Abstract
Thirty deoxynivalenol-producing F. culmorum strains, isolated from wheat grains, were incubated in vitro and analyzed for trichothecene production. Seventeen strains produced more than 1 ppm of deoxynivalenol and acetyldeoxynivalenol and were considered high-deoxynivalenol-producing strains, whereas 13 F. culmorum strains produced less than 0.07 ppm of trichothecenes and were considered low-deoxynivalenol-producing strains. For all strains, a 550-base portion of the trichodiene synthase gene (tri5) was amplified and sequenced. According to the tri5 data, the F. culmorum strains tested clustered into two groups that correlated with in vitro deoxynivalenol production. For three high-producing and three low-producing F. culmorum strains, the tri5-tri6 intergenic region was then sequenced, which confirmed the two separate clusters within the F. culmorum strains. According to the tri5-tri6 sequence data, specific PCR primers were designed to allow differentiation of high-producing from low-producing F. culmorum strains.
Trichothecenes, including deoxynivalenol, acetyldeoxynivalenol, nivalenol, and fusarenone X, are sesquiterpene toxins produced by Fusarium species, including Fusarium culmorum, which are common fungal contaminants of cereals. Trichothecenes can be found naturally worldwide on cereals (1, 9, 18, 27, 38, 45, 48, 54, 55, 59), and the consumption of these toxins is a potential problem for humans and farm animals (14, 47).
It has been established that some Fusarium species, including F. graminearum and F culmorum, are able to produce B trichothecenes, such as deoxynivalenol and acetyldeoxynivalenol, while other species are not (31). These two types of Fusarium strains (producers and nonproducers) can be distinguished on the basis of DNA polymorphism in the β-tubulin gene (43) as well as in the large ribosomal subunit or the internal transcribed spacer (21, 37, 43).
According to their trichothecene production, some Fusarium species, such as F graminearum, have been divided into two chemotypes: (i) the nivalenol chemotype, which includes isolates producing nivalenol and fusarenone X, and (ii) the deoxynivalenol chemotype, which includes isolates producing deoxynivalenol and acetyldeoxynivalenol (26, 53). Similar observations have been made for F. culmorum strains (28, 39).
In addition, it has been demonstrated that, within the same species and in the same culture conditions, toxin production by Fusarium strain may vary sharply; some strains produce large amounts of trichothecenes, whereas others produce small or undetectable amounts of trichothecenes (3, 5, 19, 28, 30, 35, 36, 39, 53, 57). Until now, no method except in vitro culture has been available to distinguish high-producing from low-producing Fusarium strains.
Several genes involved in the biosynthesis of trichothecenes have been described, most of them localized in a gene cluster. The tri5 gene encodes the trichodiene synthase, which catalyzes the first step in the biosynthesis of trichothecenes. The nucleotide sequence of the tri5 gene has been characterized in several Fusarium species (16, 22, 23). The tri6 gene encodes a protein that regulates the trichothecene biosynthesis genes (46) and has been sequenced in F. sporotrichioides (32, 46), Giberella zeae (F. graminearum) (6, 29, 32), and F. cerealis (32). For several Fusarium species, it has been shown that the tri5 (22, 23) and tri6 (32) genes were present in single copy.
In the present work, we focused on Fusarium culmorum strains producing large and small amounts of deoxynivalenol (high-producing and low-producing strains, respectively). The objectives of this study were (i) to study the genetic relationship between strains through analysis of the tri5 gene and the tri5-tri6 intergenic region and (ii) to design a PCR test to differentiate these two phenotypes.
MATERIALS AND METHODS
Isolates of Fusarium strains.
Thirty deoxynivalenol-producing F. culmorum strains isolated from cereals from different areas in France were used in this study, as presented in Table 1. Fusarium strains may also be obtained from the first author. Isolates were identified as F. culmorum by conidial morphology as described by Nelson et al. (40). Fusarium identification was confirmed by species-specific PCR-based DNA analysis with the OPT18R and OPT18F primers by the method of Schilling et al. (49). These primers were designed by sequence-characterizing randomly amplified polymorphic DNA fragments. Single-spore strains were maintained on potato dextrose agar slants at 4°C.
TABLE 1.
