlaA Walker A and B motifs. (C) TLC ATPase assays of MlaA in the presence or absence of a twofold excess of ssDNA or dsDNA reveal that ATPase activity is stimulated twofold by dsDNA but not by ssDNA at 55°C (filled bars). At 30°C, only weak activity is observed (open bars). The rate of ATP hydrolysis per MlaA active site is shown. All assays were performed in triplicate (error bars).
The ATPase activity of MlaA is stimulated by dsDNA
The conserved operon structure suggests that MlaA might act on DNA. To test whether the ATPase activity of MlaA is affected by DNA, we performed ATPase assays in the presence of DNA (Fig 3C). ssDNA has no effect on the ATPase activity of MlaA, while dsDNA stimulates the ATPase activity of MlaA approximately twofold. This stimulation suggests that the ATPase activity of MlaA might act on DNA.
MlaA binds to dsDNA with secondary structure
We tested DNA-binding activity by electrophoretic mobility shift assays. We find that MlaA strongly binds to a blunt end dsDNA oligonucleotide but only weakly interacts with ssDNA (Fig 4A). ATP binding has no strong effect on DNA binding, suggesting that ATP is not required for DNA interaction (see Supplementary Fig S1 online). To see whether MlaA needs DNA ends for binding, we performed competition experiments with circular plasmid DNA (Fig 4B). Since circular dsDNA can efficiently compete with dsDNA oligos, MlaA does not need free ends to bind to DNA.
Figure 4.
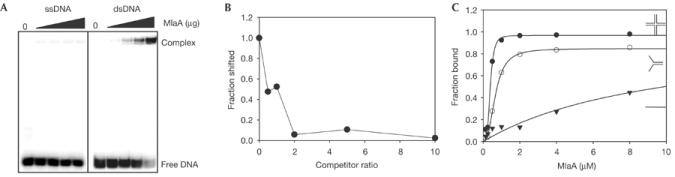
DNA-binding activity of MlaA. (A) Electrophoretic mobility shift assays show that MlaA binds to dsDNA but only weakly to ssDNA. In all, 0, 0.25, 0.5, 1 or 2 μg MlaA is incubated with 50 fmol ssDNA or dsDNA 20-mer and analysed on 6% native acrylamide gels. (B) Competition experiments using labelled dsDNA oligonucleotides and unlabelled circular dsDNA. The fraction of labelled dsDNA that is shifted by MlaA versus the competitor:probe ratio is plotted. The highly efficient competition of dsDNA oligonucleotide binding by circular plasmid dsDNA (approximately 50% at a 1:1 ratio) indicates that MlaA does not need DNA ends for efficient binding. (C) Nitrocellulose/hybond double filter binding assays with various DNA substrates (supplementary information online Table S1). The fraction of MlaA:DNA complexes (retained at nitrocellulose filters) relative to total DNA (sum of DNA retained at both nitrocellulose and hybond) versus MlaA concentration is shown. Data were fitted to f=pn/(Kd+pn), where f is the fraction of DNA bound to MlaA relative to total DNA, Kd is the dissociation constant, p is the protein concentration and n is the Hill coefficient. MlaA poorly binds ssDNA (filled triangles), but more strongly binds a dsDNA Y-substrate (open circles). The tightest binding is observed for a Holliday-junction substrate (filled circles), suggesting a preference for DNA secondary structures. dsDNA (n=2.5–4) but not ssDNA (n≈1) substrates are evidently bound cooperatively.
To obtain binding constants for various DNA substrates, we performed double-filter binding assays (see Methods). As seen before, MlaA has a strong preference for duplex DNA (Kd=0.34±0.17 μM) over ssDNA (Kd=10±7 μM). We observe, however, the strongest binding with a Holliday-junction substrate (Kd=24±10 nM), suggesting that MlaA has a preference for secondary structures. We noted cooperativity for the binding of dsDNA and Holliday junctions (Hill coefficient 2.5–4), suggesting that MlaA might act as multimers.
MlaA forms a large hexameric complex
During gel filtration chromatography we noticed that MlaA has a hydrodynamic radius corresponding to a 360-kDa protein, indicating that MlaA forms multimers (Fig 5A). To identify structural features of MlaA, we performed negative stain electron microscopy. We observed ring-shaped, hexameric particles in the electron micrographs (Fig 5B). Averaging over 1,560 particles confirmed the presence of a hexameric ring-like structure (Fig 5C). The ring has a diameter of approximately 120 Å and possesses a central hole of approximately 30 Å diameter. The electron microscopic results are in good agreement with the cooperative DNA-binding data and suggest that MlaA is a hexameric, DNA-associated AAA ATPase (Fig 5D).
