Abstract
The exon junction complex (EJC) is deposited on mRNAs as a consequence of splicing and influences postsplicing mRNA metabolism. The Mago–Y14 heterodimer is a core component of the EJC. Recently, the protein PYM has been identified as an interacting partner of Mago–Y14. Here we show that PYM is a cytoplasmic RNA-binding protein that is excluded from the nucleus by Crm1. PYM interacts directly with Mago–Y14 by means of its N-terminal domain. The crystal structure of the Drosophila ternary complex at 1.9 Å resolution reveals that PYM binds Mago and Y14 simultaneously, capping their heterodimerization interface at conserved surface residues. Formation of this ternary complex is also observed with the human proteins. Mago residues involved in the interaction with PYM have been implicated in nonsense-mediated mRNA decay (NMD). Consistently, human PYM is active in NMD tethering assays. Together, these data suggest a role for PYM in NMD.
Introduction
The exon junction complex (EJC) is a multiprotein assembly deposited by the spliceosome 20–24 nucleotides upstream of mRNA exon–exon junctions (Le Hir et al, 2000). The EJC is thought to provide a molecular link between splicing and postsplicing mRNA metabolism by influencing mRNA export, the efficiency of translation and mRNA stability (reviewed in Le Hir et al, 2003). Several of the EJC components have been identified to date and include the human proteins Y14, Mago, RNPS1, REF/Aly, UPF3 and SRm160. Mago, Y14 and possibly other EJC proteins remain associated with spliced mRNAs after their export to the cytoplasm, providing a mark for exon boundaries (Kim et al, 2001; Le Hir et al, 2001b; Lykke-Andersen et al, 2001).
In mammals, the EJC also functions in nonsense-mediated mRNA decay (NMD), a surveillance mechanism that degrades mRNAs with premature translation termination codons when present upstream of at least one exon boundary (reviewed in Le Hir et al, 2003). In particular, the EJC proteins RNPS1, Y14 and Mago have been shown to elicit NMD when tethered to an mRNA reporter downstream of a stop codon (Lykke-Andersen et al, 2001; Fribourg et al, 2003; Gehring et al, 2003). In Drosophila, Mago and Y14 are essential for cell viability but are not involved in NMD (Gatfield et al, 2003). Instead, Drosophila Mago and Y14 are essential for the localized translation of oskar mRNA during embryonic development (Micklem et al, 1997; Newmark et al, 1997; Hachet & Ephrussi, 2001). The molecular mechanisms by which Mago and Y14 are involved in different aspects of mRNA metabolism are unknown.
We and others have previously reported the structure of the Mago–Y14 heterodimer (Fribourg et al, 2003; Lau et al, 2003; Shi & Xu, 2003). A significant portion of the Mago–Y14 surface is lined by conserved residues, suggesting that they might be a site for protein–protein interactions. One of the interacting partners identified so far is the protein PYM (Forler et al, 2003). PYM, the product of the fly wibg gene, is a conserved protein with an as yet uncharacterized function and sharing no sequence similarity with other proteins. To obtain molecular insights into the interaction of Mago–Y14 and PYM, we have characterized the biochemical properties of the ternary complex and determined its structure at 1.9 Å resolution.