Trichothecene production and biomass formation by F. culmorum strains isolated from wheat
| Strain | Origin | Host | Trichothecene productiona (ppm)
|
Ergosterol production (ppm) | ||
|---|---|---|---|---|---|---|
| DON | 3-ADON | 15-ADON | ||||
| A2 | Calvados | Wheat | 1.20 | ND | ND | 898 |
| A4 | Calvados | Wheat | 1.40 | ND | ND | 990 |
| B3 | Loire Atlantique | Wheat | 24.10 | 1, 68 | ND | 1,237 |
| B5 | Loire Atlantique | Wheat | 1.41 | ND | ND | 1,457 |
| B11 | Loire Atlantique | Wheat | ND | ND | ND | 1,562 |
| C1 | Marne | Wheat | ND | ND | ND | 1,005 |
| C6 | Marne | Wheat | 3.80 | ND | ND | 916 |
| C8 | Marne | Wheat | 7.20 | 0.96 | ND | 1,124 |
| C11 | Marne | Wheat | ND | ND | ND | 1,237 |
| E1 | Seine Maritime | Wheat | 6.20 | 0.64 | ND | 1,210 |
| E2 | Seine Maritime | Wheat | 10.10 | 0.12 | ND | 1,082 |
| F11 | Somme | Wheat | ND | ND | ND | 1,151 |
| G1 | Gard | Wheat | 25.60 | 2.40 | ND | 1,654 |
| H1 | Gers | Wheat | 24.30 | 1.90 | ND | 1,594 |
| I1 | Vienne | Wheat | 47.50 | 5.60 | 0.30 | 962 |
| J11 | Loir et Cher | Wheat | ND | ND | ND | 1,202 |
| K2 | Loir et Cher | Wheat | ND | ND | ND | 1,056 |
| K11 | Loir et Cher | Wheat | ND | ND | ND | 1,094 |
| L2 | Loir et Cher | Wheat | 21.10 | ND | 17.80 | 1,353 |
| L3 | Loir et Cher | Wheat | 7.29 | 0.18 | ND | 1,268 |
| L4 | Loir et Cher | Wheat | 14.20 | ND | ND | 1,517 |
| L5 | Loir et Cher | Wheat | 3.20 | 0.08 | ND | 1,166 |
| M11 | Charente Maritime | Durum wheat | ND | ND | ND | 1,279 |
| N1 | Gard | Durum wheat | 41.02 | 4.10 | 22.34 | 938 |
| P2 | Gers | Durum wheat | ND | ND | ND | 1,805 |
| Fcbar | Haute Garonne | Wheat | 0.05 | ND | ND | 942 |
| 131O94 | Haute Garonne | Wheat | 0.06 | ND | ND | 907 |
| R964 | Haute Garonne | Wheat | 42.45 | 38.36 | ND | 1,021 |
| O594 | Haute Garonne | Wheat | 0.05 | ND | ND | 1,285 |
| Fché2 | Haute Garonne | Wheat | 0.07 | ND | ND | 1,424 |
DON, deoxynivalenol; 3-ADON and 15-ADON, 3-acetyl- and 15-acetyldeoxynivalenol, respectively. ND, not detectable.
Toxin production.
Toxin production by the Fusarium strains was conducted on autoclaved wheat grains. Wheat grains (Soissons) were moistened with sterile distilled water for 4 days at 4°C until thermodynamic water activity was maximal. Then 100 g of grain was distributed into a 500-ml Erlenmeyer flask and sterilized twice for 25 min at 110°C. Each flask was inoculated with a suspension of 2 × 105 conidia. Incubations were conducted in duplicate for each strain. Flasks were incubated at 25°C for 25 days with daily manual shaking for the first 5 days of inoculation in order to homogenize the inoculum.
Trichothecene analysis.
Wheat grains (25 g) were analyzed by gas chromatography-electron capture detection and gas chromatography-mass spectrometry as previously described (4). Standard solutions of 1 μg of deoxynivalenol, nivalenol, fusarenone X, 3-acetyldeoxynivalenol, or 15A-deoxynivalenol (provided by Sigma, St. Louis, Mo.) were also analyzed. Detection limits were 20 ng/g for nivalenol and 10 ng/g for other trichothecenes. The results were expressed in milligrams of toxin per kilogram of dry matter or as parts per million.
Ergosterol determination.
Ergosterol analysis was carried out by high-pressure liquid chromatography as previously described (7). The results were expressed as milligrams of ergosterol per kilogram of dry matter or as parts per million.
DNA preparation.
For preparation of DNA, Fusarium strains were grown in Roux flasks for 2 days at 25°C in 150 ml of malt medium. For each strain, the mycelium from four flasks was harvested, washed with sterile water, lyophilized, and stored at −20°C. DNA for PCR was extracted as described by Dellaporta et al. (10) with slight modifications. Lyophilized mycelium (≈20 mg) was extracted with 1 ml of extraction buffer (50 mM Tris [pH 8], 50 mM EDTA [pH 8], 2% Sarcosyl, 150 mM NaCl) and 1 ml of phenol for 1 h on ice. After centrifugation (10 min at 13,000 × g), the DNA in the upper aqueous phase was precipitated by addition of 1 volume of isopropanol. After centrifugation (15 min, 13,000 × g), the DNA pellets were recovered and dissolved in 300 μl of TE (10 mM Tris [pH 8], 1 mM EDTA [pH 8]) and incubated at 37°C for 1 h with 10 μl of RNase (10 mg/ml). The extracts were purified by addition of 1 volume of phenol-chloroform-isoamyl alcohol (25:24:1) and centrifuged as described above. The DNA was precipitated by addition of 1 volume of isopropanol and centrifuged. The DNA pellets were dissolved in 300 μl of sterile water.
Primers.
The primers used in this study are listed in Table 2. tox5-1 and tox5-2 allowed the amplification of a 650-bp region of the tri5 gene, including the intron (41). T1 and T2 amplified a 600-bp portion of the β-tubulin gene (42). tri6-54 was designed according to the nucleotide sequence of the tri6 gene (46), whereas N1-2, N1-2R, 4056, and 3551 were designed according to the aligned tri6-tri5 sequences of strains C1, K2, L2, and N1. Primers designed in this study were checked with Oligo4 software with the following criteria: melting temperature, limited dimer formation, and self-complementarity.
TABLE 2.
PCR primers used in this study
| Primer | Nucleotide sequence (5′ to 3′) | Target | Reference |
|---|---|---|---|
| tox5-1 | GCTGCTCATCACTTTGCTCAG | tri5 | 41 |
| tox5-2 | CTGATCTGGTCACGCTCATC | tri5 | 41 |
| T1 | AACATGCGTGAGATTGTAAGT | β-Tubulin | 42 |
| T2 | TAGTGACCCTTGGCCCAGTTG | β-Tubulin | 42 |
| tri6-54 | CTTTGATCGTGTTGCGTCTC | tri5-tri6 intergenic region | This study |
| N1-2 | CTTGTTAAGCTAAGCGTTTT | tri5-tri6 intergenic region | This study |
| N1-2R | AACCCCTTTCCTATGTGTTA | tri5-tri6 intergenic region | This study |
| 4056 | ATCCCTCAAAAACTGCCGCT | tri5-tri6 intergenic region | This study |
| 3551 | ACTTTCCCACCGAGTATTTC | tri5-tri6 intergenic region | This study |
PCR assays.