Figure 5.
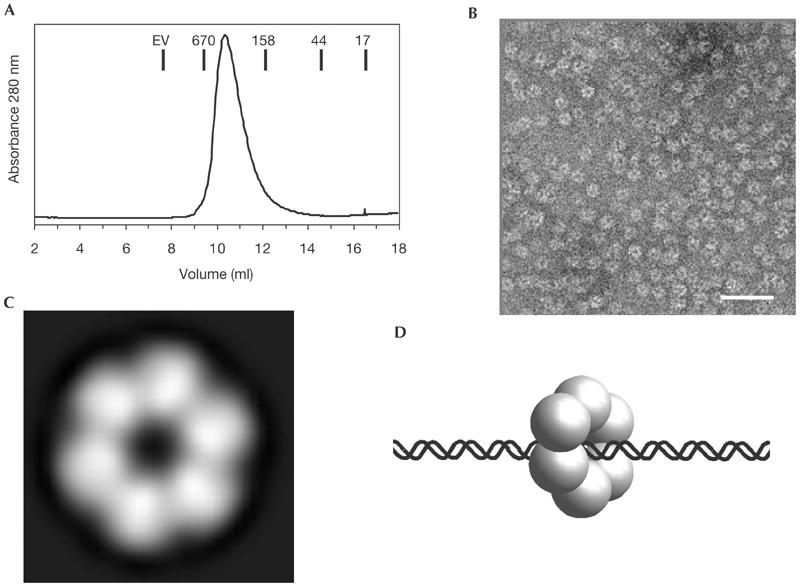
Structure of MlaA. (A) Gel filtration chromatogram showing that MlaA has a hydrodynamic radius corresponding to approximately 360-kDa molecular weight. Molecular weight standards are indicated. EV: exclusion volume. (B) Negative stain electron micrographs of MlaA show multimeric ring-shaped particles (bar: 50 nm). (C) Averaging over 1,560 electron micrograph particles shows that MlaA forms approximately 120 Å large hexameric rings with a central hole of approximately 20–30 Å diameter. This structure resembles AAA ATPases and further indicates that MlaA could be a DNA acting ATPase in DNA recombination. (D) Model for an MlaA:DNA complex with DNA threaded through the central hole.
Discussion
We have identified and characterized MlaA, a novel member of the conserved archaeal Mre11 and Rad50 operon. MlaA forms a 360-kDa large hexameric ring structure, preferentially binds dsDNA, and has an ATPase activity that is stimulated by dsDNA but not ssDNA. MlaA is evidently fused to Mre11 in M. thermoautotrophicus, further suggesting that MlaA and Mre11 functionally interact in DNA repair/recombination processes in archaea. We experimentally identified Walker A and B motifs (Figs 2, 3B). These motifs are highly conserved, suggesting that the ATPase activity of MlaA is linked to a central function of the protein. In M. thermoautotrophicus, both Walker A and B motifs are located on two open reading frames, one of which is fused to Mre11. Nevertheless, two separate Walker A and B open reading frames can assemble into a functional Walker box ATPase (Hopfner et al, 2001), suggesting that M. thermoautotrophicus MlaA is fully active despite the partitioning into two polypeptide chains.
Our biochemical ATPase and DNA-binding data and the hexameric structure suggest that MlaA is not a functional homologue of Nbs1/Xrs2, which is a subunit of the eukaryotic Mre11 complex mainly involved in mediating protein/protein interactions. In addition, we did not detect strong processive helicase activity of MlaA on either Y- or Holliday-junction substrates (see supplementary Fig S2 online). However, for technical reasons it was not possible to assay helicase activity at physiological temperatures of P. abyssii (>90°C). Thus, we cannot rule out that at physiological temperatures MlaA possesses helicase or DNA unwinding activity.
The hexameric structure of MlaA resembles AAA ATPases such as FtsK and RuvB, which are DNA translocases involved in the modulation and resolution of chromosome dimers and Holliday junctions in eubacteria. Moreover, the ATPase activity of MlaA is in the range of that of RuvB, taking into account the high physiological growth temperatures of P. abyssii (Marrione & Cox, 1996). This similar ATPase activity is consistent with the idea that MlaA translocates on DNA, possibly stimulated by additional factors. FtsK, RuvB and MlaA share similar DNA-stimulated ATPase activities and a hexameric structure, indicating that they might act on DNA by a related ATP-driven mechanism. In addition, since both RuvB and FtsK functionally interact with nucleases/resolvases, it will be interesting to see if MlaA–NurA or MlaA–Mre11 pairs also functionally interact to process DNA substrates in DSB repair and recombination.
