Abstract
PAZ PIWI domain (PPD) proteins, together with the RNA cleavage products of Dicer, form ribonucleoprotein complexes called RNA-induced silencing complexes (RISCs). RISCs mediate gene silencing through targeted messenger RNA cleavage and translational suppression. The PAZ domains of PPD and Dicer proteins were originally thought to mediate binding between PPD proteins and Dicer, although no evidence exists to support this theory. Here we show that PAZ domains are not required for PPD protein–Dicer interactions. Rather, a subregion of the PIWI domain in PPD proteins, the PIWI-box, binds directly to the Dicer RNase III domain. Stable binding between PPD proteins and Dicer was dependent on the activity of Hsp90. Unexpectedly, binding of PPD proteins to Dicer inhibits the RNase activity of this enzyme in vitro. Lastly, we show that PPD proteins and Dicer are present in soluble and membrane-associated fractions, indicating that interactions between these two types of proteins may occur in multiple compartments.
Introduction
The RNA interference (RNAi) machinery is used by eukaryotes to inhibit gene expression by sequence-specific cleavage of messenger RNA (mRNA) or translational repression (Hannon, 2002; McManus & Sharp, 2002; Schwarz & Zamore, 2002). The specificity of these processes is dependent on 21–22-nt small interfering RNAs (siRNAs) or microRNAs (miRNAs) produced from doublestranded (ds) RNAs or hairpin precursors, respectively. In some eukaryotes, the RNAi apparatus also participates in the control of chromatin structure, although it is not clear how this process occurs (Schramke & Allshire, 2003, and references therein).
Two classes of proteins are central to RNAi: members of the PAZ PIWI domain (PPD) and Dicer families. PPD proteins are required at the effector stages of RNAi (Hammond et al, 2001) and in some cases for maturation of miRNAs (Grishok et al, 2001). Family members are defined by the presence of a central PAZ domain and a carboxy terminal PIWI domain (Carmell et al, 2002). Perhaps the best-characterized proteins in RNAi pathways are the Dicer enzymes, which are dsRNAspecific RNases that cleave dsRNA into 21–25-nt base-paired fragments (Bernstein et al, 2001; Billy et al, 2001; Ketting et al, 2001). Dicer cleavage products are incorporated into PPD-containing RNA-induced silencing complexes (RISCs), which presumably mediate mRNA degradation, translational suppression or chromatin silencing (Hannon, 2002; McManus & Sharp, 2002).
Engagement of Dicer with PPD proteins is probably required for the transfer of siRNAs/miRNAs to RISCs (Zamore, 2002). Indeed, physical interactions between PPD proteins and Dicer have been demonstrated (Hammond et al, 2001; Sasaki et al, 2003), but such interactions have not been well characterized. The situation appears to be more complicated in some invertebrate metazoans in which dsRNA-binding proteins such as RDE-4 or R2D2 are thought to link Dicer enzymes to RISCs (Tabara et al, 1999; Parrish & Fire, 2001; Liu et al, 2003). Interestingly, homologues of RDE-4 and R2D2 have not been identified in humans or other mammalian species, and thus transfer of siRNAs/miRNAs to mammalian RISCs may use different mechanisms. Initially, heterotypic PAZ–PAZ interactions were thought to mediate the formation of PPD–Dicer complexes, but this seems unlikely now because a number of Dicer isoforms that do not have PAZ domains have been described (Schauer et al, 2002). In addition, a fragment of the human PPD protein EIF2C1 (AGO1) that contained the PIWI domain interacted more strongly with Dicer than a region containing only the PAZ domain (Doi et al, 2003).
In this study, we have characterized in detail the interactions between Dicer and two distantly related PPD proteins, Hiwi and AGO2. We report that direct interactions between PPD proteins and Dicer do not require PAZ domains, but are rather mediated through their PIWI and RNase III domains. Surprisingly, the binding of PPD protein to Dicer inhibits RNase activity in vitro. Finally, we found that interactions between PPD and Dicer proteins are dependent on Hsp90 activity.
Results and Discussion
Mapping of interaction domains
The nature of PPD–Dicer interactions, which is probably important for RNAi, is poorly understood. As a first step towards a more thorough understanding of this process, we mapped the regions within PPD proteins and Dicer that mediate such interactions. Two distantly related human PPD proteins, AGO2 (also known as GERp95) (Cikaluk et al, 1999) and Hiwi (Sharma et al, 2001), were chosen for these studies (Fig 1A). Along their entire lengths, the level of identity between these two proteins is approximately 22.7%.