Results And Discussion
Mago–Y14 Interacts With The N-terminal Domain Of Pym
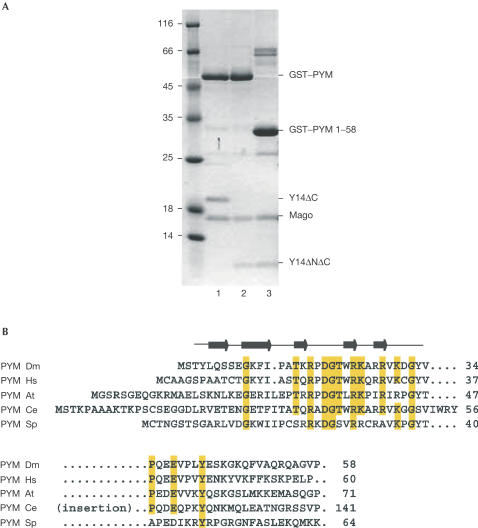
Full-length Drosophila melanogaster (Dm) PYM (residues 1–207) interacts with Mago–Y14 directly, as detected by pull-down experiments using recombinant proteins (Fig 1A). From previous structural studies (Fribourg et al, 2003; Lau et al, 2003; Shi & Xu, 2003), Mago is known to be a single structural unit, whereas Y14 folds into three distinct domains (the N-terminal domain, the RNA-binding-like domain (RBD) and a C-terminal low-complexity region). The C-terminal region of Dm Y14 is not required for Mago binding (Fribourg et al, 2003; Lau et al, 2003) nor for PYM binding (Fig 1A, lane 1). A minimal Mago–Y14 heterodimer containing only the RBD of Y14 (Mago–Y14ΔNΔC) is able to interact with full-length PYM (Fig 1A, lane 2). Guided by sequence alignments, we tested C-terminally truncated fragments of PYM and observed that PYM 1–108 (data not shown) and PYM 1–58 (Fig 1A, lane 3) retain Mago–Y14-binding properties. PYM 1–58 contains the most conserved region of the protein (Fig 1B and data not shown).
Figure 1.
The N-terminal domain of PYM is sufficient to interact with Mago–Y14. (A) Pull-down experiments in which lysates from E. coli expressing the D. melanogaster proteins indicated on the right were incubated with glutathione agarose beads. The construct Y14ΔC includes residues 1–154 and Y14ΔNΔC includes residues 67–154. (B) Sequence alignment of PYM N-terminal region from D. melanogaster (Dm), H. sapiens (Hs), Arabidopsis thaliana (At), C. elegans (Ce) and S. pombe (Sp). Secondary structure elements are shown above the sequences. Conserved residues are highlighted in yellow. The presence of an insertion in the Ce orthologue is indicated.
Drosophila full-length Mago, the Y14 RBD (67–154) and the N-terminal 58 residues of PYM were coexpressed and purified. The crystal structure of the ternary complex was determined at 1.9 Å resolution and refined to an Rfree of 24.9% and good stereochemistry (Table 1). It contains residues 3–35 of PYM, residues 67–153 of the Y14 RBD and residues 4–144 of Mago (with the exception of loops 14–19 and 38–45 that were disordered).
Table 1.
Data collection and refinement statistics
|
Data collection statistics | |
| Space group |
P43212 |
| Cell dimensions (Å) |
a=b=106.3, c=58.1 |
| X-ray source |
ESRF (ID29) |
| Resolution (Å) |
30–1.9 |
| Unique reflections |
26,797 |
| Multiplicity* |
7.5 (5.3) |
| Completeness (%)* |
99.8 (99.9) |
|
I/σ* |
5.2 (2.6) |
|
Rsym (%)* |
7.2 (28.7) |
|
Refinement statistics | |
|
Rfree (%) |
24.9 |
|
Rwork (%) |
23.5 |
| Reflection in the Rfree set |
1,047 |
| Protein residues |
392 |
| Protein atoms |
1,932 |
| Water molecules |
142 |
| Calcium ions |
3 |
| φ,ψ angles | |
| Most favoured (%) |
93.0 |
| Additionally allowed (%) |
7.0 |
| r.m.s.d. bonds (Å) |
0.003 |
| r.m.s.d. angles (deg) | 1.19 |
*Values for the outermost resolution shell (2.0–1.9 Å) are given in parentheses.
Pym Binds At A Surface Formed By Both Mago And Y14
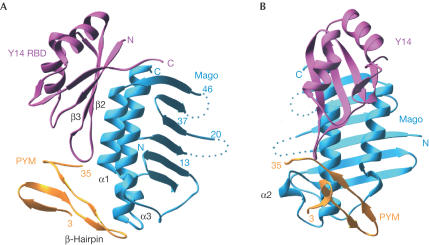
The N-terminal domain of Dm PYM binds as a small globular all-β-domain to both Mago and Y14, capping their heterodimerization interface (Fig 2). The structure of the Mago–Y14 heterodimer is very similar to that reported previously in the absence of PYM (Fribourg et al, 2003; Lau et al, 2003; Shi & Xu, 2003). Briefly, Mago consists of an antiparallel β-sheet flanked on one side by two long α-helices (α1 and α3) and a short one (α2). The α-helical surface of Mago interacts with the β-sheet surface of the Y14 RBD. More than 85% of the amino-acid residues of Mago–Y14 superpose with an overall root-mean-square deviation of less than 1.2 Å at their Cα atoms whether in the presence or absence of PYM, and whether comparing the Drosophila (Fribourg et al, 2003; Shi & Xu, 2003) or human (Lau et al, 2003) complexes. The largest differences in general are observed in Mago at the 14–19 loop, which is disordered in the present structure, and at the α2-helix. The lack of major changes in the conformation of the Mago and Y14 proteins suggests that the heterodimer acts as a rather rigid scaffold for PYM binding.
