Abstract
Bioluminescent mutants of Yersinia enterocolitica were generated by transposon mutagenesis using a promoterless, complete lux operon (luxCDABE) derived from Photorhabdus luminescens, and their production of light in the cheese environment was monitored. Mutant B94, which had the lux cassette inserted into an open reading frame of unknown function was used for direct monitoring of Y. enterocolitica cells on cheeses stored at 10°C by quantifying bioluminescence using a photon-counting, intensified charge-coupled device camera. The detection limit on cheese was 200 CFU/cm2. Bioluminescence of the reporter mutant was significantly regulated by its environment (NaCl, temperature, and cheese), as well as by growth phase, via the promoter the lux operon had acquired upon transposition. At low temperatures, mutant B94 did not exhibit the often-reported decrease of photon emission in older cells. It was not necessary to include either antibiotics or aldehyde in the food matrix in order to gain quantitative, reproducible bioluminescence data. As far as we know, this is the first time a pathogen has been monitored in situ, in real time, in a “real-product” status, and at a low temperature.
Yersinia enterocolitica thrives on refrigerated food (13). This pathogen occurs mainly in meat products (4, 7, 10), but it has also been recovered from various cheeses, such as Camembert cheese (3, 11, 16, 17, 20). To monitor pathogens in food by bioluminescence, various luxAB reporter genes (encoding the luciferase enzyme), predominantly those derived from Vibrio spp., have been used to engineer bacteria (1, 9, 18, 25). Using this approach, photon emission had to be induced by addition of n-decylaldehyde, and bacterial enumeration was mostly done by plating and subsequent counting of bioluminescent colonies (9, 18, 25).
Addition of an aldehyde can be circumvented by cloning the entire lux operon (luxCDABE) into bacterial cells. Bioluminescent cells containing the full-length lux operon of Photorhabdus luminescens have been used to monitor and track disease processes in living animals by measuring bioluminescence directly through the tissues of the animal. Successful applications have been demonstrated by monitoring bioluminescent Salmonella enterica serovar Typhimurium, Staphylococcus aureus, Streptococcus pneumoniae, and Escherichia coli infections in animals (2, 5, 6, 19). The full-length lux cassette of P. luminescens has also been utilized for real-time monitoring of the adherence of bioluminescent E. coli O157:H7 to beef carcass surface tissue in situ (24). However, in that study, the bioluminescent reporter strain harbored the lux operon on a plasmid. Hence, all experiments had to be performed in the presence of an antibiotic to maintain the lux plasmid.
To fully benefit from bioluminescence as a noninvasive tool for research in all aspects of food safety, the system must be applicable under natural conditions. This means that the particular ecosystem investigated should be influenced as little as possible. By using the whole lux operon stably integrated into the bacterial chromosome, (i) the necessity to penetrate the contaminated food matrix with an aldehyde can be avoided. Furthermore, (ii) the competitive flora is not influenced by either the aldehyde, which can be toxic at high concentrations, or by an antibiotic added to maintain the bioluminescence character of the reporter strain. Since food is often stored refrigerated, monitoring pathogens in situ at low temperatures without a temperature shift for bioluminescence measurements is also desirable. To attain this, the reporter gene cassette needs to be controlled by a promoter that is active at cold storage temperatures. As a step towards these goals, we report on a bioluminescent Y. enterocolitica strain harboring the full-length lux operon in the genome, under the control of a promoter active in foodstuff at low temperatures.
Construction of bioluminescent reporter strains.