All PCRs were conducted in 50-μl reaction mixtures containing 25 pmol of each primer (Table 2), 1.25 mM deoxynucleoside triphosphate, 2 U of Sylverstar DNA polymerase (Eurogentec, Seraing, Belgium), and 1 μl (50 to 80 ng) of fungal template. The optimized PCR temperature program (Perkin Elmer Cetus) consisted of 30 cycles of 1 min at 94°C, 1 min at 55°C, and 2 min at 72°C. Control tubes without DNA template were included in each experiment. After amplification, a 7-μl aliquot was checked by gel electrophoresis. Duplex PCRs were conducted in the conditions described above.
Sequencing of PCR products.
The PCR products were purified with a GFX gel band kit (Pharmacia Amersham Biotech) as indicated by the manufacturer. DNA templates were sequenced with unlabeled primers and the Taq dideoxy terminator cycle sequencing kit (Applied Biosystems) according to the manufacturer's instructions on a 310 DNA sequencer (Applied Biosystems). Both strands were sequenced for each PCR product (tox5-1 and tox5-2 for tri5 and T1 and T2 for β-tubulin).
Amplification, cloning, and sequencing of tri6-tri5 region.
The tri6-tri5 region from F. culmorum strains C1, K2, N1, L2, G1, and M11 was amplified by with the tri6-54 and tox5-2 primers. The PCR conditions were the same as described above except the temperature program consisted of a denaturation step (4 min, 94°C), 35 cycles of 1 min at 94°C, 1 min at 55°C, and 7 min at 72°C, and an extension step at 72°C for 5 min. The PCR product was checked by agarose gel electrophoresis, purified with a GFX gel band kit (Pharmacia Amersham Biotech), and cloned into pGEM-T with the pGEM-T A 3600 cloning kit (Promega) according to the manufacturer's instructions. Sequencing of the insert in pGEM-T was initiated by using PU and PR primers and extended, as described above, with primers corresponding to the newly sequenced regions.
DNA sequence analysis.
tri5 sequence alignments were performed with the Multalin software (http://www.toulouse.inra.fr/multalin.html) designed by Corpet (8). Unweighted maximum parsimony analyses were performed on this data set with PAUP version 4.0 (52), with heuristic search mode and 1,000 parsimony bootstrap replications.
Nucleotide sequence accession numbers.
The sequences of the tri5-tri6 intergenic regions of F. culmorum strains C1, K2, L2, N1, G1, and M11 have been deposited in GenBank under accession numbers AF480834, AF480835, AF480836, AF480837, AY134892, and AY134893, respectively.
RESULTS
F. culmorum identification.
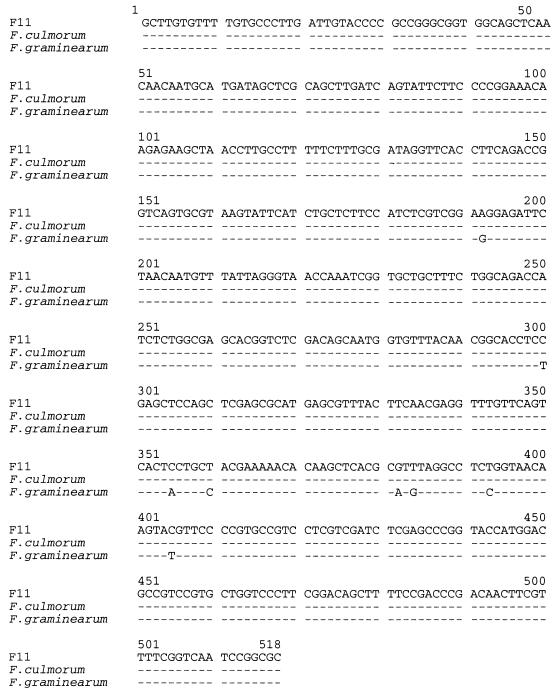
The Fusarium strains studied were isolated from commercial wheat kernels. Morphological identification of F. culmorum strains was confirmed by PCR analysis with F. culmorum-specific Scars primers: all samples had a common band of a 450-bp amplified DNA, as described by Schilling et al. (49). In addition, a portion of the β-tubulin gene of strains K2, F11, L2, J11, H1, P2, G1, and M11 was sequenced (Fig. 1). The β-tubulin region sequences were identical and were also identical to the published β-tubulin gene of F. culmorum strains NRRL 3288 and NRRL 25475 (GenBank accession numbers FCU85569 and AF006362, respectively). These data and the morphological examination confirmed that the Fusarium strains studied belonged to the same species, F. culmorum.
FIG. 1.
Sequences of β-tubulin gene region of F. culmorum strains and comparison with published data. Matches are indicated by a dash. The nucleotide sequences of the β-tubulin genes of F. culmorum strains F11, L2, H1, J1, P2, G1, K2, and M11 were identical. F. culmorum strains used for alignment were F. culmorum strains NRRL 3288 (GenBank accession number FCU85569) and F. culmorum NRRL 25475 (GenBank accession number AF006362). For these strains, the nucleotide sequences of the β-tubulin gene region studied were identical. The F. graminearum strains used for alignment were NRRL 29169 (GenBank accession number AF212751), NRRL 28434 (AF212750), NRRL 6394 (AF212748), NRRL 28720 (AF212746), NRRL 13818 (AF212745), NRRL 6101 (AF212744), NRRL 28305 (AF107876), NRRL 28302 (AF107874), NRRL 26895 (AF107860), NRRL 26158 (AF107858), NRRL 26157 (AF107857), NRRL 26156 (AF107856), NRRL 26755 (AF212743), NRRL 26754 (AF212742), NRRL 26752 (AF212741). For these strains, the nucleotide sequences of the β-tubulin gene region studied were identical.