Figure 2.
Structure of the Drosophila PYM–Mago–Y14 ternary complex. (A) View of the complex between the RBD domain of Y14 (pink), Mago (cyan) and the N-terminal domain of PYM (orange). PYM binds at the edge of the Y14 β-sheet (β2–β3 loop) and at the edge of the Mago α-helices, spanning both components of the heterodimer. Dotted lines represent the approximate path of loops in Mago that were disordered and not modelled. (B) View of the complex after a 90° rotation about the vertical axis with respect to the view in (A).
The N-terminal region of PYM (3–35) folds with a three-stranded β-sheet and a contiguous β-hairpin, and does not resemble other known structures from database searches using the program DALI (Holm & Sander, 1993). Although the crystallized construct contains 25 additional C-terminal residues, these are disordered in the structure and do not contribute to Mago–Y14 binding. Sequence comparison shows the presence of a 65-residue-long insertion at this domain boundary in the Caenorhabditis elegans homologue (Fig 1B). Thus, the structure and sequence comparison data define residues 1–35 as the domain of PYM that interacts with Mago–Y14.
Specificity Of Recognition Between Pym And Mago–Y14
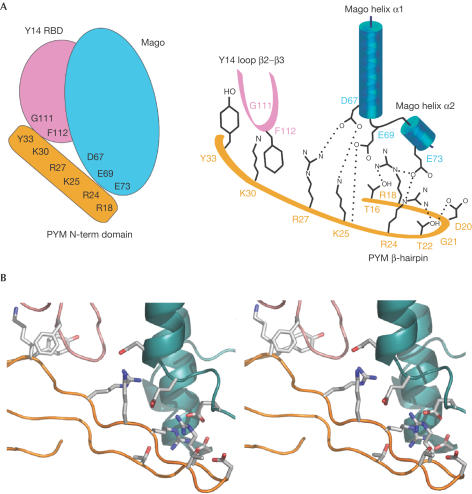
PYM binds at the α-helices of Mago with extensive electrostatic interactions and at the β2–β3 loop of Y14 with hydrophobic interactions (Fig 3). Several solvent-mediated contacts appear to strengthen the interaction, as at least 40 water molecules are found at the interface.
Figure 3.
PYM binds Mago–Y14 with extensive interactions. (A) Schematic view of the PYM–Mago–Y14 complex (left panel) and schematic diagram highlighting the key residues involved in the interaction (right panel). Positively charged residues of PYM interact with negatively charged residues of Mago helices α1 and α2. In addition, PYM interacts with the β2–β3 loop of Y14 by means of hydrophobic contacts. Colours are as in Fig 2. Hydrogen bonds are shown with dotted lines. (B) Stereo representation of the structure and of the interacting residues in a similar orientation as in Figs 2A,3A.
PYM docks with positively charged residues (Arg18PYM, Arg24PYM, Lys25PYM and Arg27PYM) to the acidic surface of the Mago α-helices (Asp67Mago, Glu69Mago, Glu73Mago and Asp116Mago) (Fig 3). Particularly well conserved are the interactions contributed by the β-hairpin portion of PYM (Figs 1B, 3). Conserved residues within the β-hairpin also include amino acids that have a structural role in constraining the fold of the hairpin by a combination of intramolecular hydrogen bonds (Thr16PYM, Asp20PYM and Thr22PYM) and flexible main-chain conformations (Pro19PYM and Gly21PYM) (Fig 3).