Figure 1.
PIWI domains interact with Dicer. (A) Schematic representation of AGO2 and Hiwi GST-fusion proteins used for binding studies. PIWI domains, PIWI-box (Pb) and PAZ domains are indicated. (B) GST and GST–PPD fusion proteins were co-expressed with CFP–Dicer in HEK293T cells. Proteins were purified on glutathione–sepharose beads, resolved by SDS–PAGE, and visualized by immunoblotting with antibodies to GST and GFP.
To establish whether either of the two PPD signature domains is sufficient for binding to Dicer, the PAZ and PIWI domains of AGO2 and Hiwi were co-expressed as GST-fusions together with CFP–Dicer in HEK293T cells followed by isolation on glutathione–sepharose beads. Bound fractions were analysed by SDS–PAGE and immunoblotting with antibodies (Abs) to GST or GFP (Fig 1B). Stable interactions between CFP–Dicer and the AGO2 or Hiwi PAZ domains were not detected; however, the PIWI domains of AGO2 and Hiwi efficiently bound to CFP–Dicer (Fig 1B). During the preparation of this work, a study was published showing that a region of AGO1 that includes the PIWI domain bound to Dicer (Doi et al, 2003). However, this study did not address whether this was the case for other PPD proteins nor did it identify the region of Dicer that binds to AGO1.
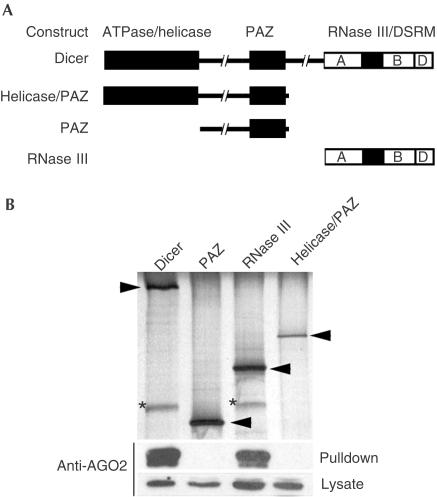
To determine which Dicer domain interacts with PPD proteins, plasmids encoding GST fused to Dicer or its subdomains (Fig 2A) were transfected into HEK293T cells stably overexpressing AGO2, and lysates were prepared and subjected to glutathione–sepharose pulldown assays. These experiments clearly show that the PAZ domain of Dicer is not required for binding to AGO2 (Fig 2B). Rather, AGO2 bound only to full-length Dicer or its carboxy terminal region, a region that lacks the PAZ domain but contains the dsRNA-binding domain (DSRM) and RNase III domains (Fig 2A). Together, these results indicate that PAZ domains are not involved in interactions between PPD and Dicer proteins.
Figure 2.
The Dicer RNase III domain is required for binding to AGO2. (A) Schematic representation of Dicer and GST-fusion proteins used for binding studies. The three regions of Dicer are indicated: ATPase/helicase, PAZ and RNase III domains. The RNase III domain is further subdivided to indicate the presence of two RNase III motifs A and B and a dsRNA-binding domain (DSRM; D). (B) Plasmids encoding GST or GST fused to domains of Dicer were transfected into a cell line overexpressing AGO2. Following GST pulldowns, proteins were visualized by silver staining (top panel) and immunoblotting (bottom panels). Arrowheads point to the silverstained GST-fusion proteins, whereas AGO2 is indicated by asterisks.
