Abstract
Toll-like receptors (TLRs) mediate recognition of microbial components. Despite activation of a shared set of signal transduction molecules, the biological effects of certain TLR agonists differ considerably. In macrophages and dendritic cells, stimulation by the prototypical stimuli CpG-DNA (TLR9), lipopolysaccharide (LPS; TLR4) and lipoteichoic acid (LTA; TLR2) resulted in striking differences in expression of IL-12. However, these stimuli induced similar amounts of the common proinflammatory cytokine TNFα. Surprisingly, an IL-12p40 promoter reporter construct was activated equally by CpG-DNA, LPS and LTA. Examinations of the chromatin structure of the endogenous IL-12p40 promoter revealed that nucleosome remodelling contributed to differential IL-12 induction. Upon stimulation, nucleosome architecture was changed to provide increased access to the IL-12p40 promoter. In dendritic cells, a differential induction of nucleosome remodelling at the IL-12p40 promoter was observed upon triggering with different TLR agonists. These results identify nucleosome remodelling as an additional restriction point in differential TLR signalling.
Introduction
Detection of pathogens through the innate immune system critically involves Toll-like receptors (TLRs). TLRs recognize major bacterial components such as lipopolysaccharide (LPS), lipoteichoic acid (LTA) or bacterial CpG-DNA. Receptor activation by different TLR agonists leads to the activation of a shared signalling pathway. However, the biological effects of different TLR agonists are quite diverse, suggesting additional TLR-specific signalling mechanisms.
An example of specific immunostimulation is the peculiar capacity of TLR9 to induce TH1 immune responses. Interleukin-12 (IL-12) is one of the most important cytokines of innate immunity that shapes the subsequent adaptive immune response and especially influences TH1/TH2 commitment. IL-12 is composed of two subunits (p35, p40) of which p40 is highly regulated in antigen-presenting cells (APCs). It has been revealed that IL-12 gene expression is tightly controlled at the transcriptional level. Control elements within the IL-12p40 promoter have been identified by reporter gene assays (Murphy et al, 1995; Plevy et al, 1997). In this respect, a Rel/NFκB element was found to be most important for promoter activity stimulated by IFNγ and LPS. In addition, proteins belonging to the IFN regulatory factor (IRF) family, such as IFN consensus sequence binding protein (ICSBP) and IRF-1, have a significant role in regulating IL-12 expression (Salkowski et al, 1999; Wang et al, 2000).
Recently, an additional mechanism of regulation has been reported. It has been shown that the endogenous IL-12p40 promoter is assembled in four tightly positioned nucleosomes. Nucleosomes are the higher-order structure of chromatin and represent an obstacle to the binding of transcription factors. Nucleosome 1 within the p40 gene promoter encompasses the critical Rel and C/EBP elements. Upon cellular activation, chromatin modifications take place and in the case of the IL-12p40 promoter this results in remodelling of the structure of the first nucleosome (Weinmann et al, 1999). As a consequence, the locus is more accessible and now allows binding of the respective transcription factors. The results of footprinting technology also corroborate these findings (Becker et al, 2001). However, the impact of IL-12p40 nucleosome remodelling on the outcome of various TLR-dependent stimuli is unknown.
Results
Differential induction of IL-12 by CpG-DNA, LPS or LTA
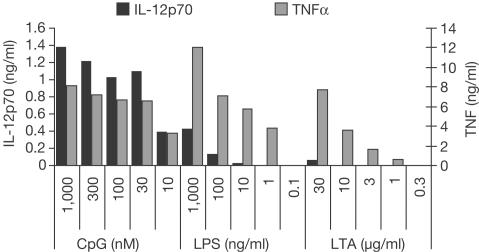
In order to examine differences in the cytokine response patterns of different TLRs, we tested the capacity of prototypical TLR ligands to induce IL-12 in dendritic cells. For comparison, we also determined TNFα, which has been described as a target gene induced through the common MyD88-dependent pathway for most of the TLR ligands described so far. In Fig 1, it is shown that CpG-DNA, LPS and LTA induced TNFα in a dose-dependent way. However, CpG-DNA was far more effective in the equivalent secretion of IL-12p70 as compared to LPS. Moreover, LTA was not capable of inducing significant amounts of IL-12. Similar results were also observed with peritoneal macrophages and RAW264.7 macrophages, albeit with lower IL-12 induction (data not shown). For the following experiments, we decided to use concentrations of the different TLR stimuli that induced similar amounts of TNFα. Differences in IL-12 protein secretion were also reflected by different IL-12p40 mRNA induction as examined by quantitative reverse transcription (RT)-PCR (data not shown).