Trichothecene production.
Trichothecene production by the 30 F. culmorum strains studied is presented in Table 1. Seventeen strains produced more than 1 ppm of deoxynivalenol and were considered high-producing strains, whereas 13 F. culmorum strains produced less than 0.07 ppm of deoxynivalenol and were considered low-producing strains. The ergosterol content was used as a measure of the fungal biomass and ranged from 898 to 1,805 mg/kg, which indicates significant growth and is in accordance with previous data (2, 33).
The lack of correlation between toxin production and ergosterol level shows that the variation in the toxigenic potential of F. culmorum strains is not explained simply by growth differences.
No detectable differences in morphological characteristics could be observed between the two types of deoxynivalenol-producing strains.
Comparison of tri5 gene sequences.
For all strains, a 650-bp PCR product was obtained with the tox5-1 and tox5-2 primers as described by Niessen and Vogel (41). A 550-base portion of the tri5 gene, which represent 55% of he tri5 gene coding region, was sequenced on both strands (Fig. 2). This sequence showed high homology (93 to 99%) to the published tri5 gene sequences of G. zeae, a Fusarium species closely related to F. culmorum (GenBank accession numbers AF336365, AF336366, AF359361, and GZU22464). As previously described (22, 41), a 60-nucleotide intron region ranging from nucleotide 288 to nucleotide 347 was observed.
FIG. 2.
Alignment of the tri5 gene region of high and low deoxynivalenol producers. -, same sequence; *, deletion; bold letter, tri5 intron. The nucleotide sequences of the tri5 genes of F. culmorum strains A2, A4, B3, B5, C6, C8, E1, E2, G1, H1, I1, L2, L3, L4, L5, N1, and R964 were identical. The nucleotide sequences of the tri5 genes of F. culmorum strains B11, C1, C11, F11, J11, K2, K11, M11, P2, Fc bar, 131094, 0594, and Fché2 were identical.
According to the tri5 data, the F. culmorum strains clustered into two groups that correlated with in vitro deoxynivalenol production. The sequence variations between the two types of tri5 gene (35 of 550 bp, 6.36%) were concentrated in the intron region, which carried 40% of the sequence differences. In particular, an 8-nucleotide deletion was observed for the low-producing F. culmorum strains. Besides the intron region, differences among strains were mainly restricted to isolated nucleotide substitutions.
Comparison of tri5-tri6 intergenic region.
In order to confirm the clustering of high-producing and low-producing F. culmorum strains, the tri5-tri6 intergenic region was further sequenced for three high-deoxynivalenol-producing strains (G1, L2, and N1) and three low-deoxynivalenol-producing strains (C1, K2, and M11). Sequence analysis clearly confirmed the two separate clusters within the F. culmorum strains corresponding exactly to in vitro deoxynivalenol production (Table 3).
TABLE 3.
Comparison of tri5-tri6 intergenic sequences of six F. culmorum strains
| Strain | Difference (%) vs F. culmorum strain:
|
||||
|---|---|---|---|---|---|
| C1 | M11 | L2 | N1 | G1 | |
| K2 | 0.42 | 0.56 | 13.19 | 13.44 | 13.33 |
| C1 | 0.24 | 13.15 | 13.21 | 13.11 | |
| M11 | 13.59 | 13.86 | 13.57 | ||
| L2 | 0.22 | 0.07 | |||
| N1 | 0.19 | ||||
Amplification with specific primers.
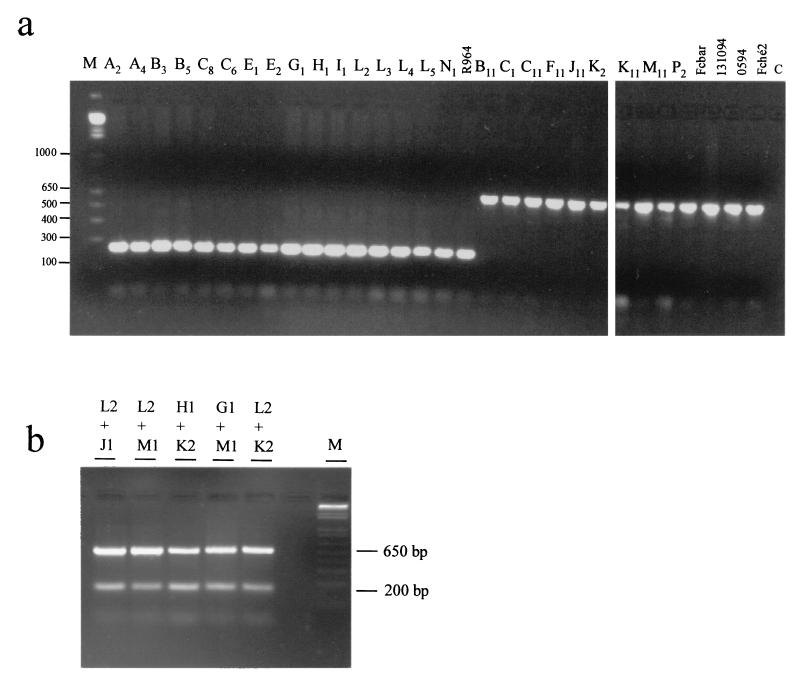
According to the tri5-tri6 sequences, PCR primers were designed in order to differentiate the high-producing from the low-producing F. culmorum strains. Amplification with N1-2 and N1-2R yielded a 200-bp fragment for the high-producing strains, whereas no amplification was observed for the low-producing strains. Conversely, with the 4056 and 3551 primers, amplification yielded a 650-bp fragment for the low-producing strains, whereas no amplification was observed for the high-producing strains (Table 1). A duplex PCR with the N1-2 and N1-2R and the 4056 and 3551 primer pairs was also conducted, which resulted in differentiation of the high-producing from the low-producing F. culmorum strains (Fig. 3).
