Abstract
PAF is a five-subunit protein complex composed of Paf1, Cdc73, Leo1, Rtf1 and Ctr9, which was purified from yeast in association with RNA polymerase II and which is believed to function in transcription elongation. However, no direct proof exists for this yet. To assay whether PAF is required in elongation, we determined the in vitro transcription-elongation efficiencies of mutant cell extracts using a DNA template containing two G-less cassettes. paf1Δ or cdc73Δ cell extracts showed reduced transcription-elongation efficiencies (16–18% of the wild-type levels), whereas leo1Δ and rtf1Δ showed wild-type levels. In vivo transcription efficiency was diminished in the four mutants analysed, as determined by their abilities to transcribe lacZ. Our work provides molecular evidence that PAF has a positive role in transcription elongation and is composed of at least two functionally different types of subunits (Paf1-Cdc73 and Leo1-Rtf1).
Introduction
Eukaryotic RNA polymerase II (RNAPII)-mediated transcription is a highly coordinated process with three major steps: initiation, elongation and termination. During elongation, specific factors associate with RNAPII to help to overcome situations derived from transient pausing or to traverse through nucleosome structures (Hartzog, 2003; Shilatifard et al, 2003). Eukaryotic factors with a function in transcription elongation are TFIIS, TFIIF, FACT/Spt16-Pob3 and DSIF/Spt4-Spt5 found in yeast and humans as well as the yeast 19S subunit of the proteasome and human Elongin, 11–19 lysine-rich leukemia (ELL) and NELF (Shilatifard et al, 2003). However, there are a number of factors that associate with RNAPII in vivo, for which different experimental evidence suggests a function in transcription elongation, but no direct proof of that function exists yet. One such factor is the PAF complex from Saccharomyces cerevisiae (Krogan et al, 2002; Pokholok et al, 2002).
PAF is a five-subunit complex containing Paf1, Cdc73, Ctr9, Rtf1 and Leo1 (Krogan et al, 2002; Mueller & Jaehning, 2002; Squazzo et al, 2002). Although initially implicated in transcription initiation, there is evidence suggesting a functional role for PAF in elongation. Thus, PAF is physically and genetically connected to the Spt4-Spt5 and Spt16-Pob3 elongation factors (Krogan et al, 2002; Squazzo et al, 2002) and localizes to transcriptionally active open reading frames, as determined by chromatin immunoprecipitation (ChIP) (Krogan et al, 2002; Pokholok et al, 2002). Also, deletions of the PAF genes confer 6-azauracil (6-AU) sensitivity in certain genetic backgrounds, a hallmark phenotype for mutations in transcription-elongation factors (Costa & Arndt, 2000; Krogan et al, 2002; Squazzo et al, 2002). Recently, PAF has been connected to histone H3 methylation. In active genes, the Lys 4 and Lys 36 residues of histone H3 are methylated by the action of the Set1 and Set2 methyltransferases, respectively (Briggs et al, 2001; Santos-Rosa et al, 2002; Strahl et al, 2002). It has been shown that the interaction of Set1 with RNAPII, its recruitment to actively transcribed chromatin and the Set1-mediated methylation of histone H3 are dependent on PAF (Krogan et al, 2003a; Ng et al, 2003). Similarly, in vivo histone methylation by Set2 is PAF-dependent (Krogan et al, 2003b).
Paf1 and Cdc73 were initially pulled down together with Hpr1 and Ccr4 as protein associated with RNAPII (Chang et al, 1999). Hpr1 is a component of the THO complex that functions at the interface between transcription elongation, mRNA metabolism and mRNA export, and which is required for efficient transcription elongation and for preventing transcription-associated genetic instability (Chávez et al, 2000; Rondón et al, 2003b). The observation that paf1Δ and cdc73Δ mutants had hyper-recombination phenotypes between direct repeats similar to THO mutants, although to a lesser extent, and that paf1Δ hpr1Δ double mutants are lethal (Chang et al, 1999) suggests a priori a functional relationship between the PAF and THO complexes.
In this study, we show that PAF is required for efficient transcription elongation, as determined by in vitro assays of whole-cell extracts (WCEs). Interestingly, whereas paf1Δ and cdc73Δ mutations strongly reduced transcription-elongation efficiency in vitro, leo1Δ and rtf1Δ had no effect. All mutants are impaired in lacZ transcription in vivo, the effect being stronger in paf1Δ and cdc73Δ mutants. This in vivo transcription defect was similar to that of the THO mutants, but, in contrast, was not linked to hyper-recombination. Our work provides molecular evidence that PAF has a positive role in transcription elongation and is formed by two functionally different subunits (Paf1-Cdc73 and Rtf1-Leo1).