A pool of random luxCDABE fusions was generated in Y. enterocolitica NCTC 10460 by mating this strain with E. coli S17-1 λpir containing a pUT-mini-Tn5 luxCDABE-Kmr plasmid, as described by Winson et al. (28). E. coli S17-1λpir containing pUT-mini-Tn5 luxCDABE-Kmr was grown at 37°C in Luria-Bertani (LB) broth (10 g of tryptone per liter, 5 g of yeast extract per liter, 5 g of NaCl per liter [pH 7.4]), supplied with 50 μg of ampicillin per ml, and Y. enterocolitica NCTC 10460 was grown in LB broth at 30°C without any antibiotics. Both strains were grown to an optical density at 600 nm (OD600) of 0.5. For mating, 100 μl of each culture was mixed, 800 μl of a 10 mM MgSO4 solution was added, and the culture was incubated at room temperature for 5 min. The cells were pelleted (5 min at 3,000 rpm at room temperature) in a model 113 centrifuge (Sigma Laborzentrifugen, Osterode am Harz, Germany), resuspended in 100 μl of LB broth before being spread onto LB agar plates (without antibiotics), and incubated overnight at 30°C. The following morning, each lawn of cells was removed from the plate surface by being carefully scraped off. These cells were then resuspended in 13% glycerol, snap-frozen in five 1-ml aliquots, each in liquid N2, and stored at −70°C. One frozen aliquot was resuspended in approximately 10 ml of LB broth to an OD600 of 0.3. Subsequently, 100 μl of this suspension was plated on LB selection plates (145 mm in diameter) containing 200 μg of kanamycin per ml and 20 μg of chloramphenicol per ml. Kanamycin selects for transpositional events, and chloramphenicol selects for Y. enterocolitica. After 24 h at 30°C, the plates were examined for colonies that produced high levels of bioluminescence.
Selection of a suitable reporter strain.
Since the Tn5 lux transposon integrates randomly into the Yersinia chromosome, transposants were screened for strongly expressed bioluminescence in the cheese environment after overnight incubation at 30°C. Six of the brightest mutants were chosen for further experiments. They were grown in brain heart infusion (BHI) broth (Merck) supplemented with 2.5% NaCl (10°C, 48 h). A 0.3-ml portion of a 100-fold dilution was applied to the surface of purchased Camembert cheeses (80 g; 45% fat [dry weight]). Light emission was measured by a photon-counting, intensified charge-coupled device camera (model C2400-75H; Hamamatsu Photonics, Hamamatsu City, Japan). After the film was exposed for 1 min (restrictor position at 0.95), the images were processed with an Argus 20 image processor (Hamamatsu). Photon emission was quantified by the Living-Image software package version 4.0 (Xenogen Corporation, Alameda, Calif.). The measurements were performed over 7 days at 10°C. Viable cell counts were enumerated by homogenizing 20 g of the Camembert cheese surface with 180 ml of 1.75% trisodium citrate-dehydrate solution (pH 7.5), using a stomacher (model Lab-Blender 400; Kleinfeld Labortechnik, Hannover, Germany). Serial 10-fold dilutions of these suspensions were plated directly on cefsulodin-irgasan-novobiocin (CIN; Oxoid) agar (30°C, 24 h) (3, 12, 13). Strain B94 showed the highest photon output per CFU (data not shown) and was chosen for further experiments. Inserting the transposon in strain B94 did not influence the growth rate (data not shown).
Microheterogeneity of Yersinia on cheese.
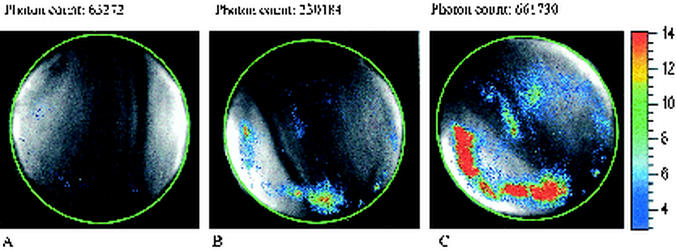
Using the intensified charge-coupled device camera, it is possible to determine the location of the bioluminescent Yersinia on the cheese surface. The distribution of the bioluminescent cells was quite irregular (Fig. 1). Although the entire cheese surface was initially contaminated with Yersinia, certain areas appeared to become more highly colonized than others. This was also observed for E. coli on beef carcasses (24) and reflects the potential of this approach to characterize the local microheterogeneity of pathogen distribution in food.
FIG. 1.