FIG. 3.
Duplex PCR amplification patterns of high- and low-deoxynivalenol-producing strains of F. culmorum (a) with one Fusarium strain per PCR and (b) with two Fusarium strains per PCR. Lane M, size markers; lane C, control (without DNA). Amplification with primer pair N1-2 and N1-2R yielded a ≈200-bp fragment; amplification with primer pair 4056 and 3551 yielded a ≈650-bp fragment. Sizes are shown on the left of panel a in base pairs.
DISCUSSION
Trichothecene biosynthesis may be regulated by environmental conditions such as temperature (20, 56), water activity (12, 20), substrate composition (13, 44), etc. Nevertheless, it is also well established that, within the same species and under optimal conditions of growth, some strains produce large amounts of trichothecenes, whereas other strains produce small or undetectable amounts of trichothecenes (15, 19, 24, 30, 35, 36, 57). Our results are consistent with these observations.
All the F. culmorum strains tested possessed the tri5 gene, which is in good accordance with the literature, which reports F. culmorum as a potential trichothecene-producing species (11, 41). However, the tri5 sequence data for the 30 F. culmorum deoxynivalenol-producing strains revealed low but significant intraspecific variations that strictly correlated with the in vitro deoxynivalenol production. This correlation was confirmed through the tri5-tri6 intergenic sequence analysis. To our knowledge, this is the first report of such a correlation between in vitro deoxynivalenol production and tri gene sequences.
Differences in the sequence of the tri5 gene region have been reported previously in other species of Fusarium. In the case of G. pulicaris (F. sambucinum), producing diacetoxyscirpenol, a type A trichothecene, Hohn and coworkers (24) observed a 42-nucleotide tandem repeated sequence upstream of the tri5 gene that was only present in the strains producing large amounts of diacetoxyscirpenol. However, this tandem repeat has not been observed in either the low-producing or high-producing strains.
In addition, Hohn et al. (25) identified a nucleotide motif (TNAGGCCT) in the tri5 promoter region of F. sporotrichioides that is necessary for the binding of Tri6 protein, which is involved in regulation of the tri genes, including the tri5 gene. The authors indicated that nucleotide changes in the TNAGGCCT sequences dramatically reduced Tri6 binding and consequently the expression of the tri genes. For the F. culmorum strains studied, three possible tri6-binding motifs were located upstream of the tri5 gene, with identical sequences for both high-producing and low-producing strains. These nucleotide sequences therefore could not account for the variation in deoxynivalenol production.
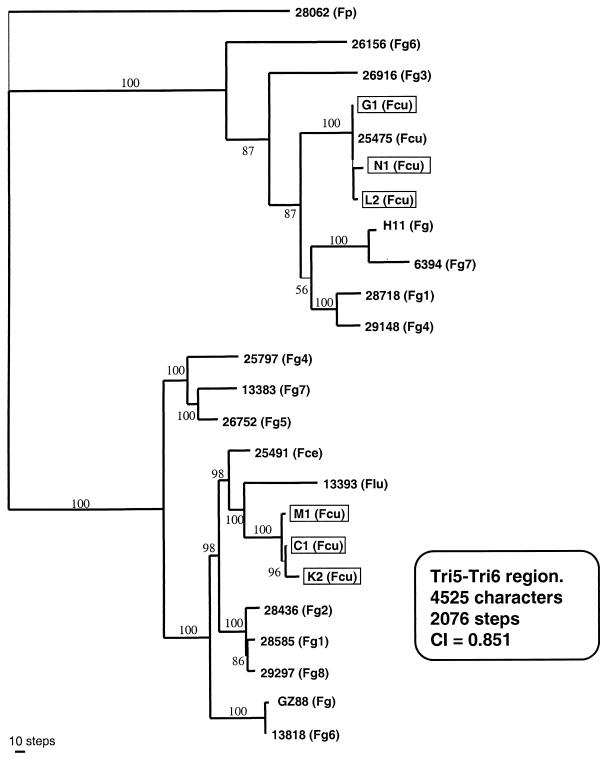
Recently, the tri gene cluster has been sequenced for an F. sporotrichioides strain (6), G. zeae (F. graminearum), and F. culmorum strains, including deoxynivalenol-producing strains and nivalenol-producing strains (6, 29, 58). According to the chemotype definition, it can be assumed that the nivalenol-producing G. zeae strains produced small or nondetectable amounts of deoxynivalenol. We compared these data with ours through a parsimony analysis of the tri6-tri5 region of these F. graminearum strains and the six F. culmorum strains used in our study (Fig. 4). Whatever the Fusarium species considered, the tree obtained formed two groups, corresponding to the high-producing and low-producing strains. Further studies are in progress to validate the PCR primers developed in our study with different deoxynivalenol-producing Fusarium species, such as F. graminearum.
FIG. 4.
Comparison of tri5-tri6 region of the six F. Culmorum strains studied with published sequences. One of the nine most parsimonious unrooted trees (differing only in the position of strains in the Fcu clade) inferred from the analysis of the tri5-tri6 region. Framed blocks indicate the strains sequenced in the present study; F. graminearum complex (Fg 1 to 8), F. lunulosporum (Flu), F. cerealis (Fce), F. culmorum (Fcu), and F. pseudograminearum (Fp) were compared. Strains 28062, 26156, 26916, 25475, H11,6394, 28718, and 29148 are described as deoxynivalenol-producing strains, whereas strains 25797, 13383, 25491, 13393, 28436, 28585, 29297, and Gz88 are nivalenol-producing strains. Bootstrap values were calculated by using 1,000 replicates. CI, confidence interval.
