Abstract
Short interfering RNAs (siRNAs) are short (21–23 nt) double-stranded RNAs that direct the sequence-specific degradation of corresponding mRNAs, resulting in suppression of gene activity. siRNAs are powerful tools for gene functional analysis in mammals. Chemically synthesized siRNAs permit transient gene repression but preclude inhibition of stable gene products as well as long-term phenotypic analyses. Permanent gene suppression can be achieved by transcribing siRNAs as stem–loop precursors from Pol III promoters. This approach, however, has a major limitation: inhibition cannot be controlled in a time- or tissue-specific manner. Thus, the approach cannot be applied to genes essential for cell survival or cell proliferation. To overcome these limitations, we have designed a CRE–lox-based strategy that allows one to repress gene activity in a time-dependent manner in cells, and in a time- or tissue-dependent manner in animals. Our approach promises to improve dramatically the procedures for functional genetics in mammals.
Introduction
RNA interference is a powerful strategy for gene inhibition in a variety of organisms (Sharp, 2001). In plants or in the worm Caenorhabditis elegans, the procedure is to introduce a doublestranded RNA corresponding to the sequence of the target mRNA into the cells. This double-stranded RNA is cleaved into short interfering RNAs (siRNAs) by a protein complex that includes the RNase Dicer (Bernstein et al, 2001). siRNAs are doublestranded RNA sequences 21–23 nucleotides (nt) long with a 3′ protruding tail of 2 nt (Zamore et al, 2000; Elbashir et al, 2001a). The antisense strand of siRNA (Martinez et al, 2002) guides another complex of proteins, the RISC complex (Hammond et al, 2000), towards the target mRNA, and induces its cleavage at a position corresponding to the centre of the siRNA sequence (Elbashir et al, 2001b,2001c). In mammalian cells, RNA interference can be triggered by short synthetic doublestranded RNA sequences mimicking the siRNAs generated by Dicer cleavage in lower organisms, without inducing the interferon response that is observed with long double-stranded RNAs in most cell types (Elbashir et al, 2001a). Synthetic siRNAs, however, have a short half-life in cells, and consequently their effect is transient. siRNAs can be constitutively transcribed as stem–loop precursors by the Pol III polymerase, and stable cell lines can be established in which the siRNA is expressed in a constitutive manner (Brummelkamp et al, 2002b; Donze & Picard, 2002; Miyagishi & Taira, 2002; Yu et al, 2002). The siRNA cassette can be inserted into a viral vector, allowing the transduction of a high proportion of cells in a population, including in cells that are otherwise difficult to transfect such as primary cell cultures (Brummelkamp et al, 2002a; Rubinson et al, 2003; Stewart et al, 2003). However, genes that are essential for cell survival or cell proliferation cannot be studied in this manner because their inhibition prevents the establishment of cell lines. Thus, there is a great need for a system that permits full exploitation of this strategy. Pol III promoters are very sensitive to sequence modifications, and it therefore appeared difficult to insert a binding site for an inducible factor such as tetracycline. Furthermore, to avoid undesired inhibition of target genes during the establishment of stable cell lines, it is of prime importance that the background expression of siRNA is kept to minimal levels. For these reasons, we have designed a CRE–lox-based approach, in which the formation of the stem–loop siRNA precursor is dependent on the expression of the CRE recombinase (Dale & Ow, 1991). We demonstrate a CRE-dependent production of siRNA, which is accompanied by a CRE-dependent inhibition of a target gene. This strategy should prove highly useful for functional genetics in tissue culture as well as in animals.