Distribution of bioluminescent Y. enterocolitica B94 on Camembert cheese directly after contamination (A), after 2 days (B), and after 3 days (C). The initial contamination level was 5 × 103 CFU/cm2. Measurement and storage were at 10°C. The total photon count of the cheese surface with a diameter of 6.5 cm is shown over each image. The scale bar on the right indicates light intensity (red is high, and blue is low).
Detection limit of strain B94.
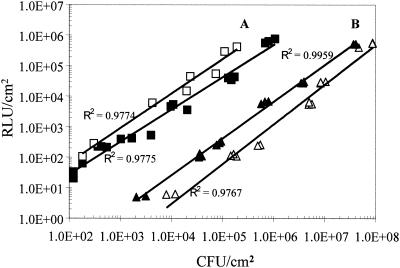
The introduction of plasmids containing luxCDABE and luxABCDE constructs derived from P. luminescens into E. coli and S. aureus (6) has demonstrated that the amount of light per cell strongly depends on the measurement temperature, lux construct, and host strain used. As shown with Y. enterocolitica B94, the significant dependence of light emission on temperature can also be observed when the luxCDABE construct is acquired by transposition. At 10°C, luciferase activity is about 15% of the one at 30°C (data not shown). Strain B94, nevertheless, is a highly sensitive reporter. To estimate the sensitivity of the method, bioluminescence of the entire cheese surface was measured. The results (Fig. 2) demonstrate a high correlation between culture-derived viable cell count and bioluminescence (r2 = 0.98). We found an in situ detection limit for Y. enterocolitica B94 on cheese of about 200 CFU/cm2.
FIG. 2.
Correlation of bioluminescence (relative light units [RLU]) and culture regimen-dependent viable cell count (CFU per square centimeter) measured at 10°C. The A lines show the correlation on cheese after 14 days of storage (open squares) and during the first 3 days after inoculation (closed squares). The B lines show the correlation for Y. enterocolitica on BHI agar with 2.5% salt (closed triangles) and without salt (open triangles) on the first 5 days. The background luminescence of cheese and BHI agar has been subtracted. Immediately after the light intensity was measured, the plates or cheeses were used to determine the viable counts (see text).
Light emission of Y. enterocolitica B94 is partly dependent on the growth phase.
Figure 2 shows that the bioluminescence per cell of strain B94 increases on the cheese surface over time. We therefore compared the bioluminescence of strain B94 when this bacterium is grown at various temperatures between 4 and 30°C to an OD600 of 0.15 (log phase) and an OD600 of 1.1 (stationary phase) (Fig. 3). Cultures of both growth temperatures were measured at the same temperature (10°C). In the logarithmic growth phase, the expression of light per cell appeared to be little affected by the growth temperature, indicating a temperature-independent regulation of the promoter in exponentially growing cultures. At higher temperatures, stationary phase resulted in a significant reduction of light emission (Fig. 3). The latter phenomenon corresponds with the observations of other researchers working with bacteria at temperatures between 28 and 40°C and using different lux systems derived from marine and terrestrial bacteria in various gram-negative as well as gram-positive transformants (5, 14, 15, 26, 27). It has been reported that upon entrance into stationary phase and during starvation, there is a decrease in luciferase activity that corresponded to a decrease in the metabolic activity of the population while the number of culturable cells remained relatively stable (26). In our study, the stationary-phase cultures were not measured at the very beginning of this growth phase (Fig. 3). Nevertheless, at 23 and 30°C, they still yield a photon count of about 10% that of the logarithmic cultures. These findings are in contrast with those of Francis and coworkers (5), who found a very steep decline of bioluminescence to nearly zero within 1 to 3 h after the cells entered stationary phase. They are, however, consistent with those of Unge et al. (26) and Marincs et al. (14), who reported a gradual growth phase-dependent light reduction. Observing similar differences with a number of lactococcal promoters, Waterfield et al. (27) demonstrated that the rate of decrease of bioluminescence depends on both the growth phase of the culture and the strength of the promoter, with low-activity promoters displaying a more rapid decay. Hence, our data may indicate that the lux cassette of Y. enterocolitica B94 is regulated by a rather strong promoter.
FIG. 3.