Figure 1.
Differential induction of IL-12p70 by TLR-dependent stimuli. Bone-marrow-derived dendritic cells were stimulated with CpG-DNA, LPS or LTA as indicated for 24 h. TNFα and IL-12p70 were determined in supernatants by ELISA.
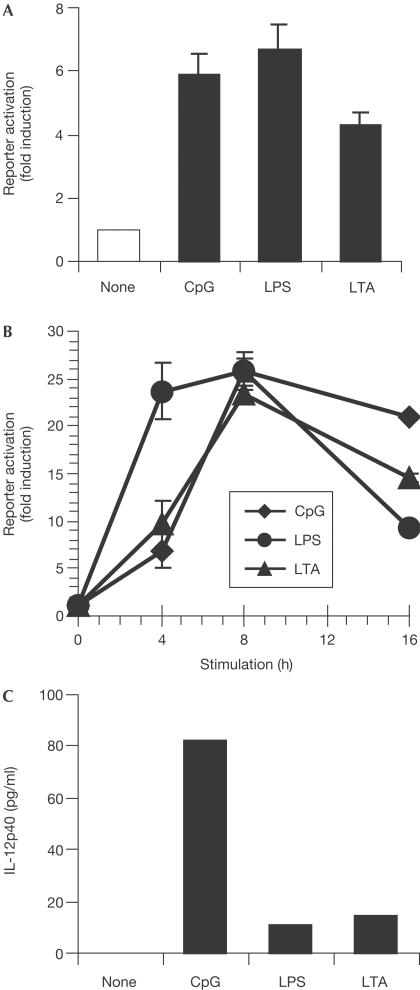
Comparable activation of IL-12p40 reporter constructs
To examine whether the observed differences in p40 induction in response to different TLR agonists are due to differential activation of required transcription factors, we made use of p40 reporter gene assays. Surprisingly, although RAW264.7 macrophages only showed minor IL-12p40 protein secretion after stimulation with LTA, a luciferase reporter plasmid of the IL-12p40 promoter (−703/+54) was efficiently induced by LTA in transient transfection assays (Fig 2A). Moreover, no major differences in the activation of this reporter by CpG-DNA, LPS and LTA were observed. Similar results were obtained when cells with a stably integrated construct were used (Fig 2B). Again, all three stimuli were capable of activating the reporter construct. In contrast, only CpG-DNA induced secretion of endogenous IL-12p40 protein in the same cells (Fig 2C). Thus, the endogenous IL-12p40 promoter locus must be controlled by additional mechanisms.
Figure 2.
IL-12p40 promoter activation by TLR agonists. (A) RAW264.7 cells were transiently transfected with an IL-12p40 promoter reporter plasmid. After 24 h, cells were stimulated with 100 nM CpG-DNA, 100 ng/ml LPS or 30 μg/ml LTA for 8 h. Subsequently, reporter gene activation was measured and is expressed relative to nonstimulated cells. Three independent transfections were performed. (B) RAW264.7 cells stably expressing the IL-12p40 promoter reporter plasmid were stimulated as above for the indicated time periods. (C) The same cells as in (B) were assayed for endogenous IL-12p40 protein after 24 h of stimulation.
