Abstract
Our understanding of the genetics of species of the best-studied hyperthermophilic archaea, Pyrococcus spp., is presently limited by the lack of suitable genetic tools, such as a stable cloning vector and the ability to select individual transformants on plates. Here we describe the development of a reliable host-vector system for the hyperthermophilic archaeon Pyrococcus abyssi. Shuttle vectors were constructed based on the endogenous plasmid pGT5 from P. abyssi strain GE5 and the bacterial vector pLitmus38. As no antibiotic resistance marker is currently available for Pyrococcus spp., we generated a selectable auxotrophic marker. Uracil auxotrophs resistant to 5-fluoorotic acid were isolated from P. abyssi strain GE9 (devoid of pGT5). Genetic analysis of these mutants revealed mutations in the pyrE and/or pyrF genes, encoding key enzymes of the pyrimidine biosynthetic pathway. Two pyrE mutants exhibiting low reversion rates were retained for complementation experiments. For that purpose, the pyrE gene, encoding orotate phosphoribosyltransferase (OPRTase) of the thermoacidophilic crenarchaeote Sulfolobus acidocaldarius, was introduced into the pGT5-based vector, giving rise to pYS2. With a polyethylene glycol-spheroplast method, we could reproducibly transform P. abyssi GE9 pyrE mutants to prototrophy, though with low frequency (102 to 103 transformants per μg of pYS2 plasmid DNA). Transformants did grow as well as the wild type on minimal medium without uracil and showed comparable OPRTase activity. Vector pYS2 proved to be very stable and was maintained at high copy number under selective conditions in both Escherichia coli and P. abyssi.
Pyrococcus species are anaerobic hyperthermophilic archaea that grow optimally at 100°C, with a maximum at around 105°C. They belong to the Thermococcales order (genera Pyrococcus and Thermococcus), the most important order (with 25 species currently described) of the second archaeal phylum, the Euryarchaeota (52). All of them are heterotrophic sulfur reducers, fermenting mostly proteinaceous substrates (some species are able to use oligo- and polysaccharides) in the presence or absence of elemental sulfur (S0) as the terminal electron acceptor (leading to H2S production). Several laboratories have chosen Pyrococcus spp. among other anaerobic hyperthermophiles as model organisms because these versatile organisms are relatively easy to handle and can be grown to high cell density for biochemical analyses. These microorganisms are of major interest in basic research as a model for investgating diverse aspects of life under extreme conditions and because of their evolutionary significance; they also have a great biotechnological potential because of the large variety of thermostable enzymes that they possess.
The genomes of three of them, Pyrococcus horikoshi (22) (http://www.bio.nite.go.jp/), Pyrococcus abyssi (http://www.genoscope.cns.fr/Pab/), and Pyrococcus furiosus (http://www.genome.utah.edu/) have been completely sequenced, and the data are publicly available. Although this unique situation for archaeal representatives allowed meaningful analyses of the speciation mechanisms at the genome level (27) and led to the identification of the first archaeal replication origin region, oriC (33), bioinformatic studies are limited, as typically about half of the open reading frames in these genomes remain without known or predicted function. Genetic techniques, representing some of the most powerful investigative tools in biology, will certainly play an important role in elucidating the function of these genes and in unraveling their mechanisms of modulated expression. Thus, it is now a priority to develop suitable gene transfer systems for this particular class of microorganisms.
For Archaea, such genetic tools were first developed in the extreme halophiles (essentially, Halobacterium and Haloferax spp.) (9, 10, 12) and later for mesophilic methanogens of the genera Methanococcus and Methanosarcina (43, 49). These genetic systems benefit now from efficient transformation methods and diverse genetic tools (including shuttle vectors and integrative expression vectors, composite transposons, and reporter genes) and have led to investigations on the expression of genes involved in the synthesis of bacteriorhodopsin (29) and gas vesicles (19, 39) in halophiles and in hydrogen metabolism, nitrogen fixation, and flagellin assembly in methanogens (for a review, see references 26 and 45).
In contrast, genetic methods for the hyperthermophilic archaea are still at early stages of development, mainly because of the unique technological barriers imposed by the thermophilic and, in some case, the strictly anaerobic nature of these organisms (for a review, see reference 38). Indeed, most progress has been obtained so far within the genus Sulfolobus, whose members are sulfur-metabolizing aerobes belonging to the Crenarchaeota phylum. Many of them were shown to possess extrachromosomal elements, conjugative plasmids, and viruses (51), such as the virus SSV1, which served as the basis for the construction of the first generation of shuttle vectors (6, 40). In addition, a variety of analogue-resistant and auxotrophic mutants (18, 21, 25) and a system of exchange and recombination of chromosomal markers by conjugation have been described (17).
Our model organism, P. abyssi, was isolated from a deep-sea hydrothermal vent at 2,000 m depth in the North Fiji Basin (southwest Pacific) (16). It is typical of other pyrococci: it grows fast (doubling time of 35 min) under optimal conditions (96 to 100°C, pH 6.8, 3% NaCl), and growth is stimulated by the presence of elemental sulfur. An important characteristic of P. abyssi is its ability to grow well on a completely defined medium consisting of a mixture of amino acids and vitamins (47). This property allowed us to isolate the first auxotrophic mutants by plating on medium solidified with the thermostable gellum gum called Gelrite (15, 46, 48).
Another important feature is the presence of a small cryptic plasmid, pGT5 (3.5 kb), in the type strain P. abyssi GE5 (13). It is the first described and so far the only plasmid of Thermococcales to be studied at the molecular level (14). In particular, it was shown that this plasmid replicates via a rolling-circle mechanism and that this process is specifically initiated by a Rep protein encoded by the largest open reading frame (ORF) of pGT5 (29, 30). This plasmid was fused with pUC19 to create a potential shuttle vector between Escherichia coli and both P. furiosus and Solfolobus acidocaldarius. The chimerical plasmid could be introduced in both organisms by CaCl2 treatment followed by heat shock (1). Later, this construct was completed by introduction of a potential selectable marker (as discussed below). The resulting shuttle vector was shown to be maintained, though in low copy number, in liquid cultures of these archaea for several generations after transformation (3).
A genetic marker to score for successful transformation events is mandatory for the set-up of an efficient cloning vector. Genes conferring resistance to growth-inhibitory compounds (generally an antibiotic) are the most convenient selectable markers and have been widely used in mesophilic bacteria but are scarce in mesophilic archaea. The choice is even more restricted for hyperthermophilic archaea, mainly because both the candidate antibiotic and the corresponding resistance-conferring gene product have to be thermostable. At the time we started this work, only two thermostable genetic markers were available: a thermostable derivative of an E. coli hygromycin phosphotransferase that had allowed selection of Sulfolobus solfataricus transformants on plates (6, 7), and an alcohol dehydrogenase (adh gene) from S. solfataricus that conferred resistance to butanol in liquid cultures of transformants in S. acidocaldarius and Pyrococcus furiosus (1, 3).
Early tests that we performed with Pyrococcus strains showed that P. abyssi strains are naturally resistant to high level of hygromycin (MIC > 300 μg/ml), which precluded the use of this antibiotic for selection of transformants. Moreover, transformation of P. abyssi with the vector pAG21 (and similar constructs) carrying the S. solfataricus adh marker (3) did not permit the selection of transformants on plates (S. Lucas and G. Erauso, unpublished data).
As an alternative, we searched other genetic markers such as those resulting in auxotrophy. The orotate phosphoribosyltransferase (OPRTase) gene (pyrE) and the orotidine 5′-monophosphate decarboxylase gene (pyrF), involved in pyrimidine biosynthesis, have unique characteristics. Strains deficient in one of these genes become resistant to 5-fluoroorotic acid (5-FOA), which inhibits growth of the wild-type strains. Thus, 5-FOA has been used successfully for positive isolation of uracil auxotrophs in many microorganisms, and their pyrE and pyrF genes have been used as genetic markers (19, 35, 50). We have chosen this approach to develop a selection system for our model organism, P. abyssi.
