Summary
In silico identification of eukaryotic selenoprotein genes
Keywords: comparative genomics, selenocysteine, selenoprotein, UGA
The dual function of the UGA codon poses a serious challenge for the annotation of genomes. Although UGA usually signals the termination of protein synthesis, it can also be decoded as selenocysteine (Sec), which is incorporated into a small but important group of proteins that are known as selenoproteins. Standard gene-analysis programs cannot predict whether a UGA codon encodes Sec or Stop. Bioinformatic tools for recognizing selenoproteins in complementary DNA (cDNA) databases are available, but they are not effective when analysing genome sequences. In a recent paper in EMBO reports, Castellano et al (2004) describe a new in silico strategy for identifying selenoprotein genes in eukaryotic genomes. Their findings expand our knowledge of the taxonspecific distribution of selenoproteins and raise provocative questions about the evolution of Sec.
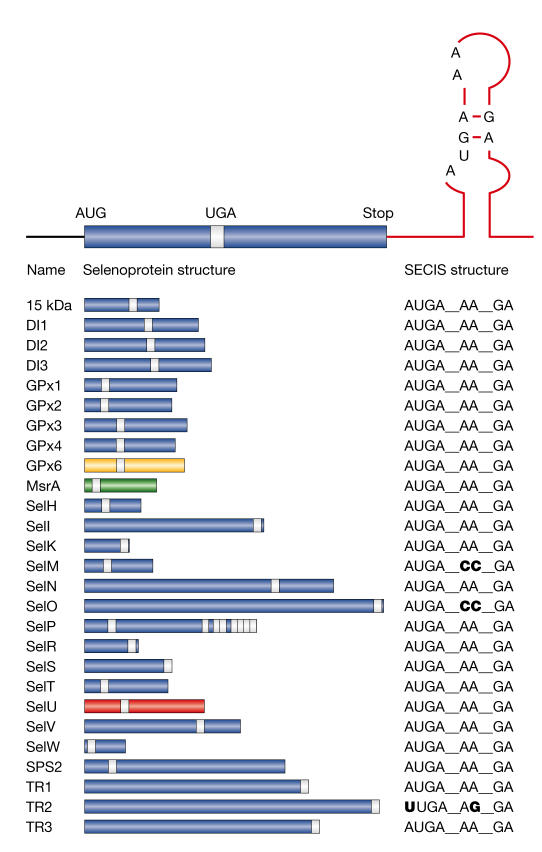
Sec is often present at the active sites of oxidoreductases, where it confers a catalytic advantage over cysteine (Cys). The insertion of Sec into the polypeptide chain during translation requires several trans-acting proteins, which synthesize Sec–tRNASec and deliver it to the ribosome (Hatfield & Gladyshev, 2002; Lescure et al, 2002; Driscoll & Copeland, 2003). The alternative decoding of UGA as Sec depends on cis-acting sequences in the transcript. The Sec-insertion sequence (SECIS) element is a stable stem–loop structure that is found in the coding region (prokaryotes) or 3′ untranslated region (archaebacteria and eukaryotes) of selenoprotein messenger RNAs (mRNAs) (Martin & Berry, 2001). The eukaryotic SECIS is conserved across species, although noncanonical SECIS elements have been reported (Fig 1).
Figure 1.
Eukaryotic selenoprotein genes identified so far. Selenoprotein messenger RNAs (mRNAs) harbour a Sec-insertion sequence (SECIS) element in their 3′ untranslated region that triggers the recoding of the UGA codon into Sec (the location of which is shown in the coding sequence by a white bar). Deviations from the canonical SECIS are shown in bold. The taxon-specific distributions of some selenoproteins are indicated as follows: glutathione peroxidase 6 (GPx6; yellow) is found in humans and pigs, but not in rodents (Kryukov et al, 2003); methionines-sulphoxide reductase A (MsrA; green) is found only in the green alga Chlamydomonas reinhardtii (Novoselov et al, 2002); selenoprotein U (SelU) (red) is found in fish, chickens, sea urchins, a green alga and a diatom (Castellano et al, 2004), but not in higher eukaryotes. DI, iodothyronine deiodinase; TR, thioredoxin reductase.
The evolution of the UGA/Sec codon is not well understood. Selenoproteins are found in all three lines of descent, but they have a scattered distribution. There are no selenoproteins in yeast or higher plants. In prokaryotes and archaebacteria, selenoproteins have been identified in only a limited number of species (Bock, 2001). Notably, there is almost no overlap between the prokaryotic and eukaryotic selenoproteomes. In eukaryotes, the number of selenoprotein genes varies between organisms, with 25 in humans, but only 3 in Drosophila melanogaster and 1 in Caenorhabditis elegans (Kryukov et al, 2003). Mammalian selenoproteins usually have lower eukaryotic orthologues in which Sec is replaced by Cys.
Historically, selenoproteins have been identified by purifying a protein and cloning its cognate cDNA. More recent in silico approaches have a greater potential to exhaustively identify selenoprotein genes in a genome. Several groups have developed computer algorithms that use the SECIS element as a signature for mammalian selenoproteins (Kryukov et al, 1999; Lescure et al, 1999). This approach is practical for screening cDNA databases but not complex genome sequences. The algorithms have been refined to search for SECIS-containing genes with in-frame TGA codons (Castellano et al, 2001; Kryukov et al, 2003), but a clear limitation of this strategy is the inability to detect genes that contain a noncanonical SECIS element.