Deoxynivalenol-producing Fusarium strains are commonly isolated from cereals (34). It has been demonstrated by using naturally infected barleys that de novo Fusarium growth and deoxynivalenol production is a potential problem during malting (17, 50). In addition, several studies indicate that deoxynivalenol is not destroyed during brewing (50, 51). As a consequence, development of a rapid detection method for strains producing large amounts of deoxynivalenol would be of great practical importance.
Acknowledgments
We thank D. Marion and K. Elmorjani for valuable discussions on the manuscript.
REFERENCES
- 1.Abbas, H. K., C. J. Mirocha, R. A. Meronuck, J. D. Pokorny, S. L. Gould, and T. Kommedahl. 1988. Mycotoxins and Fusarium spp. associated with infected ears of corn in Minnesota. Appl. Environ. Microbiol. 54:1930-1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberts, J. F., W. C. A. Gelderblom, P. G. Thiel, W. F. O. Marasas, D. J. Van Schalkwyk, and Y. Berhend. 1990. Effects of temperature and incubation period on production of fumonisin B1 by Fusarium moniliforme. Appl. Environ. Microbiol. 56:1729-1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Atanassov, Z., C. Nakamura, N. Mori, C. Kaneda, H. Kato, Y. Z. Jin, T. Yoshizawa, and K. Murai. 1994. Mycotoxin production and pathogenicity of Fusarium species and wheat resistance to Fusarium head blight. Can. J. Bot. 72:161-167. [Google Scholar]
- 4.Bakan, B., L. Pinson, B. Cahagnier, D. Melcion, E. Sémon, and D. Richard-Molard. 2001. Toxigenic potential of Fusarium culmorum strains isolated from French wheat. Food Addit. Contam. 18:998-1003. [DOI] [PubMed] [Google Scholar]
- 5.Blaney, B. J., and R. L. Dodman. 1988. Production of the mycotoxins zearalenone, 4-deoxynivalenol, and nivalenol by isolates of Fusarium graminearum groups 1 and 2 from cereals in Queensland. Aust. J. Agric. Res. 39:21-29. [Google Scholar]
- 6.Brown, D. W., S. P. McCormick, N. J. Alexander, R. H. Proctor, and A. E. Desjardins. 2001. A genetic and biochemical approach to Study trichothecene diversity in Fusarium sporotrichioides and Fusarium graminearum. Fungal Genet. Biol. 32:121-133. [DOI] [PubMed] [Google Scholar]
- 7.Cahagnier, B., L. Lesage, and D. Richard-Molard. 1993. Mould growth and conidiation in cereal grains as affected by water activities. Lett. Appl. Microbiol 17:7-13. [Google Scholar]
- 8.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalcero, A., A. Torres, M. Etcheverry, S. Chulze, and E. Varsavsky. 1997. Occurrence of deoxynivalenol and Fusarium graminearum in Argentinian wheat. Food Addit. Contam. 14:11-14. [DOI] [PubMed] [Google Scholar]
- 10.Dellaporta, S., J. Wood, and J. Hicks. 1983. A plant DNA minipreparation. Plant Mol. Biol. Rep. 1:19-21. [Google Scholar]
- 11.Edwards, S., S. Pirgozliev, M. Hare, and P. Jenkinson. 2001. Quantification of trichothecene-producing Fusarium species in harvested grain by competitive PCR to determine efficacies of fungicides against Fusarium head blight of winter wheat. Appl. Environ. Microbiol. 67:1575-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El-Maghraby, O. M. O., I. A. El-Kady, and S. Soliman. 1995. Mycoflora and Fusarium toxins of three types of corn grains in Egypt with special reference to production of trichothecene-toxins. Microbiol. Res. 150:225-232. [DOI] [PubMed] [Google Scholar]
- 13.Engelhardt, G., M. Schuster, J. Lepschy, and P. R. Wallnöfer. 1986. Production of mycotoxins by Fusarium species isolated in Germany. Z. Lebensm. Unters Forsch. 182:123-126. [DOI] [PubMed] [Google Scholar]
- 14.Eriksen, G. S., and J. E. Alexander. 1998. Fusarium toxins in cereals — a risk assessment. Nordic Council of Ministers, Copenhagen, Denmark.