Results and Discussion
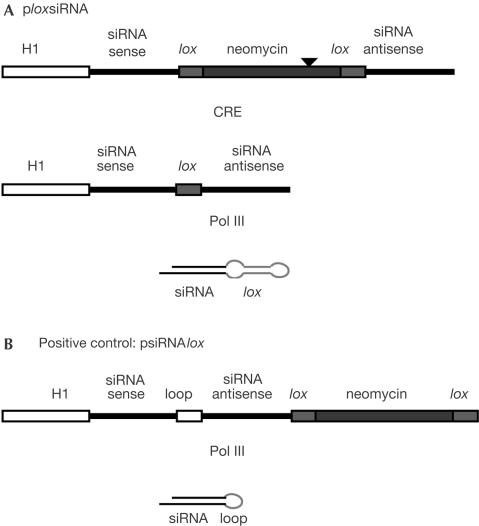
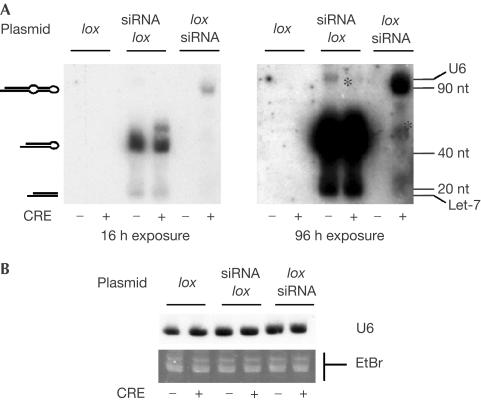
Our strategy for CRE-dependent production of siRNAs is described in Fig 1. The siRNA sequence is under the control of a Pol III promoter (H1), as described previously (Brummelkamp et al, 2002b). The sense strand is separated from the antisense strand by a neomycin cassette that is flanked by two lox sites. In the absence of CRE, a stretch of four T's in the neomycin cassette (black arrow-head in Fig 1A) terminates Pol III transcription, thereby preventing transcription of the antisense strand and resulting in an incomplete transcript. In the presence of CRE, recombination eliminates the neomycin cassette, allowing the read-through of the entire siRNA. The remaining intercalated lox sequence acts as a loop, which is necessary for formation of the siRNA and proper processing of the stem–loop precursor (Brummelkamp et al, 2002b). The strategy was tested using green fluorescent protein (GFP) as the target reporter gene with a previously described siRNA sequence (Brummelkamp et al, 2002b). A plasmid in which the entire siRNA was inserted upstream from the lox neomycin cassette, allowing the constitutive expression of siRNA, was used as a positive control; the backbone vector (pLox) was used as a negative control. Expression and processing of the stem–loop precursor was monitored by northern blot (Fig 2). As expected, a precursor of approximately 50 nt was observed for the constitutive expression vector psiRNAlox, and this precursor was processed into a 20 nt species. Formation of these products did not require the expression of the CRE recombinase, as expected. With ploxsiRNA, in the absence of CRE, no product could be detected in the range of sizes analysed. In the presence of CRE, however, a major band corresponding to the 90 nt precursor, and a 20 nt processing product could be detected. The level of this stem-lox precursor was lower than that observed with the constitutive vector (compare siRNA lox and lox siRNA in Fig 2A). This is probably due to the kinetics of accumulation of the precursor, which requires a recombination event that is not needed for the constitutive vector. However, the ratio between stem–loop precursor and product appeared to be roughly similar for the two plasmids, suggesting that the lox sequence did not significantly impair precursor processing.
Figure 1.
Strategy for siRNA production and plasmids used in this study. Black arrowhead: position of the Pol III stop codon in plox siRNA.
Figure 2.
CRE-dependent production of siRNA. COS cells were transfected with the indicated vectors, and RNAs were analysed using a probe complementary to the siRNA antisense strand. (A) A short and a long exposure of the same northern blot are shown. The positions of the precursors and processing products are indicated on the left, and the positions of an RNA ladder as well as that of cellular noncoding RNAs (U6 and Let-7) are indicated on the right; the asterisk (*) indicates bands that we interpret as corresponding to read-through products or processing intermediates. (B) The blot was hybridized with a U6 probe (U6) or RNA was stained with ethidium bromide (EtBr) to control for the amount of total RNA in each lane.