Results
Defective transcription in paf1 and cdc73 WCEs
To determine whether PAF has a positive role in transcription elongation, we used our newly reported in vitro assay for the analysis of transcription elongation (Rondón et al, 2003a). Briefly, the efficiency of transcription elongation of WCEs is assayed in a DNA template, carrying two G-less cassettes of different sizes (84 and 376 nt), which is transcribed in vitro from the hybrid GAL4–CYC1 promoter. The 84-nt G-less cassette is immediately downstream of the promoter and the 376-nt cassette is 1.48 kb away from it. Treatment of the in vitro-produced transcript with RNase T1 degrades all G-containing mRNA sequences, rendering the two G-less cassettes intact (Fig 1A). Transcription-elongation efficiency is measured as the ratio between the 367- and 84-nt G-less RNA fragments.
Figure 1.
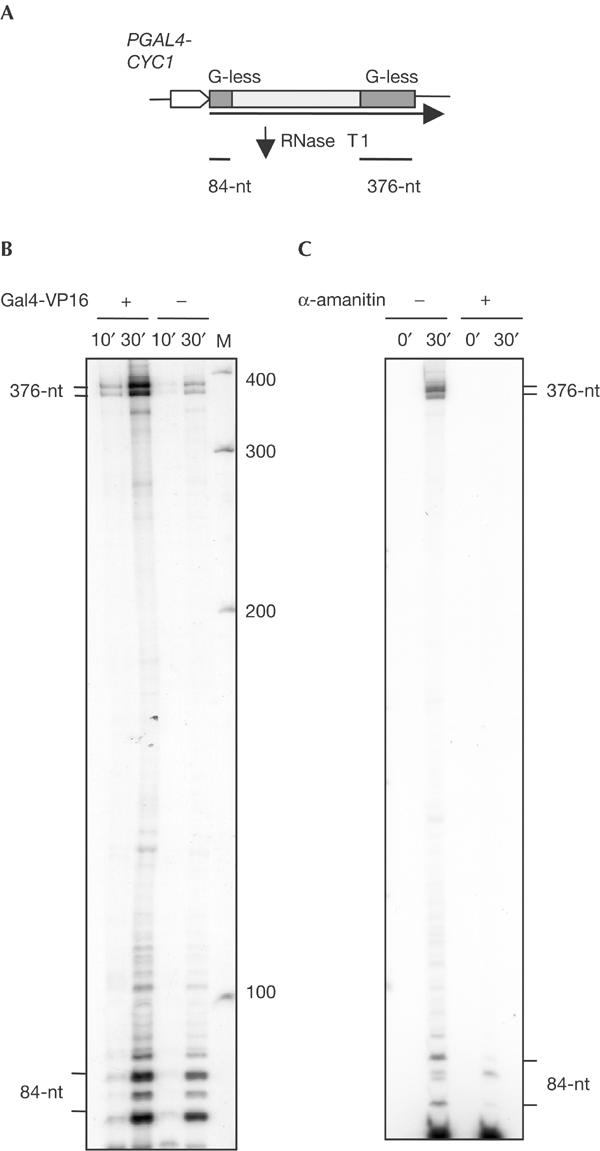
Characterization of the in vitro transcription assays in wild-type WCEs. (A) Scheme of the double G-less cassette system used for the analysis of in vitro transcription elongation. RNase T1 treatment of the mRNA renders two fragments corresponding to the G-less cassettes. (B) In vitro transcription assay of WCEs from BY4741 (WT) with or without addition of the Gal4-VP16 activator to the reaction. Each reaction was stopped after 10 or 30 min, treated with RNase T1 and run in 6% PAGE. Two bands from each G-less cassette were obtained probably due to incomplete action of RNase T1. (C) In vitro transcription assay of WCEs from BY4741 (WT) treated or not with α-amanitin. Each reaction was stopped after 10 or 30 min. The 376- and 84-nt-long fragments produced by RNAPII are α-amanitin sensitive, but fragments below 84 nt are α-amanitin insensitive; therefore, they are not the result of RNAPII transcription.