The N-terminal domain of PYM ends with an extended stretch that wraps around the β2–β3 loop of Y14 (Figs 2A,3). The β2–β3 loop is the most conserved part of Y14. It contributes a set of invariant residues for heterodimerization with Mago and another set for hydrophobic interactions with PYM (Phe112Y14 with the aliphatic side chain of Lys30PYM, and Gly111Y14 with Tyr33PYM; Fig 3).
Interaction Between Pym And Mago–Y14 Is Conserved
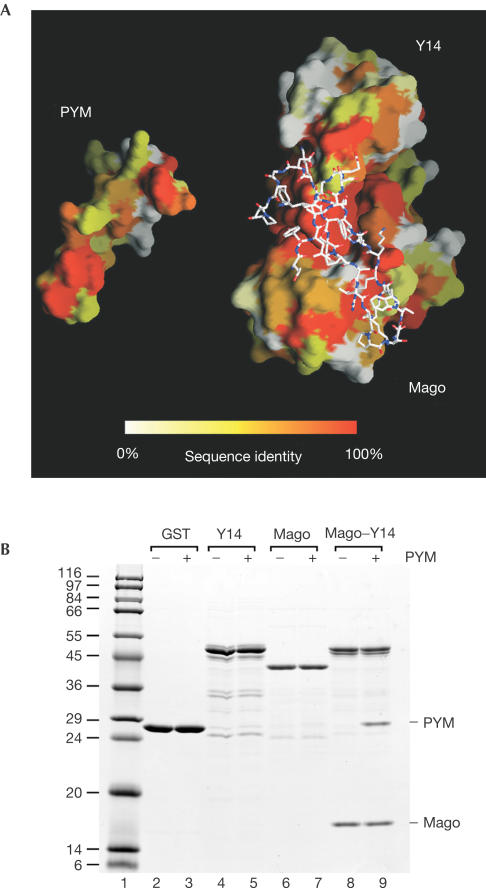
Drosophila PYM is recognized by Mago–Y14 by means of conserved surface residues spanning the entire interaction surface (Fig 4A). The conservation of the interactions suggests that formation of this trimeric complex is likely to be conserved from Schizosaccharomyces pombe to human (none of the three proteins is encoded by the Saccharomyces cerevisiae genome). We tested the formation of the Homo sapiens (Hs) ternary complex, whose components share 63, 88 and 33% sequence identity with the corresponding Dm Y14, Mago and PYM proteins. In pull-down experiments, untagged Hs PYM copurifies with glutathione S-transferase (GST)–Hs Y14–Mago (Fig 4B, lane 9), indicating that the interaction is conserved. In agreement with the structural data, Hs PYM does not interact with either Y14 or with Mago alone (Fig 4B, lanes 5 and 7).
Figure 4.
The PYM–Mago–Y14 complex is conserved across species. (A) The interaction surfaces of D. melanogaster PYM and the Mago–Y14 heterodimer have been opened up relative to the view in Fig 2A. The two surfaces are coloured according to sequence conservation, ranging from orange for conserved residues to white for variable residues. On the right, the atomic model of PYM is shown bound to the surface of Mago–Y14. (B) Lysates prepared from E. coli expressing untagged H. sapiens (Hs) PYM were incubated with glutathione agarose beads coated with GST, GST–Hs Y14, GST–Hs Mago or GST–Hs Y14–Mago dimers.