The efficiency of the siRNAs for gene inhibition was next assessed in transient transfection assays (Fig 3). As expected, the constitutive vector psiRNAlox inhibited GFP protein expression independently of CRE expression. In the case of the ploxsiRNA plasmid, no inhibition was observed in the absence of CRE. In the presence of CRE, GFP was inhibited. The level of inhibition increased with time: after 48 h (Fig 3A), inhibition was only partial as compared with the psiRNAlox vector, in good correlation with siRNA accumulation (Fig 2); it was greater after 60 h (Fig 3B), although some GFP was still detectable. No inhibition was observed with a ploxsiRNA vector directed against an irrelevant sequence (data not shown), indicating that the inhibition was gene specific.
Figure 3.
CRE-dependent inhibition of GFP in transiently transfected cells. Cells were transfected as in Fig 2, and extracts were analysed 48 h (A) or 60 h (B) later by western blot using anti-GFP or anti-tubulin antibodies. These experiments were reproduced 2–6 times. d2GFP, unstable form of GFP.
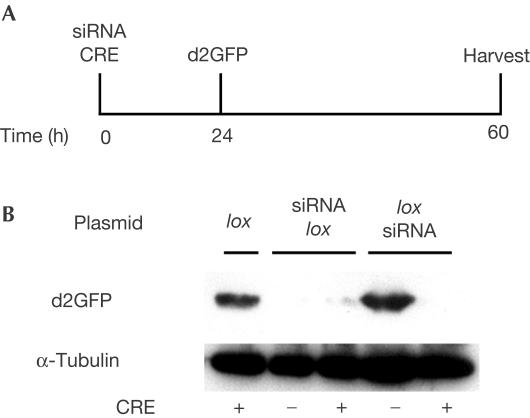
Lack of complete inhibition is probably due to the fact that, whereas GFP accumulates soon after transfection, the siRNA product accumulates at a slower rate. Consistent with this hypothesis, GFP was undetectable in two-step transfections in which the siRNA was allowed to form for 24 h before transfection of the GFP expression vector (Fig 4). Taken together, these results indicate that lox-containing siRNAs formed from ploxsiRNA, although the levels are not high (as monitored by northern blot), are indeed efficient for gene inhibition (Fig 2).
Figure 4.
CRE-dependent inhibition of GFP in twostep transfections. (A) Experimental design. Cells were first transfected with ploxsiRNA, psiRNAlox or plox, together with the CRE expression vector pMC CRE or the empty vehicle vector pMC for the controls, and then again 24 h later with pCMV-GFP. (B) Western blot analysis. These experiments were reproduced twice.
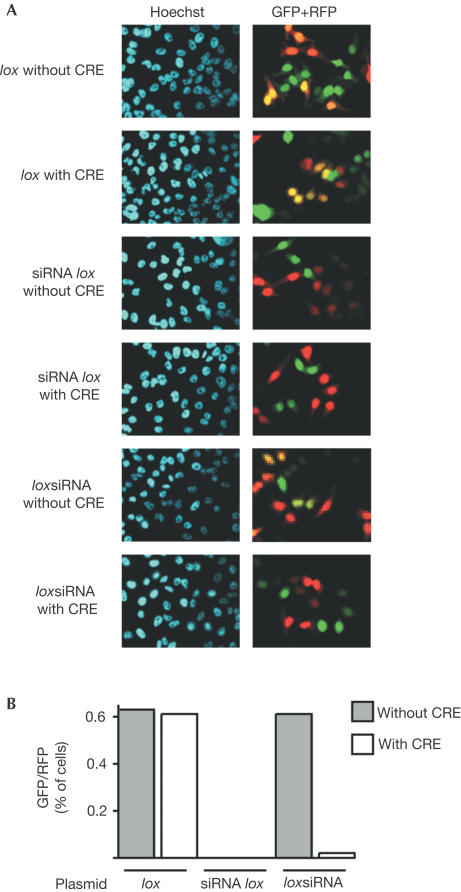
The effect of the siRNAs was analysed on a reporter gene expressed from an integrated sequence, using a cell line in which GFP expression is inducible. Cells were transfected with the siRNA- and CRE-expressing vectors, together with an RFP expression vector to distinguish transfected cells, and GFP expression was subsequently induced. GFP expression was observed in about 30% of the cells 24 h after induction, and in 60% of the cells 48 h after induction. Similar proportions were observed in control-transfected (RFP-positive) cells that received the plox empty vehicle vector (Fig 5A,B). In contrast, essentially no GFP expression was detected in transfected cells that received the constitutive psiRNAlox vector, irrespective of CRE expression. With the ploxsiRNA vector, a ‘normal' proportion (around 30% at day 1 and 65% at day 2) of GFP-positive cells among the transfected cells was observed in the absence of CRE, but this proportion dropped to less than 5% in the presence of CRE.