In this article, we report the isolation and characterization of uracil auxotrophic mutants of P. abyssi strain GE9. We describe the construction of a shuttle vector, pYS2, based on the pyrococcal plasmid pGT5 and harboring the pyrE gene of Sulfolobus acidocaldarius as a selective marker. We show that we could reproducibly transform P. abyssi cells with a polyethylene glycol (PEG)-mediated spheroplast method and that the vector pYS2 complemented pyrE auxotrophs, leading to transformants that could be selected on plates. Furthermore, we show that pYS2 replicates efficiently and is very stable in both E. coli and P. abyssi, demonstrating its utility as a shuttle vector.
MATERIALS AND METHODS
Bacterial and archaeal strains, media, and growth conditions.
E. coli strains TOP10 (Invitrogen) and SURE Kmr (Stratagene) were used for vector construction and propagation and for cloning of the pyrE gene from P. abyssi, respectively. E. coli strains were grown at 37°C in a medium containing 5 g of yeast extract, 5 g of tryptone, and 10 g of NaCl per liter (37). Ampicillin was used at 100 μg/ml and kanamycin at 50 μg/ml in both liquid and solid media.
P. abyssi strains were from our laboratory culture collection. P. abyssi type strain GE5 (16) was used as the source of plasmid pGT5, and P. abyssi strain GE9 (31) is a prototrophic strain used as the wild type, from which all the auxotrophs described in this study were derived. They were cultivated at 90°C (unless otherwise specified) with strict anaerobic procedures as previously described (16). Two types of media were used for the propagation of P. abyssi strains. Both had the same mineral base, which had the following composition (per liter of deionized water): 30 g of sea salts (Sigma), 3.3 g of PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)], 2 ml of a 5% (wt/vol) K2HPO4 solution, 1 ml of a 2% (wt/vol) CaCl2 · 2H2O solution, 1 ml of a 25 mM FeCl3 solution, and 1 ml of a 10 mM Na2WO4 solution. Resazurin (0.1 mg/liter) was added as redox indicator, and the pH was set to 6.8.
A defined medium, TAA, contained in addition 0.1 g each of the 20 natural amino acids as the sole carbon and energy source and 10 ml of a vitamin solution (4). It was sterilized by filtration to prevent degradation of thermolabile amino acids. This medium was used for general propagation, plating, and preparation of competent cells. A rich medium, YP, consisted of the mineral base supplemented with 0.1% (wt/vol) yeast extract and 0.4% (wt/vol) tryptone. The latter medium was sterilized by autoclaving; it was used for the regeneration of spheroplasts (see Transformation procedures, below). The media were dispensed into sterile Hungate tubes or glass bottles containing elemental sulfur at 1% (wt/vol) and closed with butyl rubber stoppers. After the gas phase had been replaced by N2 (100 kPa), anaerobiosis was achieved by injection of 1/100th volume of a 3% (wt/vol) Na2S · 9H2O solution.
Plating of P. abyssi and isolation of uracil auxotrophs.
Plating of P. abyssi was done essentially as described (15) with some modifications (W. Zillig, personal communication). Briefly, solid media were prepared by including 0.8% (wt/vol) Phytagel (Gellan gum; Sigma) and poured, 30 ml per 9-cm-diameter plastic petri dish. The polysulfide solution which previously served as the sulfur source was replaced with a saturating suspension (10 g per 100 ml of deionized water) of colloidal sulfur (Riedel-de Haën; distributed by Sigma-Aldrich); 100 μl of this suspension and 100 μl of inoculum were added to 800 μl of overlay medium containing 0.25% (wt/vol) Phytagel in a glass tube kept at 90°C in a heating block. The mixture was quickly homogenized with a Vortex mixer and immediately poured and evenly spread on a plate containing the supporting gel. Culture dilutions were prepared in anaerobic TAA medium. When needed, uracil was added both to the supporting gel and in the soft overlay at a concentration of 50 μg/ml from a filter-sterilized anaerobic stock solution.
For the isolation of uracil auxotrophs, 6-cm-diameter petri dishes were employed, and plates consisted of two layers of supporting gel: a first layer consisting of 10 ml of TAA medium at 0.8% (wt/vol) Phytagel and a second layer of 4 ml of TAA medium at 0.8% (wt/vol) Phytagel containing in addition uracil (50 μg/ml) and 5-FOA (800 to 1,200 μg/ml) (Sigma). A preweighed amount of 5-FOA was dissolved directly into the medium while hot to reached the desired concentration. The soft overlay was made by mixing 75 μl of the colloidal sulfur suspension and 75 μl of inoculum with 600 μl of hot (90°C) TAA medium at 0.25% (wt/vol) Phytagel containing uracil (50 μg/ml) and 5-FOA at the same concentration (800 to 1,200 μg/ml) as that of the second layer of supporting gel. All plating manipulations were performed in an anaerobic chamber (Coy Laboratory Products, Grass Lake, Mich.) under a 93% N2-7% H2 atmosphere. After inoculation, plates were incubated in home-made stainless steel anaerobic jars at 85°C under a 99% N2-1% H2S (100-kPa) atmosphere. Colonies were typically visible after 48 to 72 h. Uracil auxotrophs resistant to 5-FOA were further purified by three rounds of streaking on selective plates. Reversion frequencies were determined by plating serial dilutions onto TAA medium devoid of uracil.
Growth measurements.
Growth in Hungate tube was monitored by measurement of the optical density at 600 nm with a Spectronic 20 spectrophotometer equipped to hold 18-mm-diameter tubes, with a calibration curve. Direct cell counts were performed with a Thoma counting chamber (0.01-mm depth; Weber, London, England) viewed under a phase-contrast microscope. Viable-cell counts were made in triplicate by plating serial dilutions as described above. Plating efficiency was determined by dividing the number of CFU by the direct cell count number and multiplying the result by 100.
Transformation procedures.
P. abyssi uracil auxotrophs PA-13 and PA-20 were used as recipient strains for all transformation experiments with the pYS2 vector. Attempts were made to transform P. abyssi strains by a modified CaCl2 procedure previously reported to be successful for transformation of both P. furiosus and S. acidocaldarius (3) but did not produce any transformants on plates. Electroporation of P. abyssi in high-resistance buffer was also tried, essentially as described for Sulfolobus spp. (39) except that both 0.05 M sucrose and 10% glycerol were tested as the electroporation buffer. The whole procedure was also tested under anaerobic conditions following the precautions described for electroporation of Methanococcus species (34). Several combinations were tested by varying the electrical parameters and the composition of the electroporation buffer under aerobic and anaerobic conditions. However, again none of these efforts were successful (Table 1). One possible explanation for the failure of both methods is that the transformation efficiency was below the detection threshold of our plating procedure (Table 1).
TABLE 1.
Transformation methods tested on P. abyssi mutants PA9-13 and PA9-20 with pYS2
| Transformation method | Survivala (%) | Transformation frequencyb | Transformation efficiencyc |
|---|---|---|---|
| CaCl2 | NDd | <1.0 × 10−8e | <1.0e |
| Electroporationf | 9 ± 2 | <1.0 × 10−8e | <1.0e |
| PEG/whole cellsg | 32 ± 6 | 2.3 × 10−8 ± 1.4 × 10−8 | 4 ± 3 |
| PEG/spheroplastsg | 10 ± 4 | 1.5 × 10−6 ± 0.8 × 10−6 | 280 ± 168 |
Ratio of CFU obtained for cells subjected to the transformation treatment over the total CFU determined by plating on TAA medium supplemented with uracil; standard deviations are given.
Calculated as the ratio of the number of transformants over the CFU of cells that had gone through a mock spheroplasting/transformation procedure counted on TAA medium supplemented with uracil.
Number of transformants extrapolated for 1 μg of DNA.
ND, not determined.
No transformants were recovered; the detection limit is given.
Average of two independent experiments under the best conditions: in anaerobiosis, 200Ω, 25 μF, 1.5 kV.