Pym Is A Cytosolic Protein With RNA-binding Properties
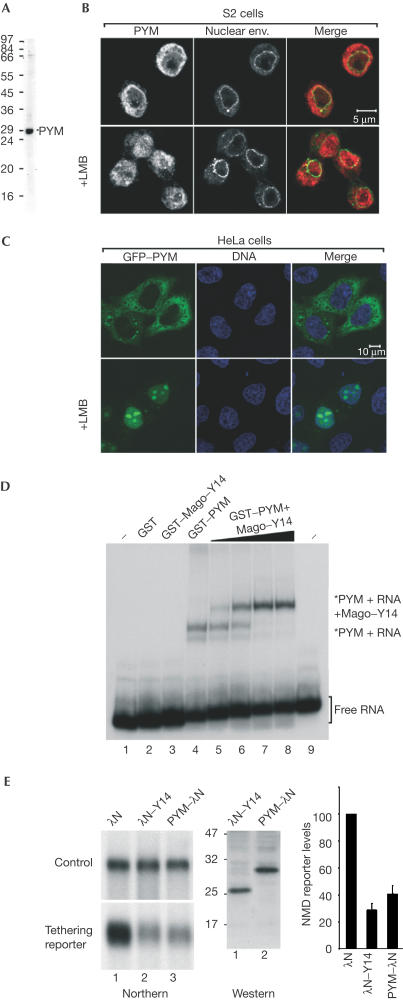
The structural and biochemical data raise the question whether PYM associates with Mago–Y14 in the nucleus or whether it is a downstream interaction. Mago and Y14 are nucleocytoplasmic shuttling proteins that localize predominantly in the nucleoplasm and in nuclear speckles (Kataoka et al, 2000, 2001; Le Hir et al, 2001a). In contrast, PYM is detected in the cytoplasm of Drosophila Schneider (S2) cells (Fig 5B, upper panels; Forler et al, 2004). The subcellular localization of PYM is conserved, as human PYM is also detected within the cytoplasm of HeLa cells transiently expressing the protein fused to green fluorescent protein (GFP–PYM) (Fig 5C, upper panels).
Figure 5.
PYM is a cytosolic RNA-binding protein involved in NMD. (A) Western blot with purified anti-Dm PYM antibody showing that a unique band of the right size is recognized in S2 cell extracts. (B) Immunolocalization of endogenous PYM in S2 cells. Endogenous PYM (red) is detected predominantly in the cytoplasm in untreated S2 cells, and in both the nucleoplasm and cytoplasm in cells treated for 12 h with leptomycin B (LMB). The nuclear envelope is stained with fluorescently labelled wheat germ agglutinin (WGA, green). (C) Subcellular localization of human PYM. HeLa cells were transfected with a plasmid expressing GFP–PYM. The fusion protein (green) is detected throughout the cytoplasm and excluded from the nucleoplasm. Following LMB treatment (3 h), GFP–PYM accumulates within the nucleus and the nucleolus. DNA is stained with Hoechst (blue). (D) Gel-mobility assay performed with a labelled RNA probe and the purified recombinant proteins indicated above the lanes. Dm Mago and PYM were N-terminally fused to GST. In lanes 2–4, the concentration of the recombinant proteins was 0.1 μg/μl. Lanes 5–8 show binding reactions with 0.1 μg/μl of GST–PYM supplemented with 0.005, 0.02, 0.05 and 0.1 μg/μl of GST Dm Mago–Y14 dimers, respectively. The position of the free RNA probe (lanes 1 and 9) and of the RNA-bound complexes is shown on the right. (E) NMD assay. Tethering PYM–λN to the 3′UTR of a reporter mRNA results in degradation of the reporter to a similar extent as observed when λN–Y14 is tethered. The steady-state levels of the NMD reporter in the presence of PYM or Y14 were quantified in three independent experiments, normalized to those of the transfection control and expressed as a percentage of the levels observed when the λN-peptide alone was coexpressed. The expression levels of the proteins were analysed by western blot with an anti-λN antibody.
Despite its cytoplasmic localization at equilibrium, Drosophila PYM is a shuttling protein exported from the nucleus by Crm1 (Fig 5B, lower panels; Forler et al, 2004). Crm1 is a transport receptor of the karyopherin β (importin β-like) family implicated in the nuclear export of a large number of proteins and whose activity is inhibited by leptomycin B (reviewed in Strom & Weis, 2001). When HeLa cells are treated with leptomycin B, GFP–PYM accumulates within the nucleoplasm and the nucleolus (Fig 5C, lower panels). This indicates that human PYM is also a shuttling protein exported from the nucleus by Crm1. The accumulation of human PYM within the nucleolus following leptomycin B treatment may reflect a specific interaction with ribosomal subunits or mislocalization due to unspecific interactions with nucleolar components such as ribosomal RNA.