- 15.Faifer, G., M. S. De Miguel, and H. M. Godoy. 1990. Patterns of mycotoxin production by Fusarium graminearum isolated from Argentine wheat. Mycopathologia 109:165-170. [Google Scholar]
- 16.Fekete, C., A. Logrieco, G. Giczey, and L. Hornok. 1997. Screening of fungi for the presence of the trichodiene synthase encoding sequence by hybridization to the Tri5 gene cloned from Fusarium poae. Mycopathologia 138:91-97. [DOI] [PubMed] [Google Scholar]
- 17.Flannigan, B., S. W. Day, P. E. Douglas, and G. B. McFarlane. 1984. Growth of mycotoxin-producing fungi associated with malting of barley. Dev. Food Sci. 7:52-60. [Google Scholar]
- 18.Fujisawa, T., M. Mori, and M. Ichinoe. 1992. Natural occurrence of trichothecenes in domestic wheats and barley harvested in 1990 and 1991, and production of mycotoxins by Fusarium isolates from these samples. Proc. Jpn. Assoc. Mycotoxicol. 35:37-39. [Google Scholar]
- 19.Gang, G., T. Miedaner, U. Schuhmacher, M. Schollenberger, and H. H. Geiger. 1998. Deoxynivalenol and nivalenol production by Fusarium culmorum isolates differing in aggressiveness toward winter rye. Phytopathology 88:879-884. [DOI] [PubMed] [Google Scholar]
- 20.Grennhalgh, R., G. A. Neish, and J. D. Miller. 1983. Deoxynivalenol, acetyl deoxynivalenol, and zearalenone formation by Canadian isolates of Fusarium graminearum on solid substrates. Appl. Environ. Microbiol. 46:625-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guadet, J., J. Julien, J. F. Lafay, and Y. Brygoo. 1989. Phylogeny of some Fusarium species, as determined by large-subunit rRNA sequence comparison. Mol. Biol. Evol. 6:227-242. [DOI] [PubMed] [Google Scholar]
- 22.Hohn, T., and M. Beremand. 1989. Isolation and nucleotide sequence of a sesquiterpene cyclase gene from the trichothecene-producing fungus Fusarium sporotrichioides. Gene 79:131-138. [DOI] [PubMed] [Google Scholar]
- 23.Hohn, T., and A. E. Desjardins. 1992. Isolation and gene disruption of the tox5 gene encoding trichodiene synthase in Gibberella pulicaris. Mol. Plant-Microbe Interact. 5:249-256. [DOI] [PubMed] [Google Scholar]
- 24.Hohn, T., A. E. Desjardins, and S. P. MacCormick. 1993. Analysis of tox5 gene expression in Gibberella pulicaris strains with different trichothecene production phenotypes. Appl. Environ. Microbiol. 59:2359-2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hohn, T. M., R. Krishna, and R. H. Proctor. 1999. Characterization of a transcriptional activator controlling trichothecene toxin biosynthesis. Fungal Genet. Biol. 26:224-235. [DOI] [PubMed] [Google Scholar]
- 26.Ichinoe, M., H. Kurata, Y. Sugiura, and Y. Ueno. 1983. Chemotaxonomy of Gibberella zeae with special reference to production of trichothecenes and zearalenone. Appl. Environ. Microbiol. 46:1364-1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim, J. C., H. J. Kang, D. H. Lee, Y. W. Lee, and T. Yoshizawa. 1993. Natural occurrence of Fusarium mycotoxins (trichothecenes and zearalenone) in barley and corn in Korea. Appl. Environ. Microbiol. 59:3798-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Langseth, W., A. Bernhoft, T. Rundberget, B. Kosiak, and M. Gareis. 1999. Mycotoxin production and cytotoxicity of Fusarium strains isolated from Norwegian cereals. Mycopathologia 144:103-113. [DOI] [PubMed] [Google Scholar]
- 29.Lee, T., O. Dae-Woong, K. Hye-Seon, L. Jungkwan, K. Yong-Ho, Y. Sung-Hwan, and L. Yin-Won. 2001. Identification of deoxynivalenol and nivalenol producing chemotypes of Gibberella zeae by with PCR. Appl. Environ. Microbiol. 67:2966-2972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Manka, M., A. Visconti, J. Chelkowski, and A. Bottalico. 1985. Pathogenicity of Fusarium isolates from wheat, rye and triticale towards seedlings and their ability to produce trichothecenes and zearalenone. Phytopathology 13:24-29. [Google Scholar]
- 31.Marasas, W. F. O., P. E. Nelson, and T. A. Toussoun. 1984. Toxigenic Fusarium species: identity and mycotoxicology. Pennsylvania State University Press, University Park. Pa.
- 32.Matsumoto, G., J. Wuchiyama, Y. Shingu, M. Kimura, K. Yoneyama, and I. Yamaguchi. 1999. The trichothecene biosynthesis regulatory gene from the type B producer Fusarium strains: sequence of Tri6 and its expression in Escherichia coli. Biosci. Biotechnol. Biochem. 63:2001-2004. [DOI] [PubMed] [Google Scholar]
- 33.Melcion, D., B. Cahagnier, B. Bakan, and D. Richard-Molard. 1998. Influence of temperature on fumonisin B1 production on maize grain by Fusarium proliferatum. Sci. Aliment. 18:301-311. [Google Scholar]
- 34.Miller, J. D. 1994. Epidemiology of Fusarium ear diseases of cereals, p. 19-36. In J. D. Miller and H. L. Trenholm (ed.), Mycotoxins in grain: compound other than aflatoxin. Eagen Press, St. Paul, Minn.
- 35.Miller, J. D., R. Grennhalgh, Y. Wang, and M. Lu. 1991. Trichothecene chemotypes of three Fusarium species. Mycologia 83:121-130. [Google Scholar]
- 36.Mirocha, C. J., H. K. Abbas, C. E. Windels, and W. Xie. 1989. Variation in deoxynivalenol, 15-acetyldeoxynivalenol, 3-acetyldeoxynivalenol, and zearalenone production by Fusarium graminearum isolates. Appl. Environ. Microbiol. 55:1315-1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mulè, G., A. Logrieco, G. Stea, and A. Bottalico. 1997. Clustering of trichothecene-producing Fusarium strains determined from 28S ribosomal DNA sequences. Appl. Environ. Microbiol. 63:1843-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Muller, H. M., J. Reimann, U. Schumacher, and K. Schwadorf. 1997. Occurrence of Fusarium toxins in barley harvested during five years in an area of southwest Germany. Mycopathologia 137:185-192. [DOI] [PubMed] [Google Scholar]
- 39.Muthomi, J. W., A. Schütze, H. W. Dehne, E. W. Mutitu, and E. C. Oerke. 2000. Characterization of Fusarium culmorum isolates by mycotoxin production and aggressiveness to winter wheat. Z. Pflanzenk. Pflanzen. 107:113-123. [Google Scholar]
- 40.Nelson, P. E., T. A. Toussoun, and W. F. O. Marasas. 1983. Fusarium species: an illustrated manual for identification. Pennsylvania State University Press, University Park, Pa.