Numbers given are the averages of at least three independent transformation experiments.
The first successful transformations were obtained with a whole-cell PEG-mediated transformation method similar to that described for the methanogenic archaeon Methanococcus maripaludis (43, 44). However, this method suffered from a very low transformation frequency and poor reproducibility (Table 1). Attempts to increase the transformation frequency by modifying the original protocol were unsuccessful. Therefore, transformation of spheroplasts rather than whole cells was investigated as a mean to improve the PEG-mediated transformation. We used a PEG-mediated transformation of spheroplast method similar to the one described for extremely halophilic Archaea (11). PEGs of different molecular weights (600, 1,000, 6,000, and 8,000) tested in this study were from Fluka, of the highest purity available (Molecular Biology grade), and used without further purification.
The mutants were grown at 90°C, shaken at 180 rpm, in 100-ml serum bottles containing 50 ml of TAA medium supplemented with uracil. When the cultures reached an OD600 between 0.12 and 0.15 (5 × 108 to 109 cells/ml by direct cell counting), which corresponds to late log phase to early stationary phase, they were removed from the incubator and allowed to cool for about 30 min. Residual sulfur present in the cultures was eliminated by simple decantation. Cells were harvested by centrifugation (5,000 × g for 15 min at 4°C) and from this step on were always kept on ice. Cells were gently washed in 0.2 volume of ice-cold SM 4× solution (SM 4× solution contained 80 g of NaCl, 0.8 g of KH2PO4, 0.2 g of NaBr, 0.04 g of SrCl2 · 6H2O, 0.08 g of H2BO3, 1 g of KCl, and 2 g of sodium citrate per liter) recovered by centrifugation (5,000 × g for 10 min at 4°C) and finally resuspended in 0.01 volume of spheroplasting buffer (1 M NaCl, 27 mM KCl, 50 mM Tris-HCl [pH 7.5]. and 20% sucrose) to reach a final concentration of ≈5 × 1010 cells/ml.
Spheroplasts were obtained by adding EDTA to a final concentration of 10 mM. Conversion of cells to spheroplasts, verified by direct observation with a phase-contrast microscope, was usually observed after 5 to 10 min. Spheroplasts typically prepared from a 50-ml culture grown to late log phase provided a sufficient number of viable cells for about five assays. As previously reported in the case of halophiles (11), we found that younger cells were somewhat more resistant to spheroplast formation and that stationary-phase cells became increasingly susceptible to lysis, both effects resulting in poor transformation. We found that EDTA at a final concentration of 10 mM allowed the conversion of more than 90% of the cells into spheroplasts in 5 to 10 min. Higher concentrations increased the proportion of cell lysis. Spheroplasts seemed to be relatively stable for at least 30 min following addition of EDTA.
Transforming DNA (0.5 μg) was added per 100-μl aliquot of spheroplast suspension. After 5 min of incubation at 4°C, an equal volume of a solution containing 50% PEG-600 (Fluka) and 50% spheroplasting buffer was added and gently but thoroughly mixed with the spheroplasts, and the preparations were kept on ice for 30 min. PEG was eliminated by dilution of the suspension with 100 μl of spheroplast dilution solution (SM 4× solution plus 15% [wt/vol] sucrose) and centrifugation (5,000 × g for 5 min at 4°C). Spheroplasts were finally resuspended in 100 μl of dilution solution, transferred to 5 ml of reduced YP medium in Hungate tubes, and incubated at 85°C for 1 h to allow regeneration of the cell wall.
Selection of transformants was achieved by plating aliquots onto TAA without uracil with the overlay technique described above. To determine the influence of the transformation/spheroplasting procedure on the viability of P. abyssi, appropriate dilutions of cell samples that had gone through mock transformation (without DNA) were also plated on TAA supplemented with uracil. Only ≈10% of the total CFU (counted on the nontransformed control) survived the spheroplasting treatment and regenerated to form colonies on plates. Colonies of putative transformants were subcultured in liquid TAA medium. Chemically competent E. coli cells were prepared according to Inoue (20a) and transformed by with the 5 min transformation protocol described by Pope and Kent (36).
Preparation of plasmidic and total DNA.
Plasmids were isolated from E. coli by with the alkaline lysis method (37). Large scale preparations of plasmid pYS2, to be used for transformation, were further purified by equilibrium centrifugation in a CsCl ethidium bromide gradient according to established protocols (37) or with Qiagen Tip 100 columns (Qiagen) following the manufacturer's recommendations.
Small-scale preparations of plasmid DNA from P. abyssi strains were typically made from 50-ml cultures, with either the guanidine isothiocyanate-phenol extraction method described previously (28) or the alkaline lysis protocol (37). Total DNA from P. abyssi was prepared as previously described (8). About 100 ng of such preparations was used as the template in PCR amplifications, and about 3 μg was used for restriction enzyme analyses and subsequent Southern hybridizations.
Cloning and sequence analysis of pyrE and pyrF genes of P. abyssi mutants.
The pyrE and pyrF genes were amplified by PCR from genomic DNA of the wild-type strain GE9, from the auxotrophic mutants, and from three independent transformants. Two sets of primers (oligonucleotides were from Genset S.A., Evry, France) were used. They were designed on the basis of the genome sequence of P. abyssi strain GE5 (Génoscope, Evry, France; http://www.genoscope.cns.fr/Pab/): the primers PAB-U1 (5′-CTCCCTCCAATCAACTTA-3′) and PAB-D1 (5′-AGTCGTAATGCTTCTTGA-3′) were used for pyrE amplification, and the primers PAB-U2 (5′-TCTAGCTTTGGACGTTTA-3′) and PAB-D2 (5′-AACATTTGGGAGAGCTGT-3′) were used for amplification of the pyrF gene. PCRs were performed with the Expand High Fidelity PCR kit (Roche Biochemicals), and the resulting 0.83-kb and 0.65-kb PCR products for pyrE and pyrF, respectively, were cloned into the pCR2.1-TOPO plasmid (TOPO TA cloning kit; Invitrogen). DNA sequences were determined from double-stranded templates by automated dye terminator sequencing with universal primers flanking the vector's cloning site.
Routine sequence analyses were done with the Lasergene software package (DNASTAR, Madison, Wis.). Homology searches were conducted on line with the Blast 2.0 program (2), and alignments were performed with the Clustal X program (42).
Construction of shuttle vectors.
Plasmid pGT5 (3,444 bp) was linearized with NspBII, and the resulting blunt-ended molecule was cloned into the unique StuI site of E. coli plasmid vector pLitmus38 (New England Biolabs). The intact pyrE gene of S. acidocaldarius was previously amplified from genomic DNA as a 1.5-kb SacI fragment by inverse PCR, and its nucleotide sequence and start point of transcription were determined (41). A 1.1-kb BamHI fragment consisting of the 0.6-kb pyrE coding region and about 0.25 kb of the upstream and ≈0.25 kb of the downstream sequences, was subcloned into the BamHI cloning site of the pLitmus38/pGT5 plasmid, resulting in pYS1 (7.2 kb).
The shuttle vector pYS2 was obtained by deleting a ≈1-kb NsiI-PvuII fragment of pYS1. The cohesive end generated by NsiI was converted to a blunt end by removal of the overhanging nucleotides with T4 DNA polymerase (New England Biolabs) in the presence of deoxynucleoside triphosphates. Circularization was done with T4 DNA ligase (New England Biolabs). The resulting construct, pYS2 (6.4 kb), was analyzed by restriction fragment analysis after transformation and propagation in E. coli strain SURE.
DNA analyses of P. abyssi transformants.