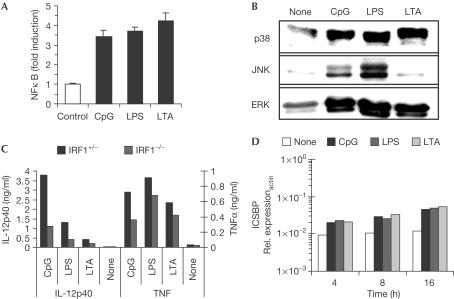
Equal activation of common transcription factors
For further insight into differences in IL-12p40 regulation after TLR triggering, we investigated the activation of transcription factors with a known role in IL-12p40 induction. First, CpG-DNA, LPS and LTA were equally effective in inducing an NFκB-dependent luciferase reporter plasmid (Fig 3A). With respect to MAP kinase activation, we only observed differences in JNK activation, with LPS being more effective and LTA showing nearly no activation (Fig 3B). However, these differences did not correlate with differential capacity to induce IL-12p40. There were no differences in the total amounts of the respective kinases (data not shown). IRF1 has also been reported to contribute to IL-12p40 secretion. Using IRF1-deficient mice, we observed a dramatic decrease in IL-12p40 secretion and also diminished TNF production (Fig 3C). However, the differences in p40 secretion upon CpG-DNA, LPS or LTA stimulation were still observed, albeit in an attenuated way. Finally, ICSBP induction was determined by quantitative RT-PCR, yet no differences between CpG-DNA, LPS and LTA triggering were obvious (Fig 3D).
Figure 3.
Signal transduction by CpG-ODN, LPS and LTA. (A) RAW264.7 cells were transiently transfected with an NFκB-dependent reporter plasmid. Cells were stimulated with 100 nM CpG-ODN, 100 ng/ml LPS or 30 μg/ml LTA for 7 h, and luciferase induction was determined. (B) RAW264.7 cells were stimulated as above for 15 min. Similar amounts of cellular extracts were blotted with phosphospecific antibodies for p38, JNK or ERK MAP kinase. (C) Peritoneal macrophages from IRF1−/− and IRF1+/− mice were stimulated as above. Supernatants were assayed for TNFα and IL-12p40. (D) RAW264.7 macrophages were stimulated as above for the indicated time periods. ICSBP mRNA expression was measured by quantitative RT-PCR.
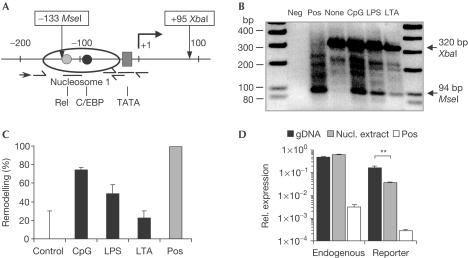
TLRs differentially reduce nucleosome remodelling
To test the hypothesis that differences in chromatin regulation contribute to the differential capacity of various TLR agonists to induce IL-12p40, we focused on inducible changes in nucleosomal architecture. To measure nucleosome remodelling, we made use of a restriction enzyme accessibility assay combined with modified ligation-mediated PCR (LM-PCR) (Fig 4A). With this approach, we were able to show that in dendritic cells the promoter is not accessible under starting conditions. However, upon stimulation, restriction enzyme accessibility increases, and differences between stimulation of CpG-DNA, LPS and LTA can be observed (Fig 4B). Although all of the stimuli were able to induce nucleosome remodelling, CpG-DNA induced more prominent changes, which corroborates the observed differences in IL-12p40 protein secretion. Using the same assay with stimulated macrophages, we only observed a remodelling upon challenge with CpG-DNA (supplementary fig S1 online).
Figure 4.
Differential nucleosome remodelling at the IL-12p40 promoter in response to TLR-dependent stimuli. (A) Schematic drawing of the structure of the IL-12p40 promoter. Half arrows indicate primer sets for two ChART PCRs. One is spanning the restriction enzyme recognition site for MseI, and the second amplifies outside this region. Straight lines indicate positions of the TaqMan probes for both these primer sets. In addition, the IL-12p40 promoter-specific primer used in the LM-PCR is indicated (arrow). (B) Bone-marrow-derived dendritic cells were examined for changes in nucleosome structure by restriction enzyme accessibility followed by LM-PCR. Cells were stimulated with 100 nM CpG-ODN, 100 ng/ml LPS or 10 μg/ml LTA for 8 h. Arrows indicate the amplicons either generated by accessibility for MseI or XbaI as a normalization. Positive controls were obtained by digestion of purified DNA with MseI (negative image). (C) Bone-marrow-derived dendritic cells were stimulated as above for 2 h, and nuclear extracts were tested for restriction enzyme accessibility by ChART assay. Remodelling is plotted in percentage of nonstimulated cells. (D) RAW264.7 cells with a stably integrated IL-12p40 reporter plasmid were tested for MseI accessibility by modified ChART assay. To this were added reverse primers that specifically hybridized either within the endogenous IL-12p40 gene or the luciferase reporter gene. Nuclear extracts of nonstimulated cells (nucl. extract) were treated with MseI and accessibility was measured as normalized expression of the specific amplicons in relation to the normalization PCR. As controls, genomic DNA (gDNA) was left untreated or completely digested (pos).