We next tested whether PYM has RNA-binding properties. We have previously shown that recombinant Mago–Y14 heterodimers do not exhibit general RNA-binding activity in gel shift assays (Fribourg et al, 2003). In contrast, recombinant PYM binds RNA directly (Fig 5D, lane 4), despite showing no sequence homology to known RNA-binding proteins. The PYM–RNA complexes can be supershifted when Mago–Y14 dimers are added to the reactions (Fig 5D, lanes 5–8), indicating that PYM can bind simultaneously to RNA and to Mago–Y14.
Pym Is A Component Of The Human Nmd Machinery
Analysis of the structure of the trimeric complex reveals that PYM binding involves the direct contribution of amino-acid residues of Mago that were previously shown to have a role in nonsense-mediated mRNA decay. In the structure of the Drosophila complex, Asp67Mago and Glu69Mago interact with Lys25PYM and Arg27PYM (Fig 3). In human Mago, a double mutation of the corresponding Asp66MagoHs and Glu68MagoHs to Arg affects NMD (Fribourg et al, 2003). This double mutation is likely to cause electrostatic repulsion with the positively charged residues of PYM, suggesting a role for PYM in NMD. To test whether PYM might be active in NMD, we used a transient transfection assay in human cells in which degradation of a reporter mRNA is elicited if a protein involved in NMD is tethered downstream of a stop codon (Lykke-Andersen et al, 2001; Fribourg et al, 2003; Gehring et al, 2003). Tethering PYM to the 3′UTR of a reporter mRNA results in its degradation as detected by northern blot analysis (Fig 5E), indicating that PYM interacts with the components of the NMD machinery.
Concluding Remarks
The interaction between Mago–Y14 and PYM is direct and conserved across species. It is surprising that whereas Mago–Y14 is predominantly nuclear, PYM localizes in the cytoplasm at equilibrium. Human PYM accumulates in the nucleoplasm and nucleolus on inhibition of the export receptor Crm1, but not in nuclear speckles as is characteristic for Mago–Y14 localization. Therefore, although we cannot exclude that PYM might interact with Mago–Y14 in the nucleus, we favour a model where the recognition is a downstream event occurring in the cytoplasm.
The molecular recognition is mediated by an intricate network of interactions between the N-terminal domain of PYM (residues 3–35, Drosophila numbering) and both Mago and Y14, reinforcing the view that Mago–Y14 functions as a single structural unit. Centrally located within the PYM-interaction surface, we find residues of Mago that were previously shown to affect NMD if mutated (Fribourg et al, 2003). The implication from the structural data that the PYM-interacting surface is important for NMD is supported by tethering experiments showing degradation of an NMD reporter when human PYM is tethered downstream of a stop codon. Thus, PYM is a component of the NMD pathway. The precise molecular mechanism by which PYM has a role in NMD is an open question for further studies.
Methods
In vitro pull-down and RNA-binding assays. Plasmids allowing the expression of Mago and Y14 (Hs and Dm, as GST fusions or untagged) were described previously (Le Hir et al, 2001a; Fribourg et al, 2003). Dm Mago and Y14 67–154 were subcloned in a bicistronic plasmid derived from the pET Novagen series (pETMC vector, gift from Christophe Romier). Plasmids for expressing PYM (Dm full length and 1–58, and Hs full length) as GST fusions or untagged were obtained by inserting the corresponding cDNAs in pGEX (Pharmacia) or pET28c (Novagen) vectors. For GST pull-down assays in Fig 1, proteins were coexpressed and 500 μl lysates in binding buffer (PBS supplemented with 0.5% Triton X-100 and 10% glycerol) were immobilized for 1 h at 4°C on 50 μl of packed glutathione agarose beads. For GST pull-down assays in Fig 4B, 5 μg of GST-tagged recombinant proteins were first immobilized on beads and then 500 μl of lysates from E. coli expressing untagged PYM were added. Beads were washed three times with 500 μl of binding buffer. Bound proteins were eluted with SDS sample buffer and analysed by SDS–PAGE, followed by Coomassie blue staining. For the RNA-binding assay shown in Fig 5A, a 77-nucleotide RNA probe was used, and binding reactions were performed as described (Fribourg et al, 2003).