Total and plasmid DNAs, intact or digested with the appropriate restriction enzymes (New England Biolabs), were separated by electrophoresis on 0.8% agarose gels containing 0.5 μg of ethidium bromide per ml and visualized under UV light with a charge-coupled device (Bio-Rad). DNA in the gel was transferred to a nylon membrane (Hybond N+; Amersham) and hybridized according to standard procedures (37). A pYS2-specific probe was prepared by recovery and purification (with the QiaexII kit; Qiagen) of the restriction fragments resulting from the double digestion of pYS2 with HindIII and BamHI, separated on an agarose gel. All purified fragments except the 1.1-kb BamHI fragment containing the pyrE gene of S. acidocaldarius were pooled and randomly prime labeled with fluorescein-11-dUTP with the ECF labeling and detection kit (Amersham). Hybridization signals were recorded on a Storm 860 scanner and analyzed with the ImageQuant Analysis software (Molecular Dynamics).
Colony hybridization analysis of P. abyssi transformants was performed by standard procedures (37) and the probe described above.
Measurement of OPRTase activity.
Cell extracts were prepared from late-exponential-phase cultures of P. abyssi strain GE9 (wild type, mutants, and transformants). After elimination of sulfur residues by simple decantation, cells were collected by centrifugation at 4°C and 8,500 rpm in a GSA Sorvall rotor for 20 min, yielding about 2 g of cell paste per liter of culture. Cells were washed by resuspension in 20 ml of TNE and concentrated by centrifugation at 4°C and 7,000 × g for 15 min. The pellets were stored at −20°C until processed. Frozen cells were thawed, resuspended in Tris-HCl buffer (0.05 M, pH 8.8), and broken by sonication for 10 min.
The OPRTase assay mix contained 10 μl of 0.05 M Tris-HCl (pH 8.8), 10 μl of 20 mM MgCl2, 10 μl of 5 mM sodium orotate, 20 μl of 6 mM phosphoribosylpyrophosphate, and 20 μl of cell extract in a total volume of 100 μl. The reaction was carried out at 70°C, and the products of the assay were analyzed by high-pressure liquid chromatography (HPLC) on Zorbax HPLC columns (Mac-MOD Analytical; detection at 254 nm) with Millennium software (Waters). Protein concentrations were determined according to the method of Bradford (5).
Assessment of plasmid stability and relative copy number.
To determine the segregation stability of the vector pYS2 in P. abyssi, a late-log-phase culture of plasmid-carrying P. abyssi mutant PA9-13 grown in TAA medium was diluted 100-fold into 50 ml of TAA medium supplemented or not with uracil (50 μg/ml) and incubated at 90°C till the stationary phase was reached (t0 + 15 h). Aliquots of these cultures were then diluted 100-fold in TAA and TAA plus uracil, respectively. The rest of the cultures were harvested, and plasmid DNA was extracted, digested with HindIII and BamHI, and analyzed by agarose gel electrophoresis. The whole procedure was repeated 10 times during 10 successive days (≈70 generations).
The relative copy number of pYS2 in PA9-13 and PA9-20 transformants was determined by comparison with that previously calculated for pGT5 in P. abyssi strain GE5 (25 ± 3 copies/chromosome) (8). Cultures of transformants carrying pYS2 and of strain GE5 containing pGT5 were grown in parallel in TAA medium and stopped at the same time in the exponential phase. The final cell density determined by direct cell counts were very similar for all cultures. Plasmid DNA was isolated as described earlier under exactly the same conditions for all samples. After digestion with HindIII and agarose gel electrophoresis, the relative copy number of pYS2 and pGT5 was calculated by comparing the intensity of the 0.9-kb HindIII band common to both plasmids and correcting for the variation in cell number used for the respective plasmid preparations. The intensities of the bands were integrated with the NIH Image program (http://rbs.info.nih.gov/nih-image/download.html).
Nucleotide sequence accession numbers.
The DNA sequences of the pyrE gene from S. acidocaldarius and from P. abyssi strain GE9 have been deposited in GenBank under accession numbers AJ459777 and AY083796, respectively.
RESULTS
Isolation of uracil-auxotrophic mutants.
P. abyssi strain GE9, previously characterized in our laboratory (31), was used as the wild type. This strain was chosen because it is naturally devoid of plasmid pGT5 (present in P. abyssi strain GE5), which we anticipated to be a potential source of vector instability in complementation experiments with a pGT5-based vector.
Spontaneous mutants resistant to 5-FOA were isolated by plating about 108 cells from late-exponential-phase cultures onto TAA plates containing 800 μg (≈2.5-fold the determined MIC) of 5-FOA and 50 μg of uracil per ml, as previously described (46). Plates of TAA medium containing the same concentrations of 5-FOA but no uracil were also inoculated with the same number of cells. No growth was observed in these control experiments. Appropriate dilutions of the cultures were plated without inhibitor in order to determine viable-cell counts and to calculate mutation frequencies per CFU. Resistant colonies appeared after 4 to 5 days of incubation at 85°C with a frequency of 2 × 10−6 ± 1 × 10−6 per CFU, comparable to that previously determined for other Thermococcales (48).
Ten independently isolated 5-FOA-resistant mutants were subsequently purified by streaking onto selective medium and checked for auxotrophy in liquid medium. Whereas all drug-resistant strains grew as well as the wild type in the presence of uracil, 7 out of the 10 mutants tested did not grow at all in uracil-free medium. All these uracil auxotrophs were stable and exhibited a high level of resistance to 5-FOA (>1,400 μg/ml). Their reversion frequency was determined by plating serial dilutions of cultures onto uracil-free medium.
Genetic characterization of uracil-auxotrophic mutants.
As we expected the mutations responsible for the observed phenotype to occur in either the pyrE gene (encoding OPRTase) or the pyrF gene (encoding orotidine 5′-monophosphate decarboxylase), we determined the complete nucleotide sequences of these genes for all the auxotrophic strains and compared them to their wild-type counterparts. Two mutants were found to harbor a large deletion in the pyrF gene, and another had no mutation in either the pyrE or the pyrF gene. Because we were interested only in pyrE mutants, these mutants are not considered in this study. Among the four simple pyrE mutants, two were selected for their lower range of reversion frequency, PA9-13 (3.5 × 10−7 ± 2 × 10−7) and PA9-20 (5 × 10−7 ± 1 × 10−7) and are described below.
The pyrE nucleotide sequences of strains GE5 and GE9 showed 95% identity. Both sequences contained an ORF of 549 bp, corresponding to the pyrE coding region, preceded by a motif (TTTAAA) centered 52 bp upstream the ATG that fits the box A consensus of archaeal promoters. The deduced amino acid sequences from the pyrE gene of strains GE5 and GE9 exhibited 98% similarity. Comparison of the wild-type GE9 pyrE sequence with that of the two auxotrophs revealed the presence of a single mutation in the coding region (no mutation was detected in the 132 bp upstream of the ATG). Although they were obtained independently, PA9-13 and PA9-20 showed the same frameshift mutation by insertion of a single base pair (T340) resulting in a stop codon (TGA348) in the novel reading frame. The corresponding polypeptide was 71 amino acid residues shorter than the wild-type OPRTase (182 amino acids). The deleterious effect of the mutations can be easily explained by the fact that the truncated polypeptide lacks most of region VLLVEDVTTTGG, highly conserved in Archaea and Bacteria. This corresponding sequence (VMLVDDVITAGT) has been shown to be the phosphoribosylpyrophosphate-binding site of the Salmonella enterica serovar Typhimurium OPRTase and to correspond to the active site of the enzyme (38).
Construction of a shuttle vector with a selectable genetic marker.
The plasmid pGT5 from P. abyssi GE5 was used as a basis for the construction of an autonomously replicating shuttle vector. Functional analysis of this plasmid permitted us to identify the essential elements required for its propagation in Pyrococcus cells and therefore to establish a strategy for the construction of a cloning vector. pGT5 contains two major ORFs which together cover 85% of the genome (14). The larger one encodes a protein of 75 kDa, Rep75, involved in the initiation of plasmid replication via a rolling-circle mechanism (29, 30). Other key elements, typical of rolling-circle replicons, the double-strand origin (dso) and single-strand origin (sso) of replication, were also identified.