To strengthen the evidence and to verify the observed differences in TLR-mediated IL-12p40 promoter remodelling, we next measured remodelling quantitatively. For this, we made use of a restriction enzyme accessibility assay quantified by real-time PCR (published as ChART). Three quantitative PCRs were set up (Fig 4A). The first PCR product spanned the expected cleavage site. A second PCR amplified a region outside the cleavage site within the promoter and finally an amplicon of the housekeeping gene β-actin was determined. The last two PCRs were used for normalization of possible differences during the DNA preparation.
Using this assay, we were able to quantify changes in nucleosome remodelling at the IL-12p40 promoter upon TLR triggering. Normalized changes in the amounts of the reporter PCR were measured and decreases were expressed as remodelling in comparison to nonstimulated cells. A complete loss of the PCR amplicon indicated full accessibility and thus 100% remodelling. In dendritic cells, nonstimulated cells showed similar or only slightly higher threshold values in the PCR reaction as with uncut genomic DNA. Thus, under starting conditions, the promoter was indeed nearly completely protected. However, upon stimulation, CpG-DNA induced a remodelling of about 74%, with LPS (49%) and LTA (22%) again being less effective (Fig 4C). Remodelling was dependent on the presence of the CpG motif, as control DNA did not induce nucleosome remodelling (supplementary fig S2 online). No significant remodelling was observed within the upstream nucleosome 2 (supplementary fig S3 online). Finally, the RAW264.7 macrophages with the stably integrated IL-12p40 reporter construct were assayed for restriction enzyme accessibility. We were able to show that indeed the endogenous locus is completely protected, whereas the integrated reporter plasmid is largely accessible (sixfold decrease upon restriction enzyme digestion) under nonstimulated conditions (Fig 4D).
Discussion
So far, cellular activation by different TLR agonists has been reported to induce and activate a set of shared intracellular signal transduction molecules (O'Neill, 2002). Despite similar signal transduction mechanisms, different TLR agonists can be distinguished in their biological effects and this could be due in part to TLR-specific signalling pathways, which have recently been described (Yamamoto et al, 2003). Of various TLRs, the specific capacity of immunostimulatory CpG-DNA to activate TH1-dominated immunity is impressive (Chu et al, 1997).
The present study extends these findings by using a comparative approach to study the prototypical ligands of TLR9, TLR4 and TLR2. In particular, we normalized the different compounds on their ability to induce TNFα, a common MyD88-dependent target gene. Indeed, all of the stimuli were able to induce this cytokine in a similar way. However, they differed markedly in their ability to induce IL-12p40. CpG-oligodeoxynucleotide (ODN) proved to be the strongest inducer of p40 expression as compared to LPS or LTA. This correlates with an earlier report (Cowdery et al, 1999) that showed a stronger increase of IL-12p40 mRNA and protein after stimulation with CpG-DNA than with LPS. Moreover, a recent report also demonstrates the inability of LTA to induce IL-12 in human cells (Hermann et al, 2002). The data clearly support the concept of TLR-specific response profiles that enable the innate immune system to generate pathogen-specific activation patterns.