Figure 5.
Inhibition of an integrated GFP transgene. HeLa 1002 cells were transfected as in Fig 2, and treated with Dox to induce GFP expression 72 h later for 24 h (A) or 48 h (B). (A) Microphotographs of cells after 72 h of Dox treatment. (B) Proportion of GFP-positive cells among the transfected, RFP-positive cells. These experiments were reproduced three times.
CRE-dependent inhibition was also achieved with a stably integrated siRNA-expressing vector directed against an endogenous gene. A ploxsiRNA directed against p53 designed as in Fig 1, or the plox empty vector, was transfected into U20S cells; stable clones were established using neomycin as a selectable marker. Selected clones were then transfected with the CRE-expressing vector or the empty vehicle vector as a control, and selected using a different selectable marker (hygromycin). CRE-dependent p53 inhibition was achieved in about 50% of the clones with the ploxsiRNAp53 vector; three examples of such clones are shown in Fig 6. No inhibition was observed when the ploxsiRNAp53 clones received the pMC empty vector, or in any of the clones stably transfected with the plox empty vector. This result demonstrates that this approach can be used to establish stably transfected cell lines in which a target endogenous gene can be downregulated at will.
Figure 6.
Inhibition of an endogenous gene from an integrated construct. U20S cells with integrated plox or ploxsiRNA as indicated were transfected with pMC CRE or pMC. p53 expression was monitored 4 weeks later.
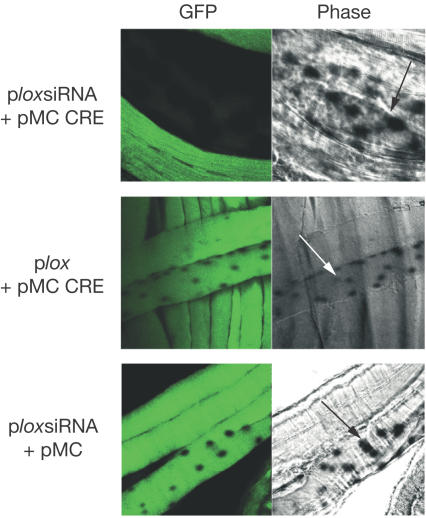
Finally, the effect of siRNA was also monitored in vivo, after electroporation of the plasmids in muscles. The tibialis anterior (TA) muscles of mice constitutively expressing GFP in a ubiquitous manner were electroporated with both the siRNA- and CRE-expressing vectors, along with a nuclear LacZ expression vector to label the nuclei of transfected fibres. The electroporated muscles were examined 12 days later. The combination of the CRE-expressing plasmid and the ploxsiRNA GFP vector resulted in a marked decrease of GFP in transfected fibres, whereas expression of CRE in the presence of the plox control vector, or transfection of the ploxsiRNA vector in the absence of CRE, did not affect GFP expression in transfected fibres (Fig 7). These results indicate that our CRE–lox-based strategy can also downregulate gene expression in live animals.
Figure 7.
CRE-dependent inhibition of GFP in vivo. The TA muscles of GFP transgenic mice were electroporated with the indicated plasmids along with an MCK-LacZ to detect transfected fibres, and analysed 12 days later for LacZ and GFP expression as described in the Methods section. The arrows point to LacZ-positive nuclei in a transfected fibre.