No clear function could be attributed to the second major ORF (ORF2), but by analogy with the modular organization of bacterial rolling-circle plasmids, we suggested that it could be involved in plasmid maintenance or copy number control (14). Since other small rolling-circle plasmids encode only a Rep protein, ORF2 might be dispensable for replication. We therefore predicted that the best site for introduction of foreign DNA would be between ORF1 and ORF2 or within ORF2. The unique NspBII site in the intergenic region downstream of ORF1 was chosen because of its location between the putative Rep75 transcriptional terminator (hairpin region RIII) and the hypothetical ORF2 promoter (14). Thus, insertion of foreign DNA in that site would apparently not disrupt any important sequence. The backbone of potential shuttle vectors was then obtained by cloning the complete pGT5 sequence linearized by NspBII into the E. coli plasmid vector pLitmus38 (a pUC18 derivative). Preliminary transformation experiments have shown that such a shuttle vector could replicate in both E. coli and Pyrococcus hosts but was unstable in E. coli and lacked a selectable marker (1).
The uracil auxotrophs PA9-13 and PA9-20 described above carried the appropriate background for selection by complementation with a wild-type pyrE gene. A potential problem with the use of a native gene as a selectable marker is the possibility of recombination between the introduced gene and the resident genomic copy. To avoid this problem, we chose the pyrE gene of another hyperthermophilic archaeon, S. acidocaldarius, the sequence of which and its transcription start site had just been established (41). The amino acid sequence deduced from the S. acidocaldarius pyrE gene exhibited 60% similarity with its P. abyssi homologue, but the two genes did not show significant identity at the nucleotide level. Although these two archaea are phylogenetically distant, their transcriptional signals are sufficiently similar to be reciprocally recognized by the respective host transcription apparatus, as indicated by previous reports on heterologous expression of Sulfolobus genes in Pyrococcus (3) and vice versa (K. Stedman, E. Martusewitsch, C. Schleper, J. van der Oost, and W. Zillig, Abstr. Int. Cong. Extremophiles 2000, abstr. L49, 2000).
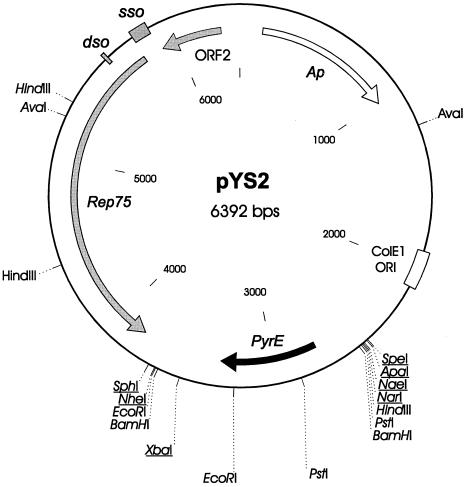
Therefore, a PCR product consisting of the S. acidocaldarius pyrE coding region and promoter sequence was introduced in the pGT5/Litmus construct, resulting in plasmid pYS1. As the size of the construct (7.2 kb, compared to 3.4 kb for pGT5) could be a potential factor of instability, we decided to reduce the pGT5 portion to the minimum required for replication in Pyrococcus spp. This was achieved by deleting most of ORF2, which we assumed to be dispensable for vector replication. The resulting construct, pYS2 (6.4 kb) (Fig. 1), was further used for transformation of the P. abyssi pyrE mutants derived from strain GE9.
FIG. 1.
Map of shuttle vector pYS2. Relevant restriction sites are shown; unique restriction sites are underlined. The white box and arrow show the E. coli pLitmus38 plasmid moiety. The gray boxes and arrows correspond to the pyrococcal pGT5 plasmid moiety (dso and sso, double-stranded and single-stranded origin of replication, respectively; Rep75, replication initiator encoded by the major ORF of pGT5; ORF2, truncated portion of the second main ORF of pGT5). The solid arrow indicates the S. acidocaldarius pyrE gene under the control of its own promoter.
PEG-mediated spheroplast transformation of P. abyssi.
The spheroplast transformation method used (see Materials and Methods) was adapted from the protocol described for halophilic archaea (11). The use of spheroplasts instead of whole cells for PEG-mediated transformation resulted in a significant improvement in the transformation frequency for both the PA9-13 and PA9-20 mutants (Table 1). Although there was significant variability in the number of transformants obtained in each experiment (which we believe is due in part to the difficulties inherent to plating these organisms), we were reproducibly able to achieve at least 102 transformants per μg of pYS2 per 109 cells of P. abyssi auxotrophs. Control experiments in which PEG was omitted never gave any transformant. PEG of various molecular weights (600, 1,000, 6,000, or 8,000) did not significantly change either the survival rate or the transformation efficiency. Therefore, whereas the presence of PEG appeared to be absolutely required for efficient transformation, its average size (within the range tested) did not appear to be an important parameter.
We also examined the effect of varying the PEG 600 concentration and found that a 25% final concentration was the best compromise between survival frequency and transformation frequency. No transformants were obtained with less than 0.05 μg of pYS2, and the best transformation frequencies were obtained with 0.5 to 1 μg of DNA. We also examined the eventuality of DNA restriction upon entry into P. abyssi cells, but we did not observe significant differences in transformation efficiencies obtained for pYS2 prepared from a P. abyssi transformant versus pYS2 prepared from E. coli strain SURE.
Demonstration of functionality of pyrE as a selectable genetic marker.
To demonstrate the ability of the S. acidocaldarius pyrE gene to complement P. abyssi pyrE mutants, we followed the growth of four independent transformants cultivated in TAA medium supplemented or not with uracil (50 μg/ml). The wild-type GE9 strain and the mutants PA9-13 and PA9-20 cultivated in the same conditions were used as controls. The four transformants tested grew equally well with or without uracil and exhibited growth rates comparable to those observed for the wild type. The maximal cell densities reached (≈109 cells/ml) for the transformants and the wild type were also similar. As expected, no growth was observed for the control cultures of the mutants in uracil-free medium. Moreover, growth of the transformants and the wild type was completely inhibited by 5-FOA at a concentration (600 μg/ml, ≈2 times the MIC), which did not affect the growth of the auxotrophs.
Accordingly, the OPRTase activities of the transformants and the wild type were comparable (Table 2), though consistently lower for the transformants (on average ≈30% lower), while the OPRTase activities of the mutants were markedly decreased (on average 28-fold reduction). As expected, orotidine 5′-monophosphate decarboxylase activities were in the same range for the transformants, the wild type, and the pyrE mutants (data not shown).
TABLE 2.
Growth tests and OPRTase activity for wild-type P. abyssi GE9, mutants PA9-13 and PA-20, and two representative transformants harboring vector pYS2
| Strain and genotype | Growth on TAA-based medium supplementeda with:
|
Avg OPRTase activityb (μM/h/mg of protein) ± SD | |||
|---|---|---|---|---|---|
| No supplement | Uracil | 5-FOA | Uracil + 5-FOA | ||
| GE9 | + | + | − | − | 0.18 ± 0.02 |
| PA9-13 (pyrE pyrF+) | − | + | − | + | <0.01 |
| PA9-13-T02 (pyrE pyrF+ [pYS2 pyrE+]c) | + | + | − | − | 0.13 ± 0.01 |
| PA9-20 (pyrE pyrF+) | − | + | − | + | <0.01 |
| PA9-20-T21 (pyrE pyrF+ [pYS2 pyrE+]) | + | + | − | − | 0.12 ± 0.02 |
TAA medium supplements were 50 μg/ml for uracil and 600 μg/ml for 5-FOA, cultures were inoculated at an initial cell density of 1 × 106 to 5 × 106 cells/ml with an exponentially growing preculture. Cell densities, were scored after 48 h of incubation at 90°C (cultures that did not grow after 48 h did not grow after longer incubation); +, ≥108 cells/ml; −, <107 cells/ml.
OPRTase activity values averages of three measurements.
The pyrE+ gene carried on pYS2 corresponds to the S. acidocaldarius wild-type pyrE gene.