PPD–Dicer interactions are direct
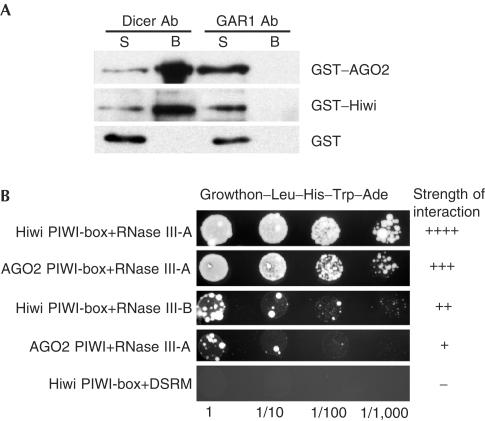
We investigated whether associations between Dicer and PPD proteins are direct by first analysing interactions between purified proteins. Human Dicer was expressed in insect cells and purified as described (Zhang et al, 2002). GST–PPD fusion proteins were expressed in COS1 cells and purified on glutathione–sepharose beads (Tahbaz et al, 2001). Protein A–sepharose beads coated with anti-Dicer Ab (Billy et al, 2001) or as a control, with Ab against human GAR1, were used for the immunoprecipitation of complexes of Dicer with GST–AGO2, GST–Hiwi or GST alone (Fig 3A). Proteins co-immunoprecipitating with Dicer were visualized by immunoblotting, using anti-GST Ab. GST–AGO2 and GST–Hiwi were retained by anti-Dicer beads but not control anti-GAR1 beads (Fig 3A, upper and middle panels) or noncoated Protein A beads (not shown). Neither anti-Dicer nor anti-GAR1 beads retained GST alone (Fig 3A, lower panel). To rule out the possibility that doublestrand or single-strand RNAs are required for stable binding of Dicer to PPD proteins, nuclease (micrococcal nuclease or a cocktail of RNases V1 and A) digestion steps were included before pulldowns and immunoblot analyses. The nuclease treatments had no measurable effects on the stability of the Dicer–PPD protein complexes (data not shown). Together, these data suggest that interactions between Dicer and PPD proteins are direct.
Figure 3.
PPD–Dicer interactions are direct. (A) Protein A–sepharose beads coated with affinity-purified Abs against Dicer or the control protein GAR1 were incubated with mixtures of purified Dicer and GST–AGO2 (upper panel), GST–Hiwi (middle panel) or GST alone (lower panel). Bead-associated (B) and unbound (S) material was subjected to SDS–PAGE and immunoblotting with anti-GST Abs. (B) Different combinations of pGBKT7 (AGO2 or Hiwi subdomains) and pGADT7 (Dicer subdomains) constructs were transformed into AH109 strain and tested for interaction by growth on selective media. Four serial dilutions were spotted for each combination. Scoring ranged from − to ++++ depending upon how well the strains grew on media lacking leucine, tryptophan, adenine and histidine. Graded examples of some PPD–Dicer interactions are shown. A complete listing of all the tested interactions is shown in supplementary table 2 online.
To eliminate the possibility that proteins co-purifying with either Dicer or GST–PPD preparations are involved in binding, we further investigated Dicer–PPD protein interactions using the yeast two-hybrid assay. Saccharomyces cerevisiae does not encode PPD or Dicer homologues and therefore, if interactions between PPD proteins and Dicer are detected in this system, it is likely that they result from direct binding. Plasmids encoding the DSRM, and the RNase III domain A or B of Dicer (see Fig 2A) fused to the activation domain of Gal-4, and plasmids encoding AGO2 and Hiwi PAZ, PIWI and PIWI-box regions (see Fig 1A) fused in-frame to the DNA-binding domain of Gal-4 were constructed. The PIWI-box is a highly conserved motif present in all PIWI domains (Cox et al, 1998). Using this method, no interactions between PAZ domains of AGO2 or Hiwi and Dicer were evident (see supplementary table 2 online). In contrast, interactions between the PIWI domains of AGO2 and Hiwi and the RNase III-A domain of Dicer were detected, although the interactions were not particularly strong. The strongest interactions occurred between the RNase III-A domain of Dicer and the PIWI-boxes of AGO2 and Hiwi (Fig 3B; see supplementary table 2 online). Relatively weak interactions were observed between the PIWI-boxes and RNase III-B. Interactions were not detected between PPD protein domains and the DSRM of Dicer (see supplementary table 2 online). Together, these results indicate that the PIWI-box of PPD proteins interacts directly with the RNase III domains of Dicer.