Taken together, these results indicate that efficient gene inhibition can be induced using the CRE–lox system in a simple and convenient manner. Such a system is an absolute requirement when analysing the function of essential genes in cells, and it should facilitate high-throughput functional genetics. It is also greatly needed for functional genetics in animals. The analysis of both the expression of the CRE-dependent siRNAs and the inhibition of a target gene demonstrated an essential advantage of our approach, which is the total absence of background expression in the absence of CRE.
In conclusion, this strategy allows one to tackle the functional analysis of essential genes. CRE can be expressed in an inducible or in a tissue-specific manner in animals, allowing one to achieve time-dependent and/or tissue-specific gene inactivation. In that sense, our approach is complementary to the Tet-inducible system that was published by van de Wetering et al (2003). Functional analysis of mammalian genomes should be made significantly easier.
Methods
Plasmid construction. Plox was constructed by inserting into the ploxNeo plasmid the Pol III promoter H1 as an Nhe1–Xba1 insert. The sequences corresponding to the siRNA sense and antisense strands were introduced as synthetic oligonucleotides using, respectively, the XbaI site or the BamH1 and Kpn1 sites. Sequences were CTAGC CCC GCAAGCTGACCCTGAAGTTCAT T (sense) and GATCC ATGAACTTCAGGGTCAGCTTGC TTTT GGTACC TAGA CCC (antisense) for GFP, and GCATGAACCGGAGGCC CAT T (sense) and GATCC ATGGGCCTCCGGTTCATGC (antisense) for p53.
Transfection. COS-7 cells were transfected with 500 ng of CMV-GFP or CMV-d2GFP (Clontech), 8 μg of pMC CRE (Gu et al, 1993) (or pMC backbone for the negative controls), and 4 μg of ploxsiRNA, psiRNAlox or plox using Polyfect (Qiagen). In some experiments, cells were transfected twice, first with 2 μg of pMC CRE or pMC backbone and 1 μg of ploxsiRNA, psiRNAlox or plox and 24 h later with 500 ng of CMV-d2GFP. Under the conditions that we have used, about 80% of the cells scored positive for GFP fluorescence. In some experiments, 300 ng of CMV luc or CMV-RFP (a kind gift from Michel Kress) was included. HeLa 1002 cells (a cell line derived from HeLa with an integrated GFP transgene inducible by doxocycline) were transfected with siRNA- and CRE-expressing plasmids as described above. Cells were treated with 1 μg/ml of doxocycline 72 h later for 24–72 h, before examination under an Axiovert fluorescence microscope.
Western blot. Extracts were analysed 60 h after transfection using standard procedures.
Northern blots. Total RNA (40 μg) was loaded onto a 15% denaturing polyacrylamide gel. RNAs were transferred onto nitrocellulose membranes by electrotransfer (45 min at 390 mA) in TBE buffer. The probe was a 32P-labelled antisense sequence corresponding to a 22 nt GFP target (5′CTAGAATGAACTTCAGGGTCAGCTTGCGGT3′).
Muscle electroporation. The MCK–nlsLacZ construct contains the coding sequence of nuclear β-galactosidase under the control of the muscle creatine kinase promoter.
Actin-GFP transgenic mice (5–10 weeks old) (Ikawa et al, 1998) were anaesthetized with 300 μl of 0.05% xylazine–1.7% ketamine in 0.9% NaCl. After skin incision, 8 μg of DNA containing 3 μg of CRE and/or siRNA expression vector and 2 μg of MCK–nlsLacZ were injected into the TA muscle using a 1 ml syringe with a 27-gauge needle. Caliper electrode plates (Q-biogen, France) were immediately applied on each side of the muscle, and a series of eight electrical pulses (2 Hz, 20 ms each) was delivered with a standard square-wave electroporator (ECM 830, Q-biogen). Electrical contact was ensured by shaving and conductive gel application.