As reversions could possibly be responsible for the Ura+ phenotype of the transformants, we verified the sequences of their genomic pyrE alleles. For each of the four transformants analyzed, the pyrE sequence was identical to that of the original auxotrophic recipient; it contained the same base pair insertion (T340) that caused a frameshift and produced an inactive enzyme. These results indicate that the pyrE gene from S. acidocaldarius present on pYS2 encodes a functional enzyme capable of conferring uracil prototrophy on pyrE mutants of P. abyssi. Finally, these results demonstrate the utility of this system for the selection of Pyrococcus transformants.
Stability and relative copy number of shuttle vector pYS2.
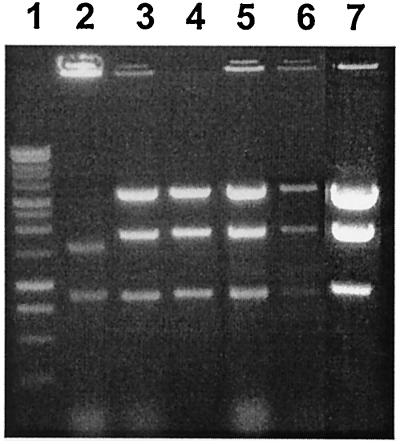
Plasmid DNAs from several independent P. abyssi transformants were extracted from subcultures grown in TAA medium. Aliquots of these preparations were used to transform E. coli, and the ampicillin-resistant clones obtained were picked and their plasmid DNA was prepared by alkaline lysis. Each DNA sample was analyzed by agarose gel electrophoresis. As illustrated in Fig. 2, plasmid DNA could be readily isolated from P. abyssi transformants in sufficient amounts to allow direct visualization on agarose gels. This result constitutes direct evidence that the vector replicates efficiently in P. abyssi cells. Moreover, the restriction endonuclease cleavage pattern of each DNA was identical to that of the original pYS2 DNA, indicating that no obvious deletions or rearrangements occurred in pYS2 during its passage in Pyrococcus cells and back in E. coli.
FIG. 2.
Stability of shuttle vector pYS2. Plasmid DNA was isolated from P. abyssi strains and from E. coli cells as described in Materials and Methods, digested with HindIII, and electrophoresed on an agarose gel. Lane 1, 1-kb ladder (Promega). Lane 2, endogenous plasmid pGT5 isolated from P. abyssi GE5. Lane 3, plasmid DNA from a PA9-20 Ura+ clone obtained after transformation with vector pYS2. Lane 4, pYS2 isolated from an Ampr clone of E. coli obtained after backtransformation with plasmid DNA from a P. abyssi transformant. Lane 5, plasmid DNA from a PA9-13 transformant after 16 h of growth in TAA medium. Lane 6, plasmid DNA from a PA9-13 transformant after 10 days of continuous growth under nonselective conditions (TAA medium plus 50 mg of uracil per ml). Lane 7, original pYS2 prepared from E. coli and used for transformation of P. abyssi mutants.
As illustrated in Fig. 2 (other data not shown), by comparing the relative intensity of the 0.9-kb HindIII band of pGT5 prepared from strain GE5 (Fig. 2, lane 1) and that of the corresponding band from pYS2 prepared from the same number of cells of P. abyssi transformants (Fig. 2, lanes 2, 4, and 6), we estimated the vector copy number to be in the same range in transformed cells as that of pGT5 in GE5, between 20 and 30 copies/chromosome (13).
A long-term growth experiment was performed with transformed P. abyssi cells to assess vector stability. A single clone containing pYS2 was cultured in TAA medium until it reached the stationary phase (typically an overnight incubation) and then diluted 1:100 in the same medium supplemented or not with uracil. Samples were taken every day and used for plasmid DNA preparation and restriction digest analysis. No change in plasmid DNA was observed after 10 days (≈70 generations) of continuous growth (not shown). Only a slight decrease in the relative amount of pYS2 was detected upon prolonged cultivation in nonselective conditions (Fig. 2, lane 6).
DISCUSSION
The development of genetic techniques for routine analysis of Pyrococcus spp. has been a long-time goal for researchers in that field. With the methods presented here, a crucial step towards this goal has been taken. For the first time, we have shown that DNA can be reproducibly introduced into Pyrococcus cells and that transformants can be directly selected on plates, a goal that had not been achieved previously (1, 3).
The shuttle vector reported here possesses a variety of features required for a cloning vector. It is very stable and replicates efficiently in both E. coli and P. abyssi. Vector pYS2 appears to be particularly stable in P. abyssi, as it could be easily recovered from transformants continuously cultured for more than 70 generations, even in the absence of selection pressure. The stability in E. coli may appear surprising, since we (Y. Zivanovic and S. Lucas, unpublished data) and others (3) previously observed that similar constructs based on the pyrococcal plasmid pGT5 combined with pUC derivatives suffered from important deletions and rearrangements in E. coli. The problem was partly solved by introducing the rom/rop gene from pBR322 in the pUC moiety in order to reduce the copy number in E. coli (3). This operation was not necessary for pYS2, as it was stable at high copy number not only in E. coli but also in P. abyssi.
Since pYS2 lacks most of the second major open reading frame (ORF2) of pGT5, it is clear that ORF2 is not required for replication in P. abyssi. It is also unlikely that ORF2 is involved in copy number control, because the vector copy number in transformants is similar to that of pGT5 in its natural host strain, P. abyssi GE5. More likely, this function is provided by the replication initiator Rep75 itself, which is able to autoregulate its activity (29). One can speculate that the deletion of ORF2 also contributed to the increased stability of the vector in E. coli. One obvious advantage of the high copy number is the facility to isolate the vector from transformant cultures in amounts sufficient to allow direct visualization on standard agarose gels. This has not been possible for other previously described plasmid-based vectors for Pyrococcus and Sulfolobus spp. (3, 6), which, because of their very low copy number, could only be detected in transformants by PCR or Southern blot and for which plasmid DNA could be recovered only after backtransformation in E. coli.
Because of its high stability and small size (6.39 kb), we assume that vector pYS2 could accommodate further insertions of foreign DNA, an additional marker and/or a candidate gene for expression in Pyrococcus spp. Their cloning will be greatly facilitated by the availability of several convenient unique restriction sites.
As an alternative to the use of antibiotic resistance markers, which are not suited for the selection of transformants in hyperthermophiles, we have demonstrated here that the pyrE gene encoding the OPRTase from S. acidocaldarius can be used for the positive selection of P. abyssi transformants on plates, by complementation of the appropriate uracil-auxotrophic recipients. The pyrE and pyrF genes are particularly useful genetic markers, as both mutant and wild-type alleles can be positively selected in most microorganisms, as they confer resistance to 5-fluoroorotic acid and bring about the return to prototrophy, respectively. Moreover, we have shown that the pYS2 vector, bearing the S. acidocaldarius pyrE gene, conferred on P. abyssi transformants the ability to grow on minimal medium as well as the wild-type strain does.
Accordingly, we found that the OPRTase activity measured in cell extracts of the transformants was comparable to that of the wild type. This was somewhat surprising since, in light of the high copy number of pYS2, we expected to measure a higher OPRTase activity. Possible explanations are that (i) although the transcriptional and translational signals of the S. acidocaldarius pyrE gene seem closely related to those generally found in Pyrococcus, they might allow only low-level expression of that gene in P. abyssi; (ii) this situation may reflect differences in the complex regulatory circuits (at the transcriptional and/or the enzymatic level) of this activity in two different cellular contexts;(iii) activity assays have shown that the optimal temperature for the S. acidocaldarius OPRTase is around 75 to 80°C (D. Charlier and D. Gigot, unpublished data). Therefore, it is possible that at 90 to 95°C, the normal temperatures for cultivation of P. abyssi, and in anaerobic conditions, the enzyme retains only partial activity. Further investigations are required to clarify this point.