Protein purification and crystallization. For crystallization studies, Dm PYM 1–58 was coexpressed as a His-tagged protein cleavable by Tev protease (pProEx-Htb, Life Technologies) together with untagged Mago and Y14 67–154 (pETMC, see above). The ternary complex was purified by Talon affinity chromatography in 20 mM Tris–HCl (pH 7.5) and 200 mM NaCl. After Tev protease cleavage, the complex was further purified by size-exclusion chromatography. The Mago–Y14–PYM complex was concentrated to 16 mg/ml and crystallized by vapour diffusion at 18°C in 100 mM HEPES (pH 7.5), 28% PEG 400 and 200 mM CaCl2. Crystals appeared overnight and were optimized by microseeding to single rod-like crystals with dimensions of 200 μm × 200 μm × 400 μm.
Structure determination. Crystals were frozen in a liquid nitrogen cryostream and diffracted to 1.9 Å resolution using synchrotron radiation at ESRF (Grenoble). Data processing was carried out with MOSFLM (CCP4, 1994). The data statistics are shown in Table 1. Phasing was performed using the program AMoRe (CCP4, 1994) and the structure of the Dm Mago–Y14 binary complex (Fribourg et al, 2003) as the search model. The model was built with the program O (Jones et al, 1991) and refined with CNS (Brünger et al, 1998) to an Rfree of 24.9% and an Rfactor of 23.5% with good stereochemistry. The model statistics are shown in Table 1. The model includes three calcium ions (from the crystallization medium) that are involved in crystal contacts. An alternate conformation is detected for the side chain of Arg109Y14.
Cellular localization. Antibodies recognizing Dm PYM were raised in rats immunized with the recombinant protein expressed in E. coli as a GST fusion. The localization of the endogenous protein was determined by indirect immunofluorescence, performed as described (Herold et al, 2003). For expression in human cells, Hs PYM cDNA was cloned as an EcoRI–BglII fragment between the EcoRI–BamHI sites of pEGFPC1 (Clontech). Leptomycin B (Sigma) was dissolved in methanol and added to the cells at a final concentration of 40 ng/ml in S2 cells and 2 ng/ml in HeLa cells.
NMD tethering assay. NMD activity was assessed by coexpression of PYM fused to the λN peptide together with a β-globin-derived reporter mRNA harbouring five boxB sites (high-affinity λN-peptide-binding sites) in the 3′UTR of the reporter as described (Gehring et al, 2003). The λN peptide was fused N-terminally to Y14, but C-terminally to PYM. Indeed, PYM protein fused N-terminally to the λN peptide was inactive in tethering assays. Transient transfections in HeLa cells, RNA extractions, and northern and western blot analysis were performed as described before (Fribourg et al, 2003).
Coordinates. The atomic coordinates and structure factors have been deposited with the Protein Data Bank (accession code 1RK8).
Acknowledgments
We are grateful to beamline scientists at the ESRF ID29 beamline (Grenoble) for assistance during data collection. We thank Christophe Romier (IGBMC-Strasbourg) for the bicistronic pETMC vector, Andrea Herold for the PYM localization data in S2 cells, Yao Wei for in vitro protein binding assay, Niels Gehring, Andreas Kulozik and Matthias Hentze for the kind gift of the reporter constructs and anti-λN peptide antibodies, and Sebastien Fribourg for critical reading of the manuscript and discussions throughout. F.B. is supported by the EU contract HRPN-CT-2002-00239.