IL-12 is apparently regulated differentially by various TLR agonists. The promoter of the IL-12p40 gene has been shown to be located in specific nucleosomes, with nucleosome 1 containing relevant transcription factor binding sites (Weinmann et al, 1999). Upon stimulation with LPS, increased accessibility of the promoter was reported, and thus nucleosome remodelling seems to be an additional mechanism regulating IL-12 induction. Here, we report that differences in IL-12p40 nucleosome remodelling occur upon stimulation with various TLR stimuli, and this correlates with differences in TLR-induced gene expression of IL-12. Although differences in remodelling induced by TLR4 and TLR9 were only moderate, the effects on IL-12 secretion were more prominent. Thus, it could be that a certain threshold level of remodelling is necessary for robust gene transcription. By now, further genes have been shown to be additionally regulated by nucleosome remodelling (Rao et al, 2001; Holloway et al, 2003). Moreover, the existence of immediately accessible genes as well as delayed accessible promoters has been demonstrated, which suggests that changes in chromatin architecture contribute to inducible inflammatory gene regulation (Saccani et al, 2001). It is of general interest that reporter gene assays, which are valuable tools for the examination of gene regulation, did not reflect physiological regulatory mechanisms. Thus, RAW264.7 macrophages which secrete IL-12p40 in low levels only in response to CpG-DNA showed an equal activation of an IL-12p40 reporter construct in response to all of the tested TLR agonists. Corroborating these findings, we found that the integrated reporter plasmid was constitutively accessible, whereas the endogenous IL-12p40 locus was protected.
An interesting observation was recently provided by Lomvardas & Thanos (2002), who were able to alter artificially the nucleosome position in the IFN-β promoter. Thereby, they changed the kinetics of IFN-β gene induction and furthermore they changed the specificity of induction in HeLa cells.
Now, the unresolved question is the nature of the stimulus that induces nucleosome remodelling. TLRs trigger an association with adaptor molecules of the MyD88 family. Recently, TICAM/TRIF has been shown to be another such adaptor in TLR4 signalling. It induces IFN-β-dependent genes, thus providing a new signal quality (Oshiumi et al, 2003). However, the observed differences in IL-12 signalling were observed quantitatively, and thus the exclusive activation of specific adaptors seems to be unlikely. In this respect, spatio-temporal components in TLR signalling could have a role. It is possible that the kinetics of activation of transcription factors differ between various TLR agonists. Perhaps the composition of NFκB dimers changes over time and this affects the activation of different genes. Indeed, TLR9 signalling differs from other TLRs by the fact that activation occurs within the endosome where TLR9 is expressed (Ahmad-Nejad et al, 2002).
With respect to the mode of activation of nucleosome remodelling, it has been shown that c-rel is not involved despite the requirement for c-rel for transcriptional activation (Weinmann et al, 2001). In addition, C/EBP proteins are also insufficient for nucleosome remodelling. Moreover, our results make it unlikely that different dimers of NFκB induce nucleosome remodelling, because of the similar activation of an NFκB reporter. Also, neither IRF1 nor ICSBP seems to be responsible for differential induction of IL-12p40. As binding of NFκB to its binding site in the promoter seems to be hindered by nucleosome 1, another mechanism must precede and induce remodelling, thus permitting access of further transcription factors.
Taken together, our data show a clear differential expression of IL-12p40 mRNA and protein after stimulation with different TLR agonists, with CpG-DNA inducing the strongest increase in IL-12 expression. This accords with a more efficient chromatin remodelling of nucleosome 1 within the IL-12p40 promoter by TLR9, which is a prerequisite for subsequent transcription initiation. Thus the data identify inducible nucleosome remodelling as an additional restriction point in TLR signalling.
Methods
Cells and culture conditions. Bone-marrow-derived macrophages and dendritic cells were isolated from BALB/c mice as described (Inaba et al, 1992). GM-CSF-differentiated dendritic cells were harvested at day 9 and were positively sorted for CD11c expression by MACS (Miltenyi BioTech, Bergisch Gladbach, Germany). IRF1−/− mice were provided by M. Lohoff (Marburg, Germany).