- 41.Niessen, M. L., and R. F. Vogel. 1998. Group specific PCR-detection of potential trichothecene-producing Fusarium species in pure cultures and cereal samples. Syst. Appl. Microbiol. 21:618-631. [DOI] [PubMed] [Google Scholar]
- 42.O'Donnell, K., and E. Cigelnik. 1997. Two divergent intragenomic rDNA ITS2 types within a monophyletic lineage of the fungus Fusarium are nonorthologous. Mol. Phylogenet. Evol. 7:103-116. [DOI] [PubMed] [Google Scholar]
- 43.O'Donnell, K., E. Cigelnik, and H. H. Casper. 1998. Molecular phylogenetic, morphological, and mycotoxin data support reidentification of the Quorn mycoprotein fungus as Fusarium venenatum. Fungal Genet. Biol. 23:57-67. [DOI] [PubMed] [Google Scholar]
- 44.O'Neill, K., A. P. Damoglou, and M. F. Patterson. 1993. Toxin production by Fusarium culmorum IMI 309344 and F. graminearum NRRL 5883 on grain substrates. J. Appl. Bacteriol. 74:625-628. [DOI] [PubMed] [Google Scholar]
- 45.Park, J. J., E. B. Smalley, and F. S. Chu. 1996. Natural occurrence of Fusarium mycotoxins in field samples from the 1992 Wisconsin corn crop. Appl. Environ. Microbiol. 62:1642-1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Proctor, R. H., T. M. Hohn, S. P. McCormick, and A. E. Desjardins. 1995. Tri6 encodes an unusual zinc finger protein involved in regulation of trichothecene biosynthesis in Fusarium sporotrichioides. Appl. Environ. Microbiol. 61:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rotter, B. A., D. B. Prelusky, and J. J. Pestka. 1996. Toxicology of deoxynivalenol (vomitoxin). J. Toxicol. Environ. Health 48:1-34. [DOI] [PubMed] [Google Scholar]
- 48.Ryu, J. C., J. S. Yang, Y. S. Song, O. S. Kwon, J. Park, and I. M. Chang. 1996. Survey of natural occurrence of trichothecene mycotoxins and zearalenone in Korean cereals harvested in 1992 with gas chromatography/mass spectrometry. Food Addit. Contam. 13:333-341. [DOI] [PubMed] [Google Scholar]
- 49.Schilling, A. G., E. M. Moller, and H. H. Geiger. 1996. Polymerase chain reaction-based assays for species-specific detection of Fusarium culmorum, Fusarium graminearum and F. avenaceum. Phytopathology 86:515-522. [Google Scholar]
- 50.Schwarz, P. B., H. H. Casper, and S. Beattie. 1995. Fate and development of naturally occurring Fusarium mycotoxins during malting and brewing. J. Am. Soc. Brew. Chem. 53:121-127. [Google Scholar]
- 51.Scott, P. M. 1996. Mycotoxins transmitted into beer from contaminated grains during brewing. J. AOAC Int. 79:875-882. [PubMed] [Google Scholar]
- 52.Swofford, D. L. 1998. PAUP: phylogenetic analysis with parsimony, version 4.0. Sinauer Associates, Sunderland, Mass.
- 53.Sydenham, E. W., W. F. O. Marasas, P. G. Thiel, G. S. Shepard, and J. J. Nieuwenhius. 1991. Production of mycotoxins by selected Fusarium graminearum and F. crookwellense isolates. Food Addit. Contam. 8:31-41. [DOI] [PubMed] [Google Scholar]
- 54.Tanaka, T., A. Hasegawa, S. Yamamoto, U. S. Lee, Y. Sugiura, and Y. Ueno. 1988. Worldwide contamination of cereals by the Fusarium mycotoxins nivalenol, deoxynivalenol and zearalenone. 1. Survey of 19 countries. J. Agric. Food Chem. 36:979-983. [Google Scholar]
- 55.Trenholm, H. L., W. P. Cochrane, H. Cohen, J. I. Elliot, E. R. Farnworth, D. W. Friend, R. M. G. Hamilton, G. A. Neish, and J. F. Standish. 1981. Survey of vomitoxin contamination of the 1980 white winter wheat crop in Ontario, Canada. J. Am. Oil Chem. Soc. 66:992A-994A. [PubMed] [Google Scholar]
- 56.Vesonder, R. F., J. J. Ellis, W. F. Kwolek, and D. J. DeMarini. 1982. Production of vomitoxin on corn by Fusarium graminearum NRRL 5883 and Fusarium roseum NRRL 6101. Appl. Environ. Microbiol. 43:967-970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker, S., S. Leath, W. Hagler, and J. Murphy. 2001. Variation among isolates of Fusarium graminearum associated with Fusarium head blight in north Carolina. Plant Dis. 85:404-410. [Google Scholar]
- 58.Ward, T., J. Bielawski, H. Kistler, E. Sullivan, and K. O'Donnell. 2002. Ancestral polymorphism and adaptive evolution in the trichothecene mycotoxin gene cluster of phytopathogenic Fusarium. Proc. Natl. Acad. Sci. USA 99:9278-9283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yoshizawa, T., and Y. Z. Jin. 1995. Natural occurrence of acetylated derivatives of deoxynivalenol and nivalenol in wheat and barley in Japan. Food Addit. Contam. 12:689-694. [DOI] [PubMed] [Google Scholar]