The cooperativity observed for MlaA DNA binding indicates that MlaA either has multiple DNA-binding sites or assembles on DNA. The latter would explain why MlaA does not need free ends to bind DNA, on the basis of a model in which MlaA forms an RuvB-like hexameric ring around DNA. In fact, multiple bands in the electrophoretic mobility shift assays could represent the binding of nonhexameric MlaA complexes.
The tight binding of Holliday junctions suggests that MlaA could have a preference for secondary structures of DNA. Thus, the ATPase activity of MlaA could be specifically involved in dealing with such structures. Secondary structure in the form of Holliday junctions and collapsed replication forks arise during homologous recombination in response to stalled replication forks. Stalled replication forks are the most prominent source of DSBs, and the Mre11 complex has been specifically linked to replication-induced DSB repair in prokaryotes and eukaryotes (Connelly et al, 1998; Maser et al, 2001). With the exception of Mre11 and Rad50, none of the enzymes that are implicated in the repair of DSBs at stalled replication forks in eubacteria, in particular RecFOR and RecBCD, are conserved in archaea. Thus, it is possible that MlaA in conjunction with the NurA nuclease could be specifically involved in such DSB repair processes.
Speculation
We propose that MlaA is a DNA-associated ATPase complex that works in conjunction with Mre11/Rad50 and NurA in DSB repair and recombination in archaea. The ATPase activity of MlaA could help specifically to convert and process recombination intermediates such as Holliday junctions or D-loop structures or stalled replication forks, lesions that are expected to frequently occur in hyperthermophilic archaea. Since archaea and eukaryotes share a similar Mre11 complex core machinery, the work presented here also suggests that functional homologues of MlaA may exist in eukaryotic genomes.
Methods
Protein expression and purification. The P. abyssii genomic DNA was a generous gift from Patrick Forterre (Universite Paris-Sud, France). We amplified the MlaA open reading frame by polymerase chain reaction from P. abyssii genomic DNA using AAAAAAAACATATGATGGAAGACAAGGGATACATAGGGAT AGTGAGG and AAAAAAAAGCGGCCGCTCAGAAGTCCACATCAGGAACGTC as primer. MlaA was cloned into pET21 (Novagen) at NdeI and NotI restriction sites. We expressed MlaA in E. coli BL21 (DE3) codon plus RIL (Stratagene) at 18°C. The cells were harvested by centrifugation, resuspended in 50 mM Tris (pH 7.5), 200 mM NaCl, 1 mM EDTA, 1 mM phenylmethyl sulphonylfluoride (Sigma), and disrupted by sonication. Cell debris and insoluble proteins were removed by centrifugation. The supernatant was incubated at 75°C for 15 min to denature E. coli proteins. Precipitated E. coli protein was removed by centrifugation. MlaA in the supernatant was precipitated by 80% ammonium sulphate, resuspended and fractioned on Superdex S300 gel filtration chromatography in the presence of 1.5 M NaCl to remove nucleic acids. The protein was further purified by hydrophobic interaction, anion exchange and high-resolution gel filtration chromatography (Pharmacia).
ATPase activity assays. We tested ATPase activity of MlaA by TLC (Carpentieri et al, 2002). A total of 500 μM of γ32P-ATP (30 Ci/mol) was incubated without and with increasing concentrations of MlaA (0–20 μM in 50 mM Tris (pH 7.5), 150 mM NaCl, 5 mM MgCl2) at 30 and 60°C for 30 min. Aliquots of 1 μl were spotted on a TLC plate (Merck), developed in 0.5 M LiCl, 1 M formic acid, dried and analysed with a Storm phosphorimager and ImageQuant Software. We tested DNA-stimulated ATPase activity analogously by TLC by preincubating 10 μM MlaA with 20 μM ssDNA 30-mer or dsDNA 30-mer for 10 min, followed by the addition of 100 μM γ32P-ATP and incubation at 55°C for 30 min.
Electrophoretic mobility shift assays. Electrophoretic mobility shift assays were performed using the oligonucleotide 5′-GGTTGCAGACCTGAGGATAG-3′ labelled at its 5′-end with 32PO4 by standard methods and used alone (ssDNA) or annealed to its complement (dsDNA). In all, 0.25, 0.5, 1 or 2 μg MlaA was incubated with 50 fmol DNA in 25 mM Hepes, pH 8.0, 50 mM NaCl, 1 mM DTT and 10% glycerol for 15 min at 20°C. We separated complexes on 6% native polyacrylamide gels in 0.5 × Tris–borate buffer and analysed by a phosphorimager.
We performed competition assays with phiX174 form II DNA (dsDNA) (New England BioLabs) by incubating 1 μg of MlaA with 50 fmol labelled ds oligonucleotide substrate as described above. In all, 0.5-, 1-, 2-, 5-, 10-fold excess of competitor (μg competitor:μg probe) was included in the reaction mix before the addition of protein. Following a 15-min incubation, we analysed the reactions on native polyacrylamide gels as described above. The percentage shift was quantified using a phosphorimager.
Filter binding assays. We assessed DNA binding using a double-filter nitrocellulose/hybond nylon binding assay (Ban et al, 1999). Protein (0–8 μM) and gel-purified DNA substrates (50 fmol, see supplementary information online) were incubated in 50 mM Tris (pH 7.5), 100 mM NaCl, 5 mM MgCl2, ±100 μM ATP for 20 min at 20°C. The nitrocellulose membrane was incubated in 0.4 M KOH and extensively rinsed with water before use. Nitrocellulose and nylon membranes were equilibrated in assay buffer and assembled on a Schleich–Schüll dot blot apparatus (nitrocellulose on top of hybond and gel blotting paper). After prewashing with 40 μl buffer, 40 μl reaction mixture were applied by vacuum, followed by 40 μl buffer to wash out unbound samples. Filter membranes were dried and analysed by a phosphorimager. Data analysis was performed with SigmaPlot (see Supplementary information Table S1 online).
Electron microscopy. Purified MlaA was applied on a carbon-coated copper grid and stained with 2% uranyl acetate (w/v). Images were recorded using a Philips CM12 electron microscope equipped with a Photometrics CCD-Camera. A total of 1,560 single molecules were extracted from the images and applied to correlation averaging methods. The only translationally aligned image stack was subjected to a classification procedure based on eigenvector–eigenvalue data analysis to estimate symmetric features. Analysis was performed with EM and Semper program packages.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref/7400037s1.doc).
Supplementary Material
Supplementary Material
Acknowledgments
We thank members of the Hopfner lab for comments. We thank W. Baumeister for support and P. Forterre for P. abyssii genomic DNA. K.P.H. is supported by the Deutsche Forschungsgemeinschaft and the EMBO Young Investigator Program. J.P.C. is supported by National Cancer Institute Grant RO1 CA87851-02 and PO1 CA92584-02.
References
- Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D (2002) FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell 108: 195–205 [DOI] [PubMed] [Google Scholar]
- Ban C, Junop M, Yang W (1999) Transformation of MutL by ATP binding and hydrolysis: a switch in DNA mismatch repair. Cell 97: 85–97 [DOI] [PubMed] [Google Scholar]
- Carney JP, Maser RS, Olivares H, Davis EM, Le Beau M, Yates JR III, Hays L, Morgan WF, Petrini JH (1998) The hMre11/hRad50 protein complex and Nijmegen breakage syndrome: linkage of double-strand break repair to the cellular DNA damage response. Cell 93: 477–486 [DOI] [PubMed] [Google Scholar]
- Carpentieri F, De Felice M, De Falco M, Rossi M, Pisani FM (2002) Physical and functional interaction between the mini-chromosome maintenance-like DNA helicase and the single-stranded DNA binding protein from the crenarchaeon Sulfolobus solfataricus. J Biol Chem 277: 12118–12127 [DOI] [PubMed] [Google Scholar]
- Connelly JC, Leach DR (2002) Tethering on the brink: the evolutionarily conserved Mre11–Rad50 complex. Trends Biochem Sci 27: 410–418 [DOI] [PubMed] [Google Scholar]
- Connelly JC, Kirkham LA, Leach DR (1998) The SbcCD nuclease of Escherichia coli is a structural maintenance of chromosomes (SMC) family protein that cleaves hairpin DNA. Proc Natl Acad Sci USA 95: 7969–7974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantinesco F, Forterre P, Elie C (2002) NurA, a novel 5′-3′ nuclease gene linked to rad50 and mre11 homologs of thermophilic Archaea. EMBO Rep 3: 537–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP (2001) The yeast Xrs2 complex functions in S phase checkpoint regulation. Genes Dev 15: 2238–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amours D, Jackson SP (2002) The Mre11 complex: at the crossroads of DNA repair and checkpoint signalling. Nature Rev Mol Cell Biol 3: 317–327 [DOI] [PubMed] [Google Scholar]
- de Jager M, van Noort J, van Gent DC, Dekker C, Kanaar R, Wyman C (2001) Human Rad50/Mre11 is a flexible complex that can tether DNA ends. Mol Cell 8: 1129–1135 [DOI] [PubMed] [Google Scholar]
- Furuse M, Nagase Y, Tsubouchi H, Murakami-Murofushi K, Shibata T, Ohta K (1998) Distinct roles of two separable in vitro activities of yeast Mre11 in mitotic and meiotic recombination. EMBO J 17: 6412–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerard E, Jolivet E, Prieur D, Forterre P (2001) DNA protection mechanisms are not involved in the radioresistance of the hyperthermophilic archaea Pyrococcus abyssii and P. furiosus. Mol Genet Genomics 266: 72–78 [DOI] [PubMed] [Google Scholar]
- Goldberg M, Stucki M, Falck J, D'Amours D, Rahman D, Pappin D, Bartek J, Jackson SP (2003) MDC1 is required for the intra-S-phase DNA damage checkpoint. Nature 421: 952–956 [DOI] [PubMed] [Google Scholar]
- Haber JE (2000) Lucky breaks: analysis of recombination in Saccharomyces. Mutat Res 451: 53–69 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Shin D, Fairley C, Tainer JA, Carney JP (2000) Mre11 and Rad50 from Pyrococcus furiosus: cloning and biochemical characterization reveal an evolutionarily conserved multiprotein machine. J Bacteriol 182: 6036–6041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopfner KP, Karcher A, Craig L, Woo TT, Carney JP, Tainer JA (2001) Structural biochemistry and interaction architecture of the DNA double-strand break repair Mre11 nuclease and Rad50-ATPase. Cell 105: 473–485 [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Putnam CD, Tainer JA (2002) DNA double-strand break repair from head to tail. Curr Opin Struct Biol 12: 115–122 [DOI] [PubMed] [Google Scholar]
- Marrione PE, Cox MM (1996) Allosteric effects of RuvA protein, ATP, and DNA on RuvB protein-mediated ATP hydrolysis. Biochemistry 35: 11228–11238 [DOI] [PubMed] [Google Scholar]
- Maser RS, Mirzoeva OK, Wells J, Olivares H, Williams BR, Zinkel RA, Farnham PJ, Petrini JH (2001) Mre11 complex and DNA replication: linkage to E2F and sites of DNA synthesis. Mol Cell Biol 21: 6006–6016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M (2000) A mechanistic basis for Mre11-directed DNA joining at microhomologies. Proc Natl Acad Sci USA 97: 6409–6414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart GS, Wang B, Bignell CR, Taylor AM, Elledge SJ (2003) MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 421: 961–966 [DOI] [PubMed] [Google Scholar]
- Tauchi H, Kobayashi J, Morishima K, van Gent DC, Shiraishi T, Verkaik NS, vanHeems D, Ito E, Nakamura A, Sonoda E, Takata M, Takeda S, Matsuura S, Komatsu K (2002) Nbs1 is essential for DNA repair by homologous recombination in higher vertebrate cells. Nature 420: 93–98 [DOI] [PubMed] [Google Scholar]
- Trujillo KM, Sung P (2001) DNA structure-specific nuclease activities in the Saccharomyces cerevisiae Rad50*Mre11 complex. J Biol Chem 276: 35458–35464 [DOI] [PubMed] [Google Scholar]
- Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J (2000) BASC, a super complex of BRCA1-associated proteins involved in the recognition and repair of aberrant DNA structures. Genes Dev 14: 927–939 [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Liu CC, Chen PL, Lee WH (2001) RINT-1, a novel Rad50-interacting protein, participates in radiation-induced G(2)/M checkpoint control. J Biol Chem 276: 6105–6111 [DOI] [PubMed] [Google Scholar]
- Zhong Q, Chen CF, Li S, Chen Y, Wang CC, Xiao J, Chen PL, Sharp ZD, Lee WH (1999) Association of BRCA1 with the hRad50–hMre11–p95 complex and the DNA damage response. Science 285: 747–750 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Material