Influence of growth temperature and growth phase on bioluminescence of Y. enterocolitica B94. The mutant was grown in BHI broth with 2.5% salt at different temperatures to an OD600 of 0.15 (5 × 107 CFU/ml) (triangles) and to an OD600 of 1.1 (4.5 × 108 CFU/ml) (diamonds). A 100-μl sample of the grown culture was spread on BHI plates preadjusted to 10°C. After 5 min at 10°C, bioluminescence was measured at 10°C. RLU, relative light units; 1.0E+02, 1 × 102.
The growth phase-dependent decrease of bioluminescence in Y. enterocolitica B94 is temperature dependent. Light emission of stationary-phase cultures of Y. enterocolitica B94 grown at 4, 10, and 15°C is strikingly different from that of cultures propagated at higher temperatures (Fig. 3). Although the photon counts of stationary-phase cultures at low temperatures are lower than those of logarithmic cultures, they do not exhibit the steep growth phase-dependent decline observed at 23 and 30°C. Instead, they are of the same magnitude as the ones detected in logarithmic cultures. We could not find any corresponding data on low-temperature measurements in the literature. Obviously, the regulation of the promoter that controls the lux operon in Y. enterocolitica B94 is temperature dependent during stationary phase, yielding stronger expression at low temperatures. This observation indicates an important function of the promoter and its original gene(s) in the cold stress response of Y. enterocolitica in the stationary phase. Furthermore, we observed an even higher relative light unit rate per cell on Camembert cheese after 14 days at 10°C than during day 1 to 3, with a detection limit around 200 CFU/cm2 (Fig. 2). These data suggest an additional upregulation of the promoter in advanced stationary-phase cultures. According to these results, the promoter and the gene(s) originally linked to it may play a role in starvation metabolism of Y. enterocolitica. These results demonstrate the suitability of mutant B94 as a sensitive reporter in long-term experiments at low temperatures, which is a setting common to food storage at refrigeration temperatures.
Light emission of Y. enterocolitica B94 is salt dependent.
Mutant B94 was grown at 10°C in BHI broth with 2.5% salt and without salt to a titer of 6 × 107 CFU/ml. Portions (0.1 ml) of a serially (tenfold) diluted culture were inoculated on precooled BHI agar (10°C). Immediately after bioluminescence was measured, Y. enterocolitica was enumerated by homogenizing all the agar on the plate (20 ± 1 g) with 180 ml of 1.75% trisodium citrate-dehydrate solution (pH 7.5) using a stomacher (model Lab-Blender 400; Kleinfeld Labortechnik). Serial 10-fold dilutions of these suspensions were plated directly on BHI agar and incubated (30°C, 24 h), and total Y. enterocolitica counts were recorded. The correlation between bioluminescence and viable cell count for cells grown on BHI agar with and without 2.5% salt is shown in Fig. 2. Clearly, the expression of luciferase is stimulated in the presence of sodium chloride. This is significant since the concentration of sodium chloride in soft cheese may well reach this concentration (23). The elevated light emission in the presence of 2.5% NaCl is detectable in the whole range of temperatures tested (4 to 37°C [data not shown]). While the presence of 2.5% salt stimulates the light emission of Y. enterocolitica B94 in BHI broth, it reduces its growth rate in this medium (data not shown), indicating stress conditions. Hence, the promoter that regulates the inserted lux cassette in strain B94 may also be involved in the salt stress response of Y. enterocolitica.
The finding that bioluminescence of mutant B94 on BHI agar was much lower than on the cheese, even in the presence of sodium chloride, was quite unexpected (Fig. 2). This could have been due to an underestimation of the viable cell count on cheese. CIN medium, however, is the most efficient medium for enumeration of Y. enterocolitica (8, 21, 22), and it appears unlikely that this approach should underestimate the cell count by a factor of 20. The difference could also be due to a repression of the promoter by BHI agar. Also, stimulation by some cheese constituents, the influence of the competitive flora, or the influence of more than one factor may play a role. This phenomenon is currently under investigation in our laboratory.