Another important finding reported here is that PEG can induce genetic transformation in Pyrococcus spp. While efficient PEG-mediated transformations have been reported for many bacteria, yeasts, and fungi (20, 23), only a few reports concerned Archaea. A simple PEG-mediated transformation method proved efficient on whole cells of Methanococcus maripaludis, yielding over 105 transformants μg−1 with an integrative vector (44). In contrast, using this method with P. abyssi, we obtained only a very few transformants per microgram of pYS2 (Table 1).
Because the use of proto/spheroplasts versus whole cells often gave higher transformation efficiencies for bacteria and archaea (11, 32, 34), we tested this solution for P. abyssi. With a protocol adapted from a previously described method for halophilic archaea (11), we have shown that more than 90% of P. abyssi cells could be converted to spheroplasts but that only 10% of them regenerated as colonies on plates. This is much below what has been reported for methanogens and halophiles (typically above 60%) (11, 32, 34). Nevertheless, the use of spheroplasts rather than whole cells improved the PEG-mediated transformation efficiency of P. abyssi about 100-fold (Table 1). Still, this is about 3 to 4 orders of magnitude below what is currently obtained for mesophilic archaea (10, 32, 49). Among the reasons generally evoked to explain poor transformation efficiencies is the presence of a restriction/modification (R/M) system in the recipient host. Although we failed to detect restriction activity in P. abyssi GE9 and did not observe significant differences in the transformation efficiency with pYS2 vector prepared from a transformant, this hypothesis cannot be completely ruled out because such a type I R/M system has recently been identified in the genome sequence of P. abyssi strain GE5 (24).
It is difficult to predict how the copy number and stability of pYS2 derivatives will be influenced by the introduction of supplementary fragments. Even so, the construction of a selectable shuttle vector that replicates efficiently and is stably maintained at high copy number in E. coli and in P. abyssi certainly opens new perspectives for the elaboration of even more suitable cloning and expression vectors. Better vectors should contribute greatly to the development of additional genetic tools for the exploration of hyperthermophilic archaea and for the expression and purification of thermophilic enzymes produced in the correct cellular environment and at high temperature.
Acknowledgments
We thank Wolfram Zillig for the gift of an anaerobic chamber and advice for plating Pyrococcus. We are grateful to Daniel Gigot for OPRTase measurements.
This work was supported by the Centre National de la Recherche Scientifique, by a grant (BIO 4CT96-0488) from the EU project “Extremophiles as Cell Factories,” by a grant from the Ministère de la Recherche (PFRMMIP), and by the Fund for Scientific Research-Flanders (FWO Vlaanderen, contract G.0069.00) and the Flanders Interuniversity Institute for Biotechnology (VIB). G.E. was supported by a grant (PRIR) from the Région Bretagne.
REFERENCES
- 1.Aagaard, C., I. Leviev, R. N. Aravalli, P. Forterre, D. Prieur, and R. A. Garrett. 1996. General vectors for archaeal hyperthermophiles: strategies based on a mobile intron and a plasmid. FEMS Microbiol. Rev. 18:93-104. [DOI] [PubMed] [Google Scholar]
- 2.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 17:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Aravelli, R. N., and R. A. Garrett. 1997. Shuttle vectors for hyperthermophilic archaea. Extremophiles 1:183-191. [DOI] [PubMed] [Google Scholar]
- 4.Balch, W. E., G. E. Fox, L. J. Magrum, C. R. Woese, and R. S. Wolfe. 1979. Methanogens: revaluation of a unique biological group. Microbiol. Rev. 43:260-296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bradford, M. M. 1976. A rapid and sensitive method for quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 6.Cannio, R., P. Contursi, M. Rossi, and S. Bartolucci. 1998. An autonomously replicating transforming vector for Sulfolobus solfataricus. J. Bacteriol. 180:3237-3240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cannio, R., Contursi, P., M. Rossi, and S. Bartolucci. 2001. Thermoadaptation of a mesophilic hygromycin B phosphotransferase by directed evolution in hyperthermophilic Archaea: selection of a stable genetic marker for DNA transfer into Sulfolobus solfataricus. Extremophiles 5:153-159. [DOI] [PubMed] [Google Scholar]
- 8.Charbonnier, F., G. Erauso, T. Barbeyron, D. Prieur, and P. Forterre. 1992. Evidence that a plasmid from a hyperthermophilic archaebacterium is relaxed at physiological temperatures. J. Bacteriol. 174:6103-6108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charlebois, R. L., W. L. Lam, S. W. Cline, and W. F. Doolittle. 1987. Characterization of pHV2 from Halobacterium volcanii and its use in demonstrating transformation of an archaebacterium. Proc. Natl. Acad. Sci. USA 84:8530-8534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cline, S. W., and W. F. Doolittle. 1987. Efficient transfection of the archaebacterium Halobacterium halobium. J. Bacteriol. 169:1341-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cline, S. W., W. L. Lam, R. L. Charlebois, L. C. Schalkwyk, and W. F. Doolittle. 1989. Transformation methods for halophilic archaebacteria. Can. J. Microbiol. 35:148-152. [DOI] [PubMed] [Google Scholar]
- 12.Cline, S. W., L. C. Schalkwyk, and W. F. Doolittle. 1989. Transformation of the archaebacterium Halobacterium volcanii with genomic DNA. J. Bacteriol. 171:4987-4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erauso, G., F. Charbonnier, T. Barbeyron, P. Forterre, and D. Prieur. 1992. Preliminary characterization of a hyperthermophilic archaebacterium with a plasmid, isolated from a north Fiji basin hydrothermal vent. C. R. Acad. Sci. 314:387-393. [Google Scholar]
- 14.Erauso, G., S. Marsin, N. Benbouzid Rollet, M. F. Baucher, T. Barbeyron, Y. Zivanovic, D. Prieur, and P. Forterre. 1996. Sequence of plasmid pGT5 from the archaeon Pyrococcus abyssi: evidence for rolling-circle replication in a hyperthermophile. J. Bacteriol. 178:3232-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Erauso, G., and D. Prieur. 1995. Plate cultivation technique for strictly anaerobic, thermophilic, sulfur-metabolizing archaea, p. 25-29. In P. A. R. E.M. Fleischmann, F. T. Robb, and H. J. Schreier (ed.), Archaea: a lab manual, vol. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 16.Erauso, G., A. L. Reysenbach, A. Godfroy, J. R. Meunier, B. Crump, F. Partensky, J. A. Baross, V. Marteinsson, G. Barbier, N. R. Pace, and D. Prieur. 1993. Pyrococcus abyssi sp. nov., a new hyperthermophilic archaeon isolated from a deep-sea hydrothermal vent. Arch. Microbiol. 160:338-349. [Google Scholar]
- 17.Ghane, F., and D. W. Grogan. 1998. Chromosomal marker exchange in the thermophilic archaeon Sulfolobus acidocaldarius: physiological and cellular aspects. Microbiology 144:1649-1657. [DOI] [PubMed] [Google Scholar]
- 18.Grogan, D. 1991. Selectable mutant phenotypes of the extremely thermophilic archaebacterium Sulfolobus acidocaldarius. J. Bacteriol. 173:7725-7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grogan, D. W. 1996. Exchange of genetic markers at extremely high temperatures in the archaeon Sulfolobus acidocaldarius. J. Bacteriol. 178:3207-3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gruber, F., J. Visser, C. P. Kubicek, and L. H. de Graaff. 1990. The development of a heterologous transformation system for the cellulolytic fungus Trichoderma reesei based on a pyrG-negative mutant strain. Curr. Genet. 18:71-76. [DOI] [PubMed] [Google Scholar]