References
- Brünger AT et al. ( 1998) Crystallographic and NMR system: a new software system for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- CCP4 ( 1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr D 50: 760–763 [DOI] [PubMed] [Google Scholar]
- Forler D, Kocher T, Rode M, Gentzel M, Izaurralde E, Wilm M ( 2003) An efficient protein complex purification method for functional proteomics in higher eukaryotes. Nat Biotech 21: 89–92 [DOI] [PubMed] [Google Scholar]
- Forler D, Rabut G, Ciccarelli FD, Herold A, Kocher T, Niggeweg R, Bork P, Ellenberg J, Izaurralde E ( 2004) RanBP2/Nup358 provides a major binding site for NXF1-p15 dimers at the nuclear pore complex and functions in nuclear mRMA export. Mol Cell Biol 24: 1155–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fribourg S, Gatfield D, Izaurralde E, Conti E ( 2003) A novel mode of RBD-protein recognition in the Y14–Mago complex. Nat Struct Biol 10: 433–439 [DOI] [PubMed] [Google Scholar]
- Gatfield D, Unterholzer L, Ciccarelli FD, Bork P, Izaurralde E ( 2003) Nonsense-mediated mRNA decay in Drosophila: at the intersection of the yeast and mammalian pathways. EMBO J 22: 3960–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Neu-Yilik G, Schell T, Hentze MW, Kulozik AE ( 2003) Y14 and hUpf3b form an NMD-activating complex. Mol Cell 11: 939–949 [DOI] [PubMed] [Google Scholar]
- Hachet O, Ephrussi A ( 2001) Drosophila Y14 shuttles to the posterior of the oocyte and is required for oskar mRNA transport. Curr Biol 11: 1166–1174 [DOI] [PubMed] [Google Scholar]
- Herold A, Teixera L, Izaurralde E ( 2003) Genome-wide analysis of nuclear export pathways in Drosophila. EMBO J 22: 2472–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Sander C ( 1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233: 123–138 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M ( 1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kataoka N, Yong J, Kim VN, Velazquez F, Perkinson RA, Wang F, Dreyfuss G ( 2000) Pre-mRNA splicing imprints mRNA in the nucleus with a novel RNA-binding protein that persists in the cytoplasm. Mol Cell 6: 673–682 [DOI] [PubMed] [Google Scholar]
- Kataoka N, Diem MD, Kim VN, Yong J, Dreyfuss G ( 2001) Magoh, a human homolog of Drosophila mago nashi protein, is a component of the splicing-depepndent exon–exon junction complex. EMBO J 20: 6424–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN, Yong J, Kataoka N, Abel L, Diem MD, Dreyfuss G ( 2001) The Y14 protein communicates to the cytoplasm the position of exon–exon junctions. EMBO J 20: 2062–2068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau CK, Diem MD, Dreyfuss G, Van Duyne GD ( 2003) Structure of the Y14–Magoh core of the exon junction complex. Curr Biol 13: 933–941 [DOI] [PubMed] [Google Scholar]
- Le Hir H, Izaurralde E, Maquat LE, Moore MJ ( 2000) The spliceosome deposits multiple proteins 20–24 nucleotides upstream of mRNA exon–exon junctions. EMBO J 19: 6860–6869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Braun IC, Forler D, Izaurralde E ( 2001a) The protein Mago provides a link between splicing and mRNA localization. EMBO Rep 2: 1119–1124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Gatfield D, Izaurralde E, Moore MJ ( 2001b) The exon–exon junction complex provides a binding platform for factors involved in mRNA export and NMD. EMBO J 20: 4987–4997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Hir H, Nott A, Moore MJ ( 2003) How introns influence and enhance eukaryotic gene expression. Trends Biochem Sci 28: 215–220 [DOI] [PubMed] [Google Scholar]
- Lykke-Andersen J, Shu MD, Steitz JA ( 2001) Communication of the position of exon–exon junctions to the mRNA surveillance machinery by the protein RNPS1. Science 293: 1836–1839 [DOI] [PubMed] [Google Scholar]
- Micklem DR, Dasgupta R, Elliott H, Gergely F, Davidson C, Brand A, Gonzales-Reyes A, St Johnston D ( 1997) The mago nashi gene is required for the polarization of the oocyte and the formation of perpendicular axes in Drosophila. Curr Biol 7: 468–478 [DOI] [PubMed] [Google Scholar]
- Newmark PA, Mohr SE, Gong L, Boswell RE ( 1997) Mago nashi mediates the posterior follicle cell-to-oocyte signal to organize axis formation in Drosophila development. Development 124: 3197–3207 [DOI] [PubMed] [Google Scholar]
- Shi H, Xu RM ( 2003) Crystal structure of the Drosophila Mago nashi–Y14 complex. Genes Dev 17: 971–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom AC, Weis K ( 2001) Importin-β-like nuclear transport receptors. Genome Biol 2: REVIEWS3008 [DOI] [PMC free article] [PubMed] [Google Scholar]