Reagents. Completely phosphorothioate-modified CpG-ODN 1668 was purchased from TIB Molbiol (Berlin). Sequences of ODNs and primers are available on request. LTA from Staphylococcus aureus was a kind gift from S. Morath (Konstanz, Germany), and was shown to be TLR2 dependent (Lehner et al, 2001). Highly purified LPS from Salmonella minnesota was kindly provided by U. Seydel (Borstel, Germany). Antibodies specific for phosphorylated MAP kinases were from Cell Signaling Technology (Frankfurt, Germany). Cytokines were measured by commercially available ELISA (Becton Dickinson, Heidelberg, Germany). mRNA expression was quantified by real-time TaqMan PCR (Applied Biosystems, Germany) by determining relative expression of target sequences to the endogenous control β-actin.
Reporter gene experiments. The IL-12p40 promoter (−703 to +54 bp) coupled to a luciferase-encoding reporter gene was a kind gift from H. Haecker (München, Germany). NFκB reporter plasmid contained six NFκB-binding sites (GGGGAATTTCC). Transient transfections were performed by electroporation of RAW264.7 cells. Stably transfected RAW264.7 cell clones were generated by co-transfection with a G418 resistance encoding plasmid. Experiments were performed with one typical clone out of four. Luciferase induction was measured with the LucLite-kit (Packard, Netherlands) in a TopCountNXT.
Restriction enzyme accessibility assay. Cells (2 × 106) were stimulated as indicated in the respective experiment. Nuclei were prepared and tested for restriction enzyme accessibility essentially as described (Weinmann et al, 1999). Digestion reaction was carried out with 25 U of MseI for 10 min at 37°C. Subsequently, DNA was purified using the DNeasy Tissue kit (Qiagen, Hilden, Germany).
Modified LM-PCR. LM-PCR was performed with the following modifications (Garrity & Wold, 1992). For normalization, 2 μg of DNA from restriction enzyme accessibility assays was additionally digested with XbaI, which cuts distal of MseI. Next, two double-stranded linker primers that generate either an MseI or an XbaI end were covalently linked to the sticky ends of restriction-enzyme-digested DNA. Ligated DNA was used in a hot-start PCR with an IL-12p40 promoter-specific primer and the universal linker primer as reverse primers. PCR products were directly analysed in 2.5% agarose gels.
ChART. To quantify nucleosome remodelling, chromatin accessibility measured by real-time PCR (ChART) assays was performed as described for the IL-2 promoter (Rao et al, 2001). DNA from restriction enzyme accessibility assays was used in three different quantitative PCR approaches using TaqMan technology. One primer set spanned the restriction sites for MseI. A second primer set amplified outside this region and a third primer set was for β-actin (Fig 4A). Both the latter primer sets were used for normalization of DNA amounts, whereas the first primer set reported accessibility of the locus. A decrease in the amount of amplicon generated by the first set indicated increased accessibility and thus nucleosome remodelling. Amounts of the reporter amplicon within the promoter were normalized to the normalization amplicon within the IL-12p40 promoter or to β-actin. Subsequently, data were calculated and plotted as percentage of nonstimulated cells.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Supplementary Figures
Acknowledgments
We thank Helene Bykow and Sandra Opper for excellent technical support. This project was supported by the Deutsche Forschungsgemeinschaft (He1452/2, He1452/4, Zi676/1) and the European Community (QLK2-CT-2000-00336).