- 20a.Inoue, H., H. Nojima, and H. Okayama. 1990. High efficiency transformation of Escherichia coli with plasmids. Gene 96:23-28. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs, K. L., and D. W. Grogan. 1998. Spontaneous mutation in a thermoacidophilic archaeon: evaluation of genetic and physiological factors. Arch. Microbiol. 169:81-83. [DOI] [PubMed] [Google Scholar]
- 22.Kawarabayasi, Y., M. Sawada, H. Horikawa, Y. Haikawa, Y. Hino, S. Yamamoto, M. Sekine, S. Baba, H. Kosugi, A. Hosoyama, Y. Nagai, M. Sakai, K. Ogura, R. Otsuka, H. Nakazawa, M. Takamiya, Y. Ohfuku, T. Funahashi, T. Tanaka, Y. Kudoh, J. Yamazaki, N. Kushida, A. Oguchi, K. Aoki, and H. Kikuchi. 1998. Complete sequence and gene organization of the genome of a hyper-thermophilic archaebacterium, Pyrococcus horikoshii OT3. DNA Res. 5:55-76. [DOI] [PubMed] [Google Scholar]
- 23.Klebe, R. J., J. V. Hariss, D. Sharp, and M. G. Douglas. 1983. A general method for polyethylene-glycol-induced genetic transformation of bacteria and yeast. Gene 25:333-341. [DOI] [PubMed] [Google Scholar]
- 24.Kobayashi, I. 2001. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 29:1362-4962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kondo, S., A. Yamagishi, and T. Oshima. 1991. Positive selection for uracil auxotrophs of the sulfur-dependent thermophilic archaebacterium Sulfolobus acidocaldarius by use of 5-fluoroorotic acid. J. Bacteriol. 173:7698-7700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lange, M., and B. K. Ahring. 2001. A comprehensive study into the molecular methodology and molecular biology of methanogenic Archaea. FEMS Microbiol. Rev. 5:553-573. [DOI] [PubMed] [Google Scholar]
- 27.Lecompte, O., R. Ripp, V. Puzos Barbe, S. Duprat, R. Heilig, J. Dietrich, J. C. Thierry, and O. Poch. 2001. Genome evolution at the genus level: comparison of three complete genomes of hyperthermophilic Archaea. Genome Res. 11:981-993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lopez-Garcia, P., and P. Forterre. 1997. DNA topology in hyperthermophilic archaea: reference states and their variation with growth phase, growth temperature, and temperature stresses. Mol. Microbiol. 23:1267-1279. [DOI] [PubMed] [Google Scholar]
- 29.Marsin, S., and P. Forterre. 1999. The active site of the rolling circle replication protein Rep75 is involved in site-specific nuclease, ligase and nucleotidyl transferase activities. Mol. Microbiol. 33:537-545. [DOI] [PubMed] [Google Scholar]
- 30.Marsin, S., and P. Forterre. 1998. A rolling circle replication initiator protein with a nucleotidyl-transferase activity encoded by the plasmid pGT5 from the hyperthermophilic archaeon Pyrococcus abyssi. Mol. Microbiol. 27:1183-1192. [DOI] [PubMed] [Google Scholar]
- 31.Marteinsson, V. T., L. Watrin, D. Prieur, J. C. Caprais, G. Raguenes, and G. Erauso. 1995. Phenotypic characterization, DNA similarities, and protein profiles of twenty sulfur-metabolizing hyperthermophilic anaerobic archaea isolated from hydrothermal vents in the southwestern Pacific Ocean. Int. J. Syst. Bacteriol. 45:623-632. [Google Scholar]
- 32.Metcalf, W. W., J. K. Zhang, E. Apolinario, K. R. Sowers, and R. S. Wolfe. 1997. A genetic system for Archaea of the genus Methanosarcina: liposome-mediated transformation and construction of shuttle vectors. Proc. Natl. Acad. Sci. USA 94:2626-2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Myllykallio, H., P. Lopez, G.-P. Lopez, R. Heilig, W. Saurin, Y. Zivanovic, H. Philippe, and P. Forterre. 2000. Bacterial mode of replication with eukaryotic-like machinery in a hyperthermophilic archaeon. Science 288:2212-2215. [DOI] [PubMed] [Google Scholar]
- 34.Patel, G., J. Nash, B. Agnew, and G. Sprott. 1994. Natural and electroporation-mediated transformation of Methanococcus voltae protoplasts. Appl. Environ. Microbiol. 60:903-907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peck, R. F., S. DasSarma, and M. P. Krebs. 2000. Homologous gene knockout in the archaeon Halobacterium salinarum with ura3 as a counterselectable marker. Mol. Microbiol. 35:667-676. [DOI] [PubMed] [Google Scholar]
- 36.Pope, B., and H. M. Kent. 1996. High efficiency 5 min transformation of Escherichia coli. Nucleic Acids Res. 24:536-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Scapin, G., C. Grubmeyer, and J. C. Sacchettini. 1994. Crystal structure of orotate phosphorybosyltransferase. Biochemistry 33:1287-1294. [DOI] [PubMed] [Google Scholar]
- 39.Schleper, C., K. Kubo, and W. Zillig. 1992. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc. Natl. Acad. Sci. USA 89:7645-7649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stedman, K. M., C. Schleper, E. Rumpf, and W. Zillig. 1999. Genetic Requirements for the Function of the Archaeal Virus SSV1 in Sulfolobus solfataricus: Construction and Testing of Viral Shuttle Vectors. Genetics 152:1397-1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thia-Toong, T. L., M. Roovers, V. Durbecq, D. Gigot, N. Glansdorff, and D. Charlier. 2002. Genes of the de novo pyrimidine biosynthesis from the hyperthermoacidophilic crenarchaeote Sulfolobus acidocaldarius: novel organization in a bipolar operon. J. Bacteriol. 184:4430-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL X-Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 24:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tumbula, D. L., T. L. Bowen, and W. B. Whitman. 1997. Characterization of pURB500 from the archaeon Methanococcus maripaludis and construction of a shuttle vector. J. Bacteriol. 179:2976-2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tumbula, D. L., R. A. Makula, and W. B. Whitman. 1994. Transformation of Methanococcus maripaludis and identification of a PstI-like restriction system. FEMS Microbiol. Lett. 121:309-314. [Google Scholar]
- 45.Tumbula, D. L., and W. B. Whitman. 1999. Genetics of Methanococcus: possibilities for functional genomics in Archaea. Mol. Microbiol. 33:1-7. [DOI] [PubMed] [Google Scholar]
- 46.Watrin, L., S. Lucas, C. Purcarea, C. Legrain, and D. Prieur. 1999. Isolation and characterization of pyrimidine auxotrophs, and molecular cloning of the pyrE gene from the hyperthermophilic archaeon Pyrococcus abyssi. Mol. Gen. Genet. 262:378-381. [DOI] [PubMed] [Google Scholar]
- 47.Watrin, L., V. Martin Jezequel, and D. Prieur. 1995. Minimal amino acid requirements of the hyperthermophilic archaeon Pyrococcus abyssi, isolated from deep-sea hydrothermal vents. Appl. Environ. Microbiol. 61:1138-1140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Watrin, L., and D. Prieur. 1996. UV and ethyl methanesulfonate effects in hyperthermophilic archaea and isolation of auxotrophic mutants of Pyrococcus strains. Curr. Microbiol. 33:377-382. [DOI] [PubMed] [Google Scholar]
- 49.Whitman, W. B., D. L. Tumbula, J. P. Yu, and W. Kim. 1997. Development of genetic approaches for the methane-producing archaebacterium Methanococcus maripaludis. Biofactors 6:37-46. [DOI] [PubMed] [Google Scholar]
- 50.Yu, J. J., Z. L. Thomas, P. W. Szaniszlo, and G. T. Cole. 1999. Isolation and confirmation of function of the Coccidioides immitis URA5 (orate phosphoribosyl transferase) gene. Gene 226:233-242. [DOI] [PubMed] [Google Scholar]
- 51.Zillig, W., H. P. Arnold, I. Holz, D. Prangishvili, A. Schweier, K. Stedman, Q. She, H. Phan, R. Garrett, and J. K. Kristjansson. 1998. Genetic elements in the extremely thermophilic archaeon Sulfolobus. Extremophiles 2:131-140. [DOI] [PubMed] [Google Scholar]
- 52.Zillig, W., and A.-L. Reysenbach. 2001. Thermococcales, p. 341-348. In R. W. Castenholz, D.R. Boone, and G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y.