Dicer activity is inhibited by PPD proteins in vitro
Dicer cleavage products (siRNAs/miRNAs) are incorporated into PPD-protein-containing RISCs that are the effectors in RNAi or translation inhibition. Presumably, interactions between PPD proteins and Dicer are required for the transfer of siRNAs or miRNAs from Dicer to RISCs (Baulcombe, 2001). Indeed, previous studies have documented the interaction between PPD proteins and Dicer (Hammond et al, 2001; Doi et al, 2003), but the effects of these interactions on Dicer activity have not been investigated. Given that PPD proteins bind to the RNase III domain of Dicer, we reasoned that this interaction could affect Dicer activity. To test this possibility, the activity of purified Dicer was assayed in the presence of GST fused to AGO2 or Hiwi. A dose-dependent decrease in Dicer activity was observed in the presence of GST–AGO2 and GST–Hiwi but not of GST alone (Fig 4). Similar results were obtained when Dicer and AGO2 were incubated together before the addition of the dsRNA substrate or when the pre-let7 RNA hairpin was used as a processing substrate instead of dsRNA (data not shown).
Figure 4.
PPD proteins inhibit the RNase activity of Dicer in vitro. (A) Effect of Hiwi and AGO2 on Dicer-mediated cleavage of dsRNA. Increasing amounts (indicated in B) of GST–Hiwi, GST–AGO2 or GST alone were added to the cleavage reactions containing 130-bp 32P-labelled dsRNA substrate. Reaction products were analysed by 8 M urea PAGE. The positions of 14-, 21- and 27-nt size markers are indicated (the 21-nt marker was only visible upon longer exposure). (B) PhosphorImager quantitation of the cleavage reactions from a typical experiment.
Hsp90 activity is required for PPD–Dicer interaction
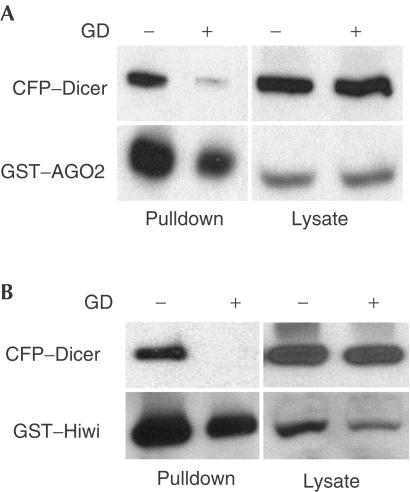
Previously, we reported that the AGO2 orthologue GERp95 interacts with an Hsp90-containing protein complex (Tahbaz et al, 2001). These interactions are thought to be important for the biogenesis of GERp95 because inhibition of Hsp90 activity with geldanamycin (GD) or radicicol resulted in the preferential degradation of nascent GERp95 and the depletion of membrane-bound pools of this protein. GD and radicicol bind specifically and stably to the ATP-binding site of Hsp90 (Prodromou et al, 1997; Stebbins et al, 1997; Schulte et al, 1998), thereby disrupting the chaperone cycle. Accordingly, these inhibitors negatively affect the activities and localizations of many Hsp90 substrates. To determine whether Hsp90 activity is important for the formation and/or stability of PPD protein–Dicer complexes, we examined the effect of GD on Dicer interactions with AGO2 and Hiwi. The association of CFP–Dicer with GST–AGO2 (Fig 5A), and GST–Hiwi (Fig 5B) was greatly inhibited by GD, indicating that Hsp90 has a role in regulating the binding of multiple PPD proteins to Dicer. The GD-dependent inhibition of Dicer–PPD interactions was confirmed by reciprocal pulldown assays using GST–Dicer expressed transiently in stable cell lines overexpressing AGO2 (data not shown). It should be noted that GD treatment caused some degradation of PPD proteins; however, during the short incubation times used here, the extent of GST–AGO2 and GST–Hiwi degradation was not sufficient to account for the near-complete GD-dependent block of Dicer binding.
Figure 5.
GD inhibits the binding of PPD proteins to Dicer. GST–AGO2 (A) or GST–Hiwi (B) were transiently co-expressed with CFP–Dicer in HEK293T cells. Cells were incubated with (+) or without (−) 5 μM GD for 2 h before lysis. Proteins were recovered on glutathione–sepharose beads and visualized by immunoblotting. CFP–Dicer was detected using a polyclonal anti-GFP Ab, whereas GST–AGO2 and GST–Hiwi were detected using anti-GST Abs.