References
- Ahmad-Nejad P, Hacker H, Rutz M, Bauer S, Vabulas RM, Wagner H (2002) Bacterial CpG-DNA and lipopolysaccharides activate Toll-like receptors at distinct cellular compartments. Eur J Immunol 32: 1958–1968 [DOI] [PubMed] [Google Scholar]
- Becker C, Wirtz S, Ma X, Blessing M, Galle PR, Neurath MF (2001) Regulation of IL-12 p40 promoter activity in primary human monocytes: roles of NF-κB, CCAAT/enhancer-binding protein β, and PU.1 and identification of a novel repressor element (GA-12) that responds to IL-4 and prostaglandin E(2). J Immunol 167: 2608–2618 [DOI] [PubMed] [Google Scholar]
- Chu RS, Targoni OS, Krieg AM, Lehmann PV, Harding CV (1997) CpG oligodeoxynucleotides act as adjuvants that switch on T helper 1 (Th1) immunity. J Exp Med 186: 1623–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowdery JS, Boerth NJ, Norian LA, Myung PS, Koretzky GA (1999) Differential regulation of the IL-12 p40 promoter and of p40 secretion by CpG DNA and lipopolysaccharide. J Immunol 162: 6770–6775 [PubMed] [Google Scholar]
- Garrity PA, Wold BJ (1992) Effects of different DNA polymerases in ligation-mediated PCR: enhanced genomic sequencing and in vivo footprinting. Proc Natl Acad Sci USA 89: 1021–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann C, Spreitzer I, Schroder NW, Morath S, Lehner MD, Fischer W, Schutt C, Schumann RR, Hartung T (2002) Cytokine induction by purified lipoteichoic acids from various bacterial species—role of LBP, sCD14, CD14 and failure to induce IL-12 and subsequent IFN-γ release. Eur J Immunol 32: 541–551 [DOI] [PubMed] [Google Scholar]
- Holloway AF, Rao S, Chen X, Shannon MF (2003) Changes in chromatin accessibility across the GM-CSF promoter upon T cell activation are dependent on nuclear factor κB proteins. J Exp Med 197: 413–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikeham S, Muramatsu S, Steinman RM (1992) Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med 176: 1693–1702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner MD, Morath S, Michelsen KS, Schumann RR, Hartung T (2001) Induction of cross-tolerance by lipopolysaccharide and highly purified lipoteichoic acid via different Toll-like receptors independent of paracrine mediators. J Immunol 166: 5161–5167 [DOI] [PubMed] [Google Scholar]
- Lomvardas S, Thanos D (2002) Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110: 261–271 [DOI] [PubMed] [Google Scholar]
- Murphy TL, Cleveland MG, Kulesza P, Magram J, Murphy KM (1995) Regulation of interleukin 12 p40 expression through an NF-κB half-site. Mol Cell Biol 15: 5258–5267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Neill LA (2002) Wanted. A molecular basis for specificity in Toll-like receptor signal transduction. Mol Cell 10: 969–971 [DOI] [PubMed] [Google Scholar]
- Oshiumi H, Matsumoto M, Funami K, Akazawa T, Seya T (2003) TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-β induction. Nat Immunol 4: 161–167 [DOI] [PubMed] [Google Scholar]
- Plevy SE, Gemberling JH, Hsu S, Dorner AJ, Smale ST (1997) Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: evidence of functional synergy between C/EBP and Rel proteins. Mol Cell Biol 17: 4572–4588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao S, Procko E, Shannon MF (2001) Chromatin remodeling, measured by a novel real-time polymerase chain reaction assay, across the proximal promoter region of the IL-2 gene. J Immunol 167: 4494–4503 [DOI] [PubMed] [Google Scholar]
- Saccani S, Pantano S, Natoli G (2001) Two waves of nuclear factor κB recruitment to target promoters. J Exp Med 193: 1351–1360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salkowski CA, Kopydlowski K, Blanco J, Cody MJ, McNally R, Vogel SN (1999) IL-12 is dysregulated in macrophages from IRF-1 and IRF-2 knockout mice. J Immunol 163: 1529–1536 [PubMed] [Google Scholar]
- Wang IM, Contursi C, Masumi A, Ma X, Trinchieri G, Ozato K (2000) An IFN-γ-inducible transcription factor, IFN consensus sequence binding protein (ICSBP), stimulates IL-12 p40 expression in macrophages. J Immunol 165: 271–279 [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Plevy SE, Smale ST (1999) Rapid and selective remodeling of a positioned nucleosome during the induction of IL-12 p40 transcription. Immunity 11: 665–675 [DOI] [PubMed] [Google Scholar]
- Weinmann AS et al. (2001) Nucleosome remodeling at the IL-12 p40 promoter is a TLR-dependent, Rel-independent event. Nat Immunol 2: 51–57 [DOI] [PubMed] [Google Scholar]
- Yamamoto M et al. (2003) Role of adaptor TRIF in the MyD88-independent Toll-like receptor signaling pathway. Science 301: 640–643 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures