Abstract
The extracellular speciation of mercury may control bacterial uptake and methylation. Mercury-polysulfide complexes have recently been shown to be prevalent in sulfidic waters containing zero-valent sulfur. Despite substantial increases in total dissolved mercury concentration, methylation rates in cultures of Desulfovibrio desulfuricans ND132 equilibrated with cinnabar did not increase in the presence of polysulfides, as expected due to the large size and charged nature of most of the complexes. In natural waters not at saturation with cinnabar, mercury-polysulfide complexes would be expected to shift the speciation of mercury from HgS0(aq) toward charged complexes, thereby decreasing methylation rates.
Sulfate-reducing bacteria are important methylators of mercury in aquatic systems (13, 19). These bacteria are found throughout reduced zones but are commonly concentrated at oxic-anoxic boundaries, where methylation rates in natural sediments and soils are often highest. The speciation of mercury has been shown to be an important determinant of its biological uptake; neutral species entering by passive diffusive transport are believed to be the major cell permeants, as charge hinders partitioning into (and hence diffusion through) a lipid bilayer (8). Passive diffusion of a neutral mercury-chloride complex, HgCl2, has been shown to occur in aerobic environments (2, 22, 27, 28), and estimates of the formation constant for neutral HgS0(aq) (14), together with field and experimental evidence (3, 5-8), indicate this to be the dominant mercury species taken up by cells at low sulfide concentrations.
Recent studies (25, 30) have demonstrated that polysulfides have a significant effect on the speciation of mercury in sulfidic waters. Several processes can lead to their formation; these include partial oxidation of hydrogen sulfide species, partial reduction of oxidized sulfur species, and the nucleophilic attack of HS− on elemental sulfur (12, 17, 34) as follows: HS− + (x − 1)/8 S8(s) = Sx2− + H+, where x is 3 to 6. The dominant dissolved mercury-polysulfide species in sulfidic water in equilibrium with cinnabar [HgS(s)] and zero-valent sulfur are inferred to be the charged complexes Hg(Sx)22− and HgSxOH− (15, 25, 31). Recent octanol-water partitioning experiments (25) have confirmed the hydrophilic nature of the dominant complexes, although an uncharged mercury-polysulfide complex (possibly HgS50, referred to here as HgSx0) also appears to form at an approximately 10 pM concentration in solutions in equilibrium with cinnabar.
The maximum uptake rate of a cell membrane permeant by passive diffusion, V (amol cell−1 day−1), can be estimated by the equation V = P · C(aq) · A, where P is the ability of the cell permeant to cross the membrane (cm s−1), C(aq) is its extracellular aqueous concentration (amol cm−3) (the intracellular concentration is neglected), and A is the area of a cell (cm2 cell−1) (28, 32). P depends on a permeant's tendency to partition into and its diffusivity within a lipid bilayer membrane; a molecule's Kow, or octanol-water partition coefficient [approximately 25 for both HgS0 and Hg(SH)20 (9) to 900 to 11,000 for HgSx0 (25)], is thus an important determinant of permeativity. An empirical relationship exists between Kow and P∗ for membrane permeants (28, 32), with P∗ defined as the theoretical limit for the ability of a molecule to permeate the membrane. P in turn can be estimated from the empirical relationship log P = log P∗ − mv · v, where mv is the size selectivity of membranes (0.0546 mol cm−3) and v is the estimated molar volume of the permeant (cm3 mol−1) (32). This approach has been used in the case of uptake of other uncharged mercury species by phytoplankton (28) and sulfate-reducing bacteria (6). Using molar volumes of 39 (considering that the actual structure is likely HgSHOH), 60, and 105 cm3 mol−1 (6, 10), the resulting P's are 0.07, 0.006, and 0.02 cm s−1 for HgS0(aq), Hg(SH)20, and HgSx0, respectively.
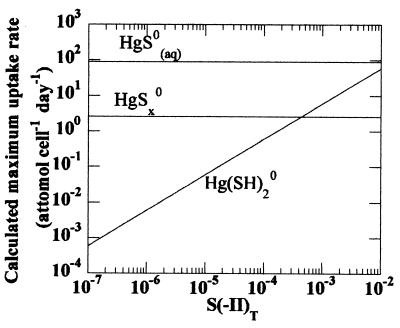
Figure 1 depicts the maximum uptake rate, V, predicted from our speciation model (25) for HgSx0, HgS0(aq), and Hg(SH)2 as a function of total sulfide concentration for solutions in equilibrium with elemental sulfur and cinnabar. The higher calculated Kow of the mercury-polysulfide complex is offset by its large size and relatively lower concentration, such that the contribution of HgSx0 to diffusive mercury uptake is expected to be small relative to that of HgS0(aq). If the hypothesis that the concentration of membrane-permeative species controls bacterial uptake is correct, mercury-polysulfide complexes would be predicted not to contribute appreciably to the biological availability of mercury species via passive uptake processes despite marked increases in the solubility of cinnabar. Rather, HgS0(aq) would play a dominant role.
FIG. 1.
The calculated maximum uptake rate of a cell membrane permeant at pH 7.3 as a function of (SII)T.