At 12 days after injection, TA muscles were dissected and fixed for 30 min with 4% paraformaldehyde in phosphate-buffered saline (PBS) and incubated for 2–3 h in 0.4 mg/ml 5-bromo-4-chloro-indolyl-β-D-galactoside, 4 mM K3Fe(CN)6, 4 mM K4Fe(CN)6 and 2 mM MgCl2 in PBS at 37°C for LacZ staining. LacZ-positive regions were further dissected under a microscope. Fluorescent and phase contrast images were acquired on a Zeiss confocal microscope (LSM510, Zeiss).
Acknowledgments
We dedicate this paper to the memory of Pr. Claude Hélène, an internationally esteemed pioneer in the field of gene inhibition. We thank V. Guasconi for her help with immunofluorescence, R. Agami for the gift of pSUPER, J.-C. François for the gift of the HeLa 1002 cell line, M. Kress for the gift of various plasmids, L.L. Pritchard for a critical reading of the manuscript and A. Hamiche for helpful discussion. This work was supported by a grant from the European fifth FP (grant QLG1-1999-00866).
References
- Bernstein E, Caudy AA, Hammond SM, Hannon GJ (2001) Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002a) Stable suppression of tumorigenicity by virus-mediated RNA interference. Cancer Cell 2: 243–247 [DOI] [PubMed] [Google Scholar]
- Brummelkamp TR, Bernards R, Agami R (2002b) A system for stable expression of short interfering RNAs in mammalian cells. Science 296: 550–553 [DOI] [PubMed] [Google Scholar]
- Dale EC, Ow DW (1991) Gene transfer with subsequent removal of the selection gene from the host genome. Proc Natl Acad Sci USA 88: 10558–10562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donze O, Picard D (2002) RNA interference in mammalian cells using siRNAs synthesized with T7 RNA polymerase. Nucleic Acids Res 30: e46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001a) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Lendeckel W, Tuschl T (2001b) RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev 15: 188–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T (2001c) Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J 20: 6877–6888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Zou YR, Rajewsky K (1993) Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell 73: 1155–1164 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Bernstein E, Beach D, Hannon GJ (2000) An RNA-directed nuclease mediates post-transcriptional gene silencing in Drosophila cells. Nature 404: 293–296 [DOI] [PubMed] [Google Scholar]
- Ikawa M, Yamada S, Nakanishi R, Okabe M (1998) ‘Green mice' and their potential usage in biological research. FEBS Lett 430: 83–87 [DOI] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T (2002) Singlestranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574 [DOI] [PubMed] [Google Scholar]
- Miyagishi M, Taira K (2002) U6 promoter-driven siRNAs with four uridine 3′ overhangs efficiently suppress targeted gene expression in mammalian cells. Nat Biotechnol 20: 497–500 [DOI] [PubMed] [Google Scholar]
- Rubinson DA et al. (2003) A lentivirus-based system to functionally silence genes in primary mammalian cells, stem cells and transgenic mice by RNA interference. Nat Genet 33: 401–406 [DOI] [PubMed] [Google Scholar]
- Sharp PA (2001) RNA interference—2001. Genes Dev 15: 485–490 [DOI] [PubMed] [Google Scholar]
- Stewart SA et al. (2003) Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA 9: 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Wetering M, Oving I, Muncan V, Pon Fong MT, Brantjes H, van Leenen D, Holstege FC, Brummelkamp TR, Agami R, Clevers H (2003) Specific inhibition of gene expression using a stably integrated, inducible small-interfering-RNA vector. EMBO Rep 4: 609–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JY, DeRuiter SL, Turner DL (2002) RNA interference by expression of short-interfering RNAs and hairpin RNAs in mammalian cells. Proc Natl Acad Sci USA 99: 6047–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Tuschl T, Sharp PA, Bartel DP (2000) RNAi: doublestranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101: 25–33 [DOI] [PubMed] [Google Scholar]