Two cellular pools of AGO2, Hiwi and Dicer
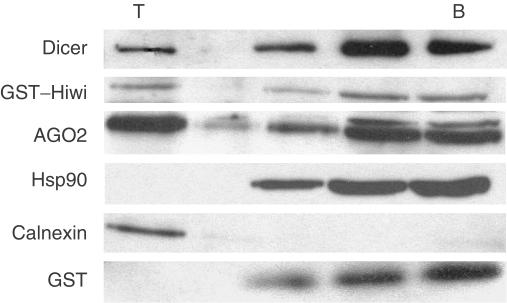
Our previous work showed that the rat orthologue of AGO2 is present in membrane-associated and cytosolic pools (Cikaluk et al, 1999; Tahbaz et al, 2001). Recent studies have shown that Dicer also localizes to the cytoplasm (Billy et al, 2001; Provost et al, 2002; Findley et al, 2003), but quantitative evidence of membrane association has not been reported. The intracellular site(s) of PPD–Dicer interactions have not been investigated, and therefore we compared the intracellular distributions of PPD proteins and Dicer in subcellular fractions. Similar to AGO2, a large pool of Dicer was present in soluble fractions, whereas a smaller but significant cohort of this RNase co-purified with membranes (Fig 6). Calnexin, an integral membrane protein of the endoplasmic reticulum, was used as a marker for the membrane fraction. Hsp90 was present only in the soluble fractions. As antibodies to Hiwi are not available, the distribution of GST–Hiwi expressed in HEK293T cells was examined. Similar to AGO2 and Dicer, GST–Hiwi was present in both soluble and membrane-associated fractions (Fig 6).
Figure 6.
Dicer and PPD proteins are present in soluble and membrane-associated pools. Homogenates of HEK293T cells transiently expressing GST–Hiwi were subjected to membrane flotation assays on discontinuous sucrose gradients. Fractions were collected from the tops (T) of gradients, TCA precipitated, resolved by SDS–PAGE and subjected to immunoblotting with Abs specific for AGO2, GST, Dicer, calnexin and Hsp90. Membrane-associated material is found at the top of the gradient, whereas soluble proteins remain in the bottom (B) three fractions.
Conclusions and Speculation
Our data indicate for the first time that interactions between Dicer and PPD proteins are direct, and that the Dicer RNase III domain is required for binding to PPD proteins. Moreover, the results from this study point towards the existence of stable Dicer–PPD complexes in multiple locations (cytosol and membranes). The existence of two pools of Dicer–PPD complexes may reflect the roles of these proteins in mRNA degradation or translation taking place on free and membrane-associated polysomes. Indeed, association of RISC complexes and siRNAs with ribosomes has been previously documented (Hammond et al, 2001; Djikeng et al, 2003). Likewise, a fraction of Dicer present in mouse P19 cell extracts sediments in the 80S region of gradients (our unpublished results), but the relevance of Dicer association with ribosomes is unclear.
Interactions between Dicer and PPD proteins (Hammond et al, 2001; Doi et al, 2003; Sasaki et al, 2003, and this work) are likely to be important for the transfer of siRNAs generated by Dicer to RISC complexes, of which PPD proteins are a part (Hammond et al, 2001; Hutvagner & Zamore, 2002; Martinez et al, 2002). Notably, by interacting with the RNase III domain of Dicer, as demonstrated here, PPD proteins are brought to the proximity of siRNAs produced by the catalytic domain of the enzyme. In this context, it is important to note that, in reactions carried out with the purified recombinant Dicer, a considerable fraction of siRNA products was found to remain associated with the enzyme (Zhang et al, 2002). Hence, the interaction with PPD proteins or RISC complexes might be expected to stimulate the activity of Dicer by facilitating the product release. However, rather unexpectedly, we found that PPD proteins inhibit the activity of the purified Dicer in vitro. It is possible that, physiologically, active Dicer molecules are not bound to PPD proteins or that Dicer–PPD protein complexes contain other components that activate Dicer or modulate the inhibitory role of PPD proteins. If the former is true, Hsp90 is a candidate that could indirectly influence Dicer activity by regulating the binding of PPD proteins.
In summary, the likely existence of two pools of Dicer–PPD complexes, together with a role for Hsp90 in PPD–Dicer associations and the inhibitory effect of PPD proteins on Dicer activity in vitro, indicates that Dicer and PPD proteins are parts of a rather complex and dynamic network of interactions important for executing siRNA and miRNA functions.
Materials and Methods
Materials. The list of materials and suppliers can be found in the supplementary information online.