For solutions at equilibrium with cinnabar, the concentration of HgS0(aq) is independent of the concentrations of sulfide species and polysulfides (the intrinsic solubility of cinnabar is estimated to be approximately 10−9.3 M). Under such conditions, the presence of polysulfides would not be anticipated to affect the availability of mercury by passive diffusion.
By measuring rates of mercury methylation in active cultures of the sulfate-reducing bacterium Desulfovibrio desulfuricans ND132 in equilibrium with cinnabar, HgS(s), in both the absence and the presence of polysulfides, we tested the above predictions. We also compared the observed methylation rates with absolute estimates of the ability to permeate the membrane.
Cultures, growth medium, and cell density determination.
D. desulfuricans strain ND132, isolated from mesohaline Chesapeake Bay (20), was cultured (at 22°C) under sulfate-reducing conditions in a bicarbonate-buffered basal salt medium (29) with 16 mM lactate and 28 mM Na2SO4. This strain will be made available upon request. Cell density was determined by direct counts using 4′,6-diamidino-2-phenylindole (Sigma) staining and epifluorescence microscopy and in some cultures by measuring optical density at 600 nm.
Synthesis and measurement of polysulfides.
Polysulfides were generated via the reaction of HS− (prepared using washed crystals of Na2S · 9H2O; Mallinckrodt) with elemental sulfur (S8, sublimed; Mallinckrodt). It is convenient to envision each polysulfide dianion as comprising one S(II) atom and (x − 1) S(0) atoms. The total dissolved zero-valent sulfur concentration can be expressed as S(0)T = 8 · [S8(aq)] + Σ(x − 1) · [(Sx2−) + (HSx−)]; note that S8(aq) is sufficiently low that its contribution to S(0)T can be neglected under these conditions. The total polysulfide concentration S(0)T was measured by the triphenylphosphine sulfide (TPPS) method (1, 11, 33). TPPS was then analyzed by high-performance liquid chromatography (33). The total concentration of hydrogen sulfide species, (H2S)T = (H2S) + (HS−) + (S2−), referred to as the sulfide concentration, was determined using the methylene blue method of Cline (21) (detection limit of approximately 1 μM).
Total dissolved mercury and methylmercury determination.
Syringe-filtered (0.02-μm-pore-size Anotop) aliquots were digested with bromine monochloride (BrCl) and analyzed by stannous chloride reduction and cold-vapor atomic fluorescence spectrometry (9, 18). Reagent blanks (0.05 to 0.2 ng of mercury) were consistent throughout a given day using the same batch of reagents. Detection limits were 0.5 to 2.0 ng/liter. Monomethyl mercury in syringe-filtered samples was determined by gas chromatography with cold-vapor atomic fluorescence spectrometry detection (26). Samples were purged after acetic acid addition to remove sulfide and extracted into dichloromethane. The detection limit for this procedure (extracting 2 ml of sample, with no preconcentration) was 40 ng/liter.
Growth of D. desulfuricans in the presence of polysulfides.
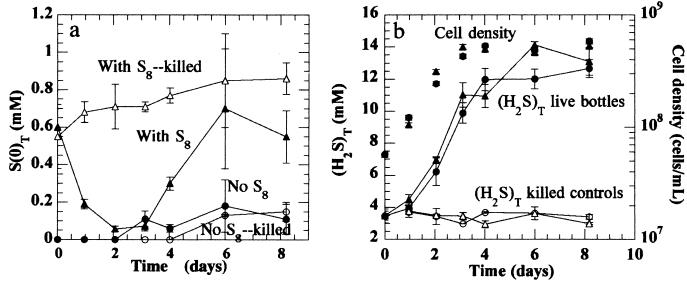
Before studying the effect of polysulfides on mercury methylation, we investigated their effect on the growth of sulfate-reducing cultures. Bottles containing medium in the presence or absence of S8 (all solids were ethanol washed and subjected to tyndallization [16]) at pH 7.3 with an initial sulfate concentration of 28 mM were equilibrated with an initial (H2S)T of 3.5 mM for 1 week prior to inoculation. After inoculation, the polysulfide concentration began to decrease immediately in bottles with live samples, with no lag time (Fig. 2a). For killed controls, the polysulfide concentration increased slightly (probably due to the additional sulfide transferred with the inoculum). The decrease in polysulfide concentration appears to be due to utilization by the organism (23). This has been observed under S(0)-reducing conditions, under which the addition of a small amount of (H2S)T, which solubilizes S(0) to polysulfides, increases the reduction rate of S(0) (24). In the experiments of Fig. 2, sulfide production from polysulfide reduction was small compared to that generated by reduction of sulfate, and the resulting increase was not significant. Sulfide production leveled off after day 4, apparently due to lactate limitation, and polysulfide concentration rebounded to its original value. Neither (H2S)T production nor bacterial growth were significantly affected by the presence of polysulfides (Fig. 2b).
FIG. 2.
S(0)T (a) and cell density and (H2S)T (b) over time in triplicate growing cultures of D. desulfuricans ND132, in pH 7.3 medium with 28 mM