We have shown here that insertion of a transposon carrying a promoterless modified lux operon downstream of a suitable host promoter allows sensitive real-time monitoring of Y. enterocolitica in situ. However, since strongly expressed promoters are often regulated by a number of environmental factors, it is necessary to select appropriate reporter strains carefully with respect to the conditions prevalent in the foodstuff which will be under investigation. The cheese samples were not artificially influenced by any chemical, antibiotic, or physical treatment in order for bioluminescence measurements to be made. Thus, bacterial numbers can be quantified accurately. Our study shows that it is possible to bridge the gap towards a real-product status assessment of pathogens in real time and in situ, even at low temperatures.
REFERENCES
- 1.Chen, J., and M. W. Griffiths. 1996. Luminescent Salmonella strains as real time reporters of growth and recovery from sublethal injury in food. Int. J. Food Microbiol. 31:27-43. [DOI] [PubMed] [Google Scholar]
- 2.Contag, C. H., P. R. Contag, J. I. Mullins, S. D. Spilman, D. K. Stevenson, and D. A. Benaron. 1995. Photonic detection of bacterial pathogens in living hosts. Mol. Microbiol. 18:593-603. [DOI] [PubMed] [Google Scholar]
- 3.Erkmen, O. 1996. Survival of virulent Yersinia enterocolitica during the manufacture and storage of Turkish Feta cheese. Int. J. Food Microbiol. 33:285-292. [DOI] [PubMed] [Google Scholar]
- 4.Falcao, D. P. 1991. Occurrence of Yersinia spp. in foods in Brazil. Int. J. Food Microbiol. 14:179-182. [DOI] [PubMed] [Google Scholar]
- 5.Francis, K. P., J. Yu, C. Bellinger-Kawahara, D. Joh, M. J. Hawkinson, G. Xiao, T. F. Purchio, M. G. Caparon, M. Lipsitch, and P. R. Contag. 2001. Visualizing pneumococcal infections in the lungs of live mice using bioluminescent Streptococcus pneumoniae transformed with a novel gram-positive lux transposon. Infect. Immun. 69:3350-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francis, K. P., D. Joh, C. Bellinger-Kawahara, M. J. Hawkinson, T. F. Purchio, and P. R. Contag. 2000. Monitoring bioluminescent Staphylococcus aureus infections in living mice using a novel luxABCDE construct. Infect. Immun. 68:3594-3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fukushima, H., K. Hoshina, R. Nakamura, and Y. Ito. 1987. Occurrence of Yersinia spp. in raw beef, pork and chicken. Zentbl. Bakteriol. Mikrobiol. Hyg. 184:50-59. [PubMed] [Google Scholar]
- 8.Hamama, A., A. el Marrakchi, and F. el Othmani. 1992. Occurrence of Yersinia enterocolitica in milk and dairy products in Morocco. Int. J. Food Microbiol. 16:69-77. [DOI] [PubMed] [Google Scholar]
- 9.Hudson, L. M., A. R. Hill, J. Chen, and M. W. Griffiths. 1997. Bioluminescence: a rapid indicator of Escherichia coli 0157:H7 in selected yogurt and cheese varieties. J. Food Prot. 60:891-897. [DOI] [PubMed] [Google Scholar]
- 10.Johannessen, G. S., G. Kapperud, and H. Kruse. 2000. Occurrence of pathogenic Yersinia enterocolitica in Norwegian pork products determined by a PCR method and a traditional culturing method. Int. J. Food Microbiol. 54:75-80. [DOI] [PubMed] [Google Scholar]
- 11.Johnson, E. A., J. H. Nelson, and M. Johnson. 1990. Microbiological safety of cheese made from heat treated milk. II. Microbiology. J. Food Prot. 53:519-540. [DOI] [PubMed] [Google Scholar]
- 12.Karaioannoglou, P., P. Koidis, D. Papageorgiou, and A. Mantis. 1985. Survival of Yersinia enterocolitica during the manufacture and storage of feta cheese. Milchwissenschaft 40:204-206. [Google Scholar]
- 13.Little, C. L., and S. Knochel. 1994. Growth and survival of Yersinia enterocolitica, Salmonella and Bacillus cereus in Brie stored at 4, 8 and 20°C. Int. J. Food Microbiol. 24:137-145. [DOI] [PubMed] [Google Scholar]
- 14.Marincs, F. 2000. On-line monitoring of growth of Escherichia coli in batch cultures by bioluminescence. Appl. Microbiol. Biotechnol. 53:536-541. [DOI] [PubMed] [Google Scholar]
- 15.Marincs, F., and D. W. White. 1994. Immobilization of Escherichia coli expressing the lux genes of Xenorhabdus luminescens. Appl. Environ. Microbiol. 60:3862-3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moustafa, M. K. 1990. Isolation of Yersinia enterocolitica from raw milk and soft cheese in Assiut city. Assiut Vet. Med. J. 23:106-109. [Google Scholar]
- 17.Nooitgedagt, A. J., and B. J. Hartog. 1988. A survey of the microbiological quality of Brie and Camembert cheese. Neth. Milk Dairy J. 42:57-72. [Google Scholar]
- 18.Ramsaran, H., J. Chen, B. Brunke, A. Hill, and M. W. Griffiths. 1998. Survival of bioluminescent Listeria monocytogenes and Escherichia coli O157:H7 in soft cheeses. J. Dairy Sci. 81:1810-1817. [DOI] [PubMed] [Google Scholar]
- 19.Rocchetta, H. L., C. J. Boylan, J. W. Foley, P. W. Iverson, D. L. Letourneau, C. L. McMillian, P. R. Contag, D. E. Jenkins, and T. R. Parr, Jr. 2001. Validation of a noninvasive, real-time imaging technology using bioluminescent Escherichia coli in the neutropenic mouse thigh model of infection. Antimicrob. Agents Chemother. 45:129-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schiemann, D. A. 1978. Association of Yersinia enterocolitica with the manufacture of cheese and occurrence in pasteurized milk. Appl. Environ. Microbiol. 36:274-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schiemann, D. A. 1982. Development of a two-step enrichment procedure for recovery of Yersinia enterocolitica from food. Appl. Environ. Microbiol. 43:14-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schiemann, D. A. 1979. Synthesis of a selective agar medium for Yersinia enterocolitica. Can. J. Microbiol. 25:1298-1304. [DOI] [PubMed] [Google Scholar]
- 23.Shaw, M. B. 1986. Modern cheesemaking: soft cheeses, vol. 1. Elsevier Applied Science, London, England.
- 24.Siragusa, G. R., K. Nawotka, S. D. Spilman, P. R. Contag, and C. H. Contag. 1999. Real-time monitoring of Escherichia coli O157:H7 adherence to beef carcass surface tissues with a bioluminescent reporter. Appl. Environ. Microbiol. 65:1738-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tomicka, A., J. Chen, S. Barbut, and M. W. Griffiths. 1997. Survival of bioluminescent Escherichia coli O157:H7 in a model system representing fermented sausage production. J. Food Prot. 60:1487-1492. [DOI] [PubMed] [Google Scholar]
- 26.Unge, A., R. Tombolini, L. Mølbak, and J. K. Jansson. 1999. Simultaneous monitoring of cell number and metabolic activity of specific bacterial populations with a dual gfp-luxAB marker system. Appl. Environ. Microbiol. 65:813-821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waterfield, N. R., R. W. Le Page, P. W. Wilson, and J. M. Wells. 1995. The isolation of lactococcal promoters and their use in investigating bacterial luciferase synthesis in Lactococcus lactis. Gene 165:9-15. [DOI] [PubMed] [Google Scholar]
- 28.Winson, M. K., S. Swift, P. J. Hill, C. M. Sims, G. Griesmayr, B. W. Bycroft, P. Williams, and G. S. Stewart. 1998. Engineering the luxCDABE genes from Photorhabdus luminescens to provide a bioluminescent reporter for constitutive and promoter probe plasmids and mini-Tn5 constructs. FEMS Microbiol. Lett. 163:193-202. [DOI] [PubMed] [Google Scholar]