Castellano et al (2004) have now developed a SECIS-independent comparative-genomics strategy for finding eukaryotic selenoprotein genes. The human (Homo sapiens) and fugu (Takifugu rubripes) genomes were compared to identify genes with in-frame TGA codons. Inter- and intra-genomic analyses identified human–fugu pairs, which were Sec–Sec, Sec–Cys or Cys–Sec. Candidate genes were analysed for the following: conservation of primary amino-acid sequence or secondary structure around the Sec/Cys residue, as an indication of protein-coding potential; orthologues in other species in which the context of the Sec/Cys residue is conserved; and the presence of a potential SECIS element. A new family of selenoprotein genes, SelU, was identified in fugu as well as in other fish, chickens, sea urchins, a green alga and a diatom. Surprisingly, SelU homologues in other species, including humans, contain Cys instead of Sec.
Theory holds that the number of selenoprotein genes increases from lower eukaryotes to vertebrates, but there have been clues to indicate that this might not be the case. The methionine-S-sulphoxide reductase (MsrA) gene encodes Sec in Chlamydomonas reinhardtii, which is a green alga, whereas vertebrate MsrA genes encode Cys (Novoselov et al, 2002). Another twist came with the discovery of the human selenoprotein, glutathione peroxidase 6 (GPx6), which has Cys homologues in rodents (Kryukov et al, 2003). Interestingly, the rat GPx6 gene contains a nonfunctional fossil SECIS element, which indicates that Sec was replaced with Cys. These results, and the discovery of the SelU gene family, strongly argue against the old hypothesis that mammals express the full spectrum of eukaryotic selenoproteins. Furthermore, it now seems likely that there are additional unknown selenoproteins in the eukaryotic world. The new comparative-genomics strategy for recognizing UGA/Sec codons will be an invaluable tool for identifying new selenoprotein genes. One potential limitation of this approach is that it is based on the conservation of the UGA/Sec codon context. Therefore, sporadic selenoproteins that are unique to a particular species, or those in which Sec is the last or penultimate amino acid, might be missed.
Does the Sec/Cys replacement represent a gain or loss of function during evolution? Sec is sensitive to oxidation, which led to the proposal that the UGA/Sec codon was lost as the environment became more oxidizing. Alternatively, the independent origin of prokaryotic and eukaryotic selenoproteins supports the hypothesis that Sec was acquired because it provided an evolutionary advantage. The identification of additional Cys-encoding genes that contain a fossil SECIS element will provide important insights into possible evolutionary pathways. Another potential reason for maintaining the UGA/Sec codon is that Sec insertion competes with termination. Many eukaryotic selenoproteins contain a Sec residue near the C-terminus (Fig 1). It can be envisioned that a truncated protein that is produced by termination at the UGA/Sec codon could act as a dominant negative or have a regulatory function in the cell. Regardless of the reason for the Sec/Cys replacement, it is clear that the field faces the challenging task of determining the biological functions of new selenoproteins and explaining their selective advantages.
Despite the functional diversity and scattered distribution of selenoproteins, many features of the Sec-incorporation mechanism are conserved across species (Hatfield & Gladyshev, 2002; Lescure et al, 2002; Driscoll & Copeland, 2003). Although Sec has a catalytic advantage over Cys, the ability to synthesize Sec-containing proteins comes with a price. Many proteins are dedicated to the synthesis and delivery of Sec–tRNASec, and it seems expensive for the cell to maintain this machinery for one amino acid. Yet, in spite of the cost, the Sec-insertion machinery is preserved in worms and flies, which express only one and three selenoproteins, respectively. As more genomes are sequenced, it will be fascinating to determine the taxonspecific distribution of not only particular selenoproteins but also the Sec-insertion machinery.

References
- Bock A (2001) in Selenium: its Molecular Biology and Role in Human Health (ed Hatfield DL), 7–22. Kluwer Academic, Norwell, Massachusetts, USA [Google Scholar]
- Castellano S, Morozova N, Morey M, Berry MJ, Serras F, Corominas M, Guigo R (2001) In silico identification of novel selenoproteins in the Drosophila melanogaster genome. EMBO Rep 2: 697–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castellano S, Novoselov SV, Kryukov GV, Lescure A, Blanco E, Krol A, Gladyshev VN, Guigo R (2004) Reconsidering the evolutionary distribution of eukaryotic selenoproteins: a novel non-mammalian family with scattered phylogenetic distribution. EMBO Rep 5: 71–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll DM, Copeland PR (2003) Mechanism and regulation of selenoprotein synthesis. Annu Rev Nutr 23 17–40 [DOI] [PubMed] [Google Scholar]
- Hatfield DL, Gladyshev VN (2002) How selenium has altered our understanding of the genetic code. Mol Cell Biol 22: 3565–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN (2003) Characterization of mammalian selenoproteomes. Science 300, 1439–1442 [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Kryukov VM, Gladyshev VN (1999) New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J Biol Chem 274: 33888–33897 [DOI] [PubMed] [Google Scholar]
- Lescure A, Fagegaltier D, Carbon P, Krol A (2002) Protein factors mediating selenoprotein synthesis. Curr Protein Pept Sci 3: 143–151 [DOI] [PubMed] [Google Scholar]
- Lescure A, Gautheret D, Carbon P, Krol A (1999) Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J Biol Chem 274: 38147–38154 [DOI] [PubMed] [Google Scholar]
- Martin GW, Berry MJ (2001) in Selenium: its Molecular Biology and Role in Human Health (ed Hatfield DL), 45–53. Kluwer Academic, Norwell, Massachusetts, USA [Google Scholar]
- Novoselov S, Rao M, Onoshko NV, Zhi H, Kryukov GV, Xiang Y, Weeks DP, Hatfield DL, Gladyshev VN (2002) Selenoproteins and selenocysteine insertion in the model plant system, Chlamydomonas reinhardtii. EMBO J 21: 3681–3693 [DOI] [PMC free article] [PubMed] [Google Scholar]