We first confirmed that the transcripts detected in these assays initiated at the GAL4–CYC1 fusion promoter, by showing that both the 367- and 84-nt G-less RNA fragments accumulated more efficiently when exogenous Gal4-VP16 activator was added to the reaction (Fig 1B). In addition, we confirmed that these transcripts were made by RNAPII, because they did not accumulate in the presence of α-amanitin concentrations at which RNAPII is specifically inhibited (Fig 1C).
We assayed the transcription-elongation capacity of WCEs of the PAF null mutants, paf1Δ, cdc73Δ, leo1Δ and rtf1Δ. As can be seen in Fig 2, after 30 min, paf1Δ and cdc73Δ cell extracts fully transcribed the 376-nt G-less cassette with 16 and 18% of the efficiency of the wild type, respectively. However, rtf1Δ and leo1Δ mutants showed wild-type transcription-elongation efficiencies. This suggests that PAF could be functionally divided into two types of subunits. For further confirmation, we determined the kinetics of transcription elongation in time-course experiments in the mutants of one subunit of each class, paf1Δ and rtf1Δ. As shown in Fig 3, the transcription-elongation efficiency of paf1Δ was dramatically decreased, reaching 7–11% of the wild-type efficiency depending on the reaction time. However, the kinetics of transcription elongation was the same in rtf1Δ and wild-type cells, with only a slight decrease after 40 min of reaction, in which rtf1Δ efficiency was 73% of the wild type. Therefore, we can conclude that PAF is required for efficient transcription elongation and is formed at least by two functionally different subunits, Paf1-Cdc73 and Leo1-Rtf1.
Figure 2.
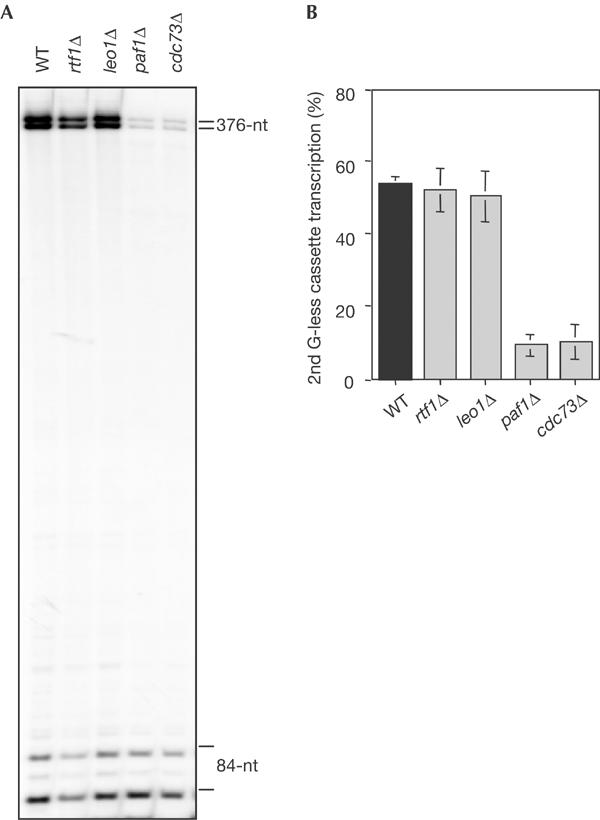
In vitro transcription assays of wild-type and PAF mutant WCEs. (A) In vitro transcription assay of WCEs from BY4741 (WT), YGL244w (rtf1Δ), YOR123c (leo1Δ), YBR279w (paf1Δ) and YLR418c (cdc73Δ) strains. Each reaction was stopped after 30 min, treated with RNase T1 and run in 6% PAGE. (B) Efficiency of transcription elongation determined as the percentage of transcripts that reached the 376-nt G-less cassette versus those covering the 84-nt cassette. Bands were quantified in a Fuji FLA3000 and normalized with respect to their C content. The mean value and standard deviations of three independent experiments are shown.
Figure 3.
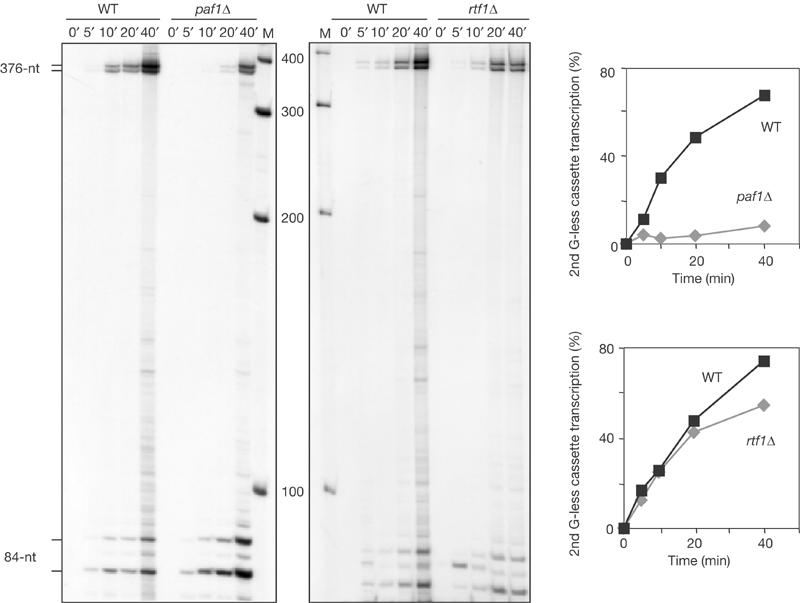
In vitro assays of time-course transcription-elongation experiments in wild-type and PAF mutant WCEs. In vitro transcription assay of 100 μg of WCEs from wild-type BY4741 and isogenic paf1Δ and rtf1Δ strains. Reactions were stopped at 0, 5, 10, 20 and 40 min, treated with RNase T1 and run in 6% PAGE. Strains and other details are as in Fig 2.
PAF mutants show in vivo transcription defects
Next, we assayed the effect of PAF mutations on gene expression in vivo. We analysed the ability of paf1Δ, cdc73Δ, rtf1Δ and leo1Δ to transcribe lacZ- and PHO5-coding regions, which differ in length and GC content, driven from the GAL1 promoter. In mutants showing elongation defects, such as spt4Δ and hpr1Δ, lacZ (long and GC-rich gene) is poorly transcribed whereas PHO5 (short and low-GC gene) is transcribed at wild-type levels (Chávez et al, 2000; Rondón et al, 2003). As can be seen in Fig 4A, in galactose-containing media, β-galactosidase activity in paf1Δ and cdc73Δ was 19% of wild-type levels but 44–51% in rtf1Δ and leo1Δ mutants, whereas acid phosphatase activity was only slightly affected. The latter can reflect a partial defect in their ability to activate the GAL1 promoter, as has been shown for paf1Δ (Shi et al, 1996). Northern analysis revealed that the accumulation of lacZ mRNA was strongly reduced in paf1Δ and cdc73Δ mutants and less strongly in leo1Δ and rtf1Δ mutants (Fig 4B), whereas PHO5 transcription was either slightly decreased or unaffected in the PAF mutants tested (Fig 4B), consistent with the enzymatic analyses. Therefore, even though efficient lacZ transcription requires the entire PAF complex, Paf1 and Cdc73 have a more prominent role in its transcription.
Figure 4.
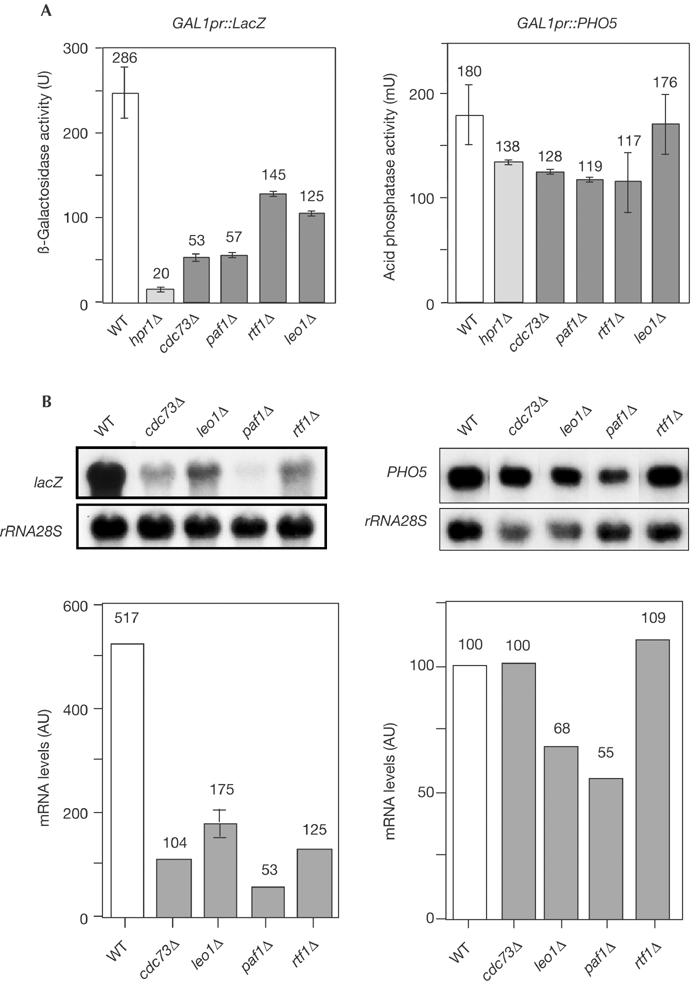
Effect of PAF mutations on gene expression. (A) ß-Galactosidase and acid phosphatase activities of wild type and isogenic paf1Δ, cdc73Δ, rtf1Δ and leo1Δ transformed with p316GAL1lacZ and pSCh202 as compared to hpr1Δ. The mean and standard deviations of two induced cultures of different transformants are shown. No detectable activity was observed in repressed cultures (2% glucose). (B) Northern analysis of lacZ and PHO5 mRNAs driven from the GAL1 promoter. Mid-log-phase cultures were induced with 2% galactose, and samples were taken after 2 h. The 3 kb BamHI–BglII lacZ fragment and an internal 589 bp 25S rRNA fragment obtained by PCR were used as 32P-labelled DNA probes. Northern blots were quantified in a Fuji FLA 3000. RNA levels are given in arbitrary units (AU) normalized with respect to the rRNA levels of each sample. Strains are as in Fig 2.
To confirm that the low mRNA levels detected by northern analysis in the PAF mutants were not due to a higher decay of lacZ mRNA, we analysed the kinetics of degradation of the lacZ mRNA. As can be seen in Fig 5, degradation curves were similar in wild type and PAF mutants.
Figure 5.
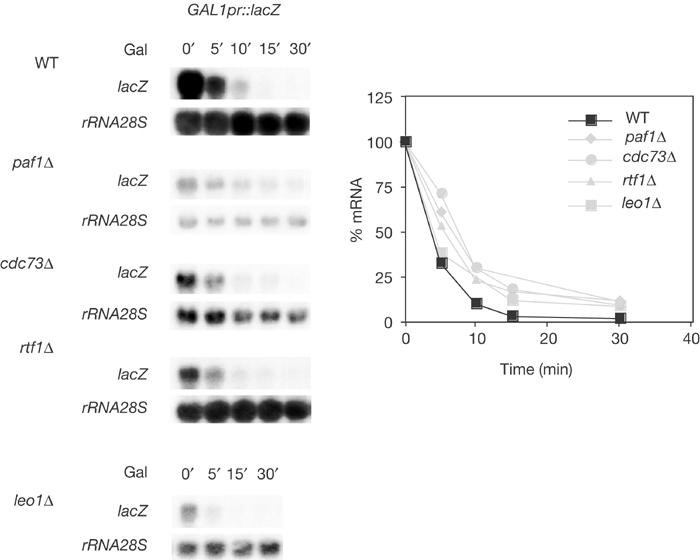
Kinetics of degradation of lacZ mRNAs in PAF mutants. (A) Northern analysis of lacZ mRNA decay in wild-type BY4741 and isogenic paf1Δ, cdc73Δ, rtf1Δ and leo1Δ cells harbouring p416GAL1lacZ. Mid-log-phase cultures were grown in 2% galactose synthetic medium for 8 h and transferred to 2% glucose synthetic medium before samples were taken. Given the low initial levels of lacZ mRNA in paf1 mutants, paf1 data were normalized with respect to the steady-state background levels. Strains used and other details are as in Fig 4.
Paf-transcription defects are not hyper-recombinogenic
Mutants of PAF have similar transcription-elongation defects as THO mutants, but with different intensities. paf1Δ and cdc73Δ mutants also increased recombination in the leu2-k::ADE2-URA3::leu2-k direct-repeats system, though to a lesser extent than THO mutants (Chang et al, 1999). Consequently, we wondered whether the transcription-elongation impairment of PAF mutants led to transcription-dependent hyper-recombination.
We reasoned that if the hyper-recombination phenotypes of paf1Δ and cdc73Δ were linked to their transcription defects, these mutants should be hyper-recombinant for direct-repeat systems such as LY and L-lacZ but not for systems such as L and L-PHO5 as was the case for THO mutants. LY and L-lacZ are based on two 0.6 kb leu2 repeats flanking pBR322 and lacZ, whereas L and L-PHO5 are based on the same repeats flanking either no sequence or PHO5. LY and L-lacZ are the only systems carrying intervening regions through which transcription is THO- or PAF-dependent. The recombination levels of paf1Δ and cdc73Δ mutants were the same in all systems analysed (Fig 6). The values were similar to wild type and different from the hpr1Δ mutant, in which recombination increased 475- and 45-fold in the LY and L-lacZ systems, respectively (Fig 4B; Chávez & Aguilera, 1997; Prado et al, 1997). Therefore, in contrast to THO mutants, the transcription defect and the previously reported hyper-recombination phenotypes of PAF mutants are not linked.
Figure 6.
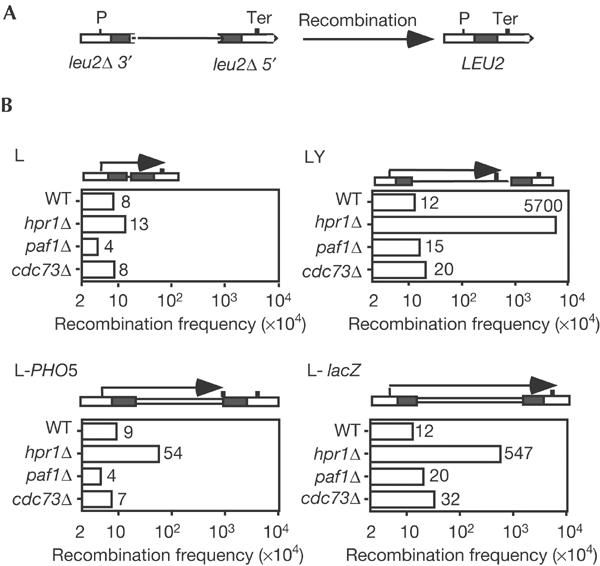
Analysis of recombination of paf1Δ and cdc73Δ. (A) Scheme of the 0.6 kb leu2-based direct-repeat recombination assays used and the deletions resulting from recombination between the repeats. (B) Recombination frequencies of strains WMCD-1B (WT), WMCD-5A (cdc73Δ) and WMP-3C (paf1Δ) transformed with plasmids pRS314-L (L), pRS314-LY (LY), pSCh204 (L-lacZ) and pSCh206 (L-PHO5). In the scheme of each repeat system, the transcripts driven from the external LEU2 promoter are indicated by arrows. The mean of 3–4 independent median frequencies, each obtained from six independent cultures, is given for each genotype.
Discussion
Our work provides molecular evidence that the PAF complex has a positive role in transcription elongation and is composed of at least two functionally different types of subunits.
Our in vitro assays showing that paf1Δ and cdc73Δ cell extracts have a reduced efficiency of transcription elongation (16–18% of the wild-type levels), whereas leo1Δ and rtf1Δ mutants are not affected, indicate that PAF is formed, in addition to the Ctr9 protein, by two types of subunits. The Paf1 and Cdc73 subunits would have a functional role in transcription elongation that can be detected in vitro, whereas Rtf1 and Leo1 would not. However, the observation that in addition to paf1Δ and cdc73Δ, the leo1Δ and rtf1Δ mutants were also impaired in in vivo lacZ transcription, although to a lesser extent, suggests that Leo1 and Rtf1 could also have a role in transcription.
It is important to note that our in vitro assays are performed with naked DNA templates. Therefore, we cannot discard the possibility that Leo1 and Rtf1 could be required for elongation through chromatin. Indeed, PAF is necessary for Set1 recruitment to the RNAPII in actively transcribed chromatin and for Lys 79 and Lys 4 methylation of histone H3 by Dot1 and Set1, respectively (Krogan et al, 2003a; Ng et al, 2003). Furthermore, Set2, a methyltransferase of histone H3 Lys 36, needs Rtf1 and Cdc73 for recruitment to actively transcribed genes and for histone methylation (Krogan et al, 2003b). In addition, co-immunoprecipitation assays show interaction between PAF and Chd1, a highly conserved chromatin remodelling factor with ATPase-dependent remodelling activity in vitro (Simic et al, 2003). This interaction could be mediated by the Spt16-Pob3 complex to which PAF and Chd1 bind (Krogan et al, 2002). Furthermore, the absence of PAF is lethal in combination with mutants affected in chromatin assembly (Formosa et al, 2002). It is possible that PAF has a role in transcription elongation through chromatin. However, our in vitro analysis, performed with naked DNA templates, indicates that PAF has a functional role in elongation that is independent of chromatin and is mediated by Paf1-Cdc73 subunits.
There also exists the possibility that Leo1 and Rtf1 had a functional role modulating the transcriptional elongation activity of the complex in response to cell signalling (stress, cell cycle, etc.), which would not be detected in vitro. The PAF complex could be a general factor involved in elongation under specific growth conditions or a specific factor involved in elongation of particular genes, for example, cell cycle- or stress-regulated genes. In agreement with this idea, paf1Δ decreases the expression of only a small subset of genes, 30% of which are cell cycle regulated (Porter et al, 2002), and paf1Δ and ctr9Δ stop cell cycle progression before budding (Koch et al, 1999).
The transcription defects of paf1Δ and cdc73Δ have also been observed in THO mutants. Copurification of Paf1 and Cdc73 with Hpr1 and the observation that paf1Δ and cdc73Δ mutants are also hyper-recombinant (Chang et al, 1999), although to a lesser extent than THO mutants, could suggest that THO and PAF complexes have similar functions. However, our study shows that this is not the case. Whereas the in vivo transcription defects of THO mutants are tightly linked to an increase in recombination, this is not the case for PAF mutants (Fig 6B), indicating that mutants of the THO and PAF complexes cause different types of transcription-elongation defects. Whereas THO is functionally related to mRNA export and THO mutations are synthetic lethal in a mex67-5 mRNA export mutant background (Jimeno et al, 2002), PAF is not related to mRNA export and paf1Δ mex67-5 mutants are viable (data not shown). This is consistent with the absence of detectable subunits of PAF in purified THO complex in silver-stained SDS–PAGE (Chávez et al, 2000; Strasser et al, 2002). Nevertheless, copurification of Paf1, Cdc73 and Hpr1 in association with RNAPII (Chang et al, 1999) is consistent with the idea that both complexes function in actively transcribed chromatin (Krogan et al, 2002; Pokholok et al, 2002; Strasser et al, 2002).
In summary, PAF has a positive role in transcription elongation and is composed, in addition to the Ctr9 subunit, of two functionally different types of subunits (Paf1-Cdc73 and Leo1-Rtf1). Further molecular analysis should contribute to the understanding of the specific role of PAF in transcription.
Materials and Methods
Yeast strains and plasmids. Yeast strains used were wild-type BY4741 and the isogenic mutants paf1Δ (YBR279w), cdc73Δ (YLR418c), rtf1Δ (YGL244w) and leo1Δ (YOR123c) (Euroscarf, Frankfurt). Strains WMCD-1B (WT), WMCD-5A (cdc73Δ) and WMP-3C (paf1Δ) were originated from genetic crosses between W303-1B and YLR418c or YBR279w. The plasmids used for expression analyses were p416GAL1lacZ (Mumberg et al, 1994) and pSCh202 (Chávez & Aguilera, 1997). For recombination assays we used pRS314-L, pRS314-LY, pSCh204 and pSCh206 (Chávez & Aguilera, 1997; Prado et al, 1997). Plasmid pRJRGAL4-VP16 (M. Ptashne, Sloan-Kettering Institute, New York) was used to purify the His6-tagged Gal4-VP16 recombinant protein from Escherichia coli. As a template for the in vitro transcription-elongation assays we used plasmid pGCYC1-402 (Rondón et al, 2003a).
Preparation of yeast WCEs and in vitro transcription assays. These were performed as described (Rondón et al, 2003). For the inhibition of RNAPII, 5 ng/μl α-amanitin was added to the transcription reaction and preincubated at 30°C for 15 min.
In vivo analysis of gene expression, mRNA decay and genetic recombination. ß-Galactosidase and acid phosphatase activities, mRNA levels and recombination frequencies were determined as described (Chávez et al, 2000; Rondón et al, 2003b).
Acknowledgments
We are grateful to J.Q. Svejstrup and M. Ptashne for plasmid gifts, R. Luna and S. Jimeno for a critical reading of the manuscript, and D. Haun for style supervision. This work was supported by grants from the Spanish Ministry of Science and Technology (BMC2000-0439) and the Human Frontier Science Program (RG1999/0075). A.G.R. was the recipient of a predoctoral training grant from the Spanish Ministry of Education.
References
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD (2001) Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 15: 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang M, French-Cornay D, Fan HY, Klein H, Denis CL, Jaehning JA (1999) A complex containing RNA polymerase II, Paf1p, Cdc73p, Hpr1p, and Ccr4p plays a role in protein kinase C signaling. Mol Cell Biol 19: 1056–1067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez S, Aguilera A (1997) The yeast HPR1 gene has a functional role in transcriptional elongation that uncovers a novel source of genome instability. Genes Dev 11: 3459–3470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chávez S, Beilharz T, Rondón AG, Erdjument-Bromage H, Tempst P, Svejstrup JQ, Lithgow T, Aguilera A (2000) A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J 19: 5824–5834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa PJ, Arndt KM (2000) Synthetic lethal interactions suggest a role for the Saccharomyces cerevisiae Rtf1 protein in transcription elongation. Genetics 156: 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Formosa T, Ruone S, Adams MD, Olsen AE, Eriksson P, Yu Y, Rhoades AR, Kaufman PD, Stillman DJ (2002) Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway. Polymerase passage may degrade chromatin structure. Genetics 162: 1557–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA (2003) Transcription elongation by RNA polymerase II. Curr Opin Genet Dev 13: 119–126 [DOI] [PubMed] [Google Scholar]
- Jimeno S, Rondón AG, Luna R, Aguilera A (2002) The yeast THO complex and mRNA export factors link RNA metabolism with transcription and genome instability. EMBO J 21: 3526–3535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koch C, Wollmann P, Dahl M, Lottspeich F (1999) A role for Ctr9p and Paf1p in the regulation of G1 cyclin expression in yeast. Nucleic Acids Res 27: 2126–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Kim M, Ahn SH, Zhong G, Kobor MS, Cagney G, Emili A, Shilatifard A, Buratowski S, Greenblatt JF (2002) RNA polymerase II elongation factors of Saccharomyces cerevisiae: a targeted proteomics approach. Mol Cell Biol 22: 6979–6992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ et al. (2003a) The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Krogan NJ et al. (2003b) Methylation of histone H3 by Set2 in Saccharomyces cerevisiae is linked to transcriptional elongation by RNA polymerase II. Mol Cell Biol 23: 4207–4218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CL, Jaehning JA (2002) Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol Cell Biol 22: 1971–1980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mumberg D, Muller R, Funk M (1994) Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res 22: 5767–5768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA (2002) Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell 9: 799–809 [DOI] [PubMed] [Google Scholar]
- Porter SE, Washburn TM, Chang M, Jaehning JA (2002) The yeast pafl-rNA polymerase II complex is required for full expression of a subset of cell cycle-regulated genes. Eukaryot Cell 1: 830–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Piruat JI, Aguilera A (1997) Recombination between DNA repeats in yeast hpr1delta cells is linked to transcription elongation. EMBO J 16: 2826–2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondón AG, García-Rubio M, González-Barrera S, Aguilera A (2003) Molecular evidence for a positive role of Spt4 in transcription elongation. EMBO J 22: 612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rondón AG, Jimeno S, García-Rubio M, Aguilera A (2003) Molecular evidence that the eukaryotic THO/TREX complex is required for efficient transcription elongation. J Biol Chem 278: 39037–39043 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T (2002) Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Shi X, Finkelstein A, Wolf AJ, Wade PA, Burton ZF, Jaehning JA (1996) Paf1p, an RNA polymerase II-associated factor in Saccharomyces cerevisiae, may have both positive and negative roles in transcription. Mol Cell Biol 16: 669–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A, Conaway RC, Conaway JW (2003) The RNA polymerase II elongation complex. Annu Rev Biochem 72: 693–715 [DOI] [PubMed] [Google Scholar]
- Simic R, Lindstrom DL, Tran HG, Roinick KL, Costa PJ, Johnson AD, Hartzog GA, Arndt KM (2003) Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J 22: 1846–1856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo SL, Costa PJ, Lindstrom DL, Kumer KE, Simic R, Jennings JL, Link AJ, Arndt KM, Hartzog GA (2002) The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J 21: 1764–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl BD et al. (2002) Set2 is a nucleosomal histone H3-selective methyltransferase that mediates transcriptional repression. Mol Cell Biol 22: 1298–1306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K et al. (2002) TREX is a conserved complex coupling transcription with messenger RNA export. Nature 417: 304–308 [DOI] [PubMed] [Google Scholar]