Yeast two-hybrid assay. Coding regions for human Dicer or PPD protein (AGO2 and Hiwi) subdomains were fused with the Gal-4 activation domain of pGADT7 and the Gal-4 DNA-binding domain of pGBKT7, respectively (see supplementary information online). The resulting plasmid constructs were transformed into S. cerevisiae strain AH109 and plated on complete minimum media (CMM) lacking leucine and tryptophan for 2–3 days at 30°C. Transformants were then streaked onto media lacking leucine, tryptophan, histidine and adenine. In cases where colonies grew in less than 7 days, these combinations were deemed as evidence of positive interactions, whereas lack of growth after 14 days was deemed as a negative interaction.
In vitro Dicer activity assays. DsRNA processing assays were performed as previously described (Zhang et al, 2002), and contained 130-bp dsRNA uniformly labelled with [α-32P]UTP, 20 ng of the recombinant human Dicer–HisC protein, and indicated amounts of GST–Hiwi, GST–AGO2 or GST.
Other procedures. Detailed methods including plasmid construction, GST-pulldowns and immunoprecipitations can be found in the supplementary information online.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Data
Acknowledgments
We thank Dr Henry Parker for help with subcloning and Margaret Hughes for cell culture support. This work was funded by a grant from the Canadian Institutes of Health Research (CIHR) (T.C.H). T.C.H. is the recipient of a Senior Medical Scholarship from the Alberta Heritage Foundation for Medical Research (AHFMR). N.T. and K.J. are supported by pre-doctoral studentship awards from CIHR and AHFMR, respectively. F.A.K. was the recipient of a long-term fellowship from the Human Frontier Science Program. Friedrich Miescher Institute is a part of the Novartis Research Foundation.
References
- Baulcombe D (2001) RNA silencing: diced defence. Nature 409: 295–296 [DOI] [PubMed] [Google Scholar]
- Bernstein E, Caudy A, Hammond S, Hannon G (2001) Role for bidentate ribonuclease in the initiation step of RNA interference. Nature 409: 363–366 [DOI] [PubMed] [Google Scholar]
- Billy E, Brondani V, Zhang H, Muller U, Filipowicz W (2001) Specific interference with gene expression induced by long, doublestranded RNA in mouse embryonal teratocarcinoma cell lines. Proc Natl Acad Sci USA 98: 14428–14433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmell MA, Xuan Z, Zhang MQ, Hannon GJ (2002) The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev 16: 2733–2742 [DOI] [PubMed] [Google Scholar]
- Cikaluk D, Tahbaz N, Hendricks L, DiMatti G, Hansen D, Pilgrim D, Hobman T (1999) GERp95, a membrane-associated protein that belongs to a family of proteins involved in stem cell differentiation. Mol Biol Cell 10: 3357–3372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox DN, Chao A, Baker J, Chang L, Qiao D, Lin H (1998) A novel class of evolutionarily conserved genes defined by piwi are essential for stem cell self-renewal. Genes Dev 12: 3715–3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djikeng A, Shi H, Tschudi C, Shen S, Ullu E (2003) An siRNA ribonucleoprotein is found associated with polyribosomes in Trypanosoma brucei. RNA 9: 802–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi N, Zenno S, Ueda R, Ohki-Hamazaki H, Ui-Tei K, Saigo K (2003) Short-interfering-RNA-mediated gene silencing in mammalian cells requires Dicer and eIF2C translation initiation factors. Curr Biol 13: 41–46 [DOI] [PubMed] [Google Scholar]
- Findley SD, Tamanaha M, Clegg NJ, Ruohola-Baker H (2003) Maelstrom, a Drosophila spindle-class gene, encodes a protein that colocalizes with Vasa and RDE1/AGO1 homolog, Aubergine, in nuage. Development 130: 859–871 [DOI] [PubMed] [Google Scholar]
- Grishok A, Pasquinelli A, Conte D, Li N, Parrish S, Ha I, Baillie D, Fire A, Ruvkun G, Mello C (2001) Genes and mechanisms related to RNA interference regulate expression of small temporal RNAs that control C. elegans developmental timing. Cell 106: 23–34 [DOI] [PubMed] [Google Scholar]
- Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ (2001) Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150 [DOI] [PubMed] [Google Scholar]
- Hannon GJ (2002) RNA interference. Nature 418: 244–251 [DOI] [PubMed] [Google Scholar]
- Hutvagner G, Zamore PD (2002) A microRNA in a multiple-turnover RNAi enzyme complex. Science 297: 2056–2060 [DOI] [PubMed] [Google Scholar]
- Ketting RF, Fischer SE, Bernstein E, Sijen T, Hannon GJ, Plasterk RH (2001) Dicer functions in RNA interference and in synthesis of small RNA involved in developmental timing in C. elegans. Genes Dev 15: 2654–2659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Rand TA, Kalidas S, Du F, Kim HE, Smith DP, Wang X (2003) R2D2, a bridge between the initiation and effector steps of the Drosophila RNAi pathway. Science 301: 1921–1925 [DOI] [PubMed] [Google Scholar]
- Martinez J, Patkaniowska A, Urlaub H, Luhrmann R, Tuschl T (2002) Singlestranded antisense siRNAs guide target RNA cleavage in RNAi. Cell 110: 563–574 [DOI] [PubMed] [Google Scholar]
- McManus MT, Sharp PA (2002) Gene silencing in mammals by small interfering RNAs. Nat Rev Genet 3: 737–747 [DOI] [PubMed] [Google Scholar]
- Parrish S, Fire A (2001) Distinct roles for RDE-1 and RDE-4 during RNA interference in Caenorhabditis elegans. RNA 7: 1397–1402 [PMC free article] [PubMed] [Google Scholar]
- Prodromou C, Roe SM, O'Brien R, Ladbury JE, Piper PW, Pearl LH (1997) Identification and structural characterization of the ATP/ADP-binding site in the Hsp90 molecular chaperone. Cell 90: 65–75 [DOI] [PubMed] [Google Scholar]
- Provost P, Dishart D, Doucet J, Frendewey D, Samuelsson B, Radmark O (2002) Ribonuclease activity and RNA binding of recombinant human Dicer. EMBO J 21: 5864–5874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T, Shiohama A, Minoshima S, Shimizu N (2003) Identification of eight members of the Argonaute family in the human genome. Genomics 82: 323–330 [DOI] [PubMed] [Google Scholar]
- Schauer S, Jacobsen S, Meinke D, Ray A (2002) DICER-LIKE1: blind men and elephants in Arabidopsis development. Trends Plant Sci 7: 482–491 [DOI] [PubMed] [Google Scholar]
- Schramke V, Allshire R (2003) Hairpin RNAs and retrotransposon LTRs effect RNAi and chromatin-based gene silencing. Science 301: 1069–1074 [DOI] [PubMed] [Google Scholar]
- Schulte TW, Akinaga S, Soga S, Sullivan W, Stensgard B, Toft D, Neckers LM (1998) Antibiotic radicicol binds to the N-terminal domain of Hsp90 and shares important biologic activities with geldanamycin. Cell Stress Chaperones 3: 100–108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz DS, Zamore PD (2002) Why do miRNAs live in the miRNP? Genes Dev 16: 1025–1031 [DOI] [PubMed] [Google Scholar]
- Sharma AK, Nelson MC, Brandt JE, Wessman M, Mahmud N, Weller KP, Hoffman R (2001) Human CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood 97: 426–434 [DOI] [PubMed] [Google Scholar]
- Stebbins CE, Russo AA, Schneider C, Rosen N, Hartl FU, Pavletich NP (1997) Crystal structure of an Hsp90–geldanamycin complex: targeting of a protein chaperone by an antitumor agent. Cell 89: 239–250 [DOI] [PubMed] [Google Scholar]
- Tabara H, Sarkissian M, Kelly WG, Fleenor J, Grishok A, Timmons L, Fire A, Mello CC (1999) The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99: 123–132 [DOI] [PubMed] [Google Scholar]
- Tahbaz N, Carmichael JB, Hobman TC (2001) GERp95 belongs to a family of signal-transducing proteins and requires Hsp90 activity for stability and Golgi localization. J Biol Chem 276: 43294–43299 [DOI] [PubMed] [Google Scholar]
- Zamore PD (2002) Ancient pathways programmed by small RNAs. Science 296: 1265–1269 [DOI] [PubMed] [Google Scholar]
- Zhang H, Kolb FA, Brondani V, Billy E, Filipowicz W (2002) Human Dicer preferentially cleaves dsRNAs at their termini without a requirement for ATP. EMBO J 21: 5875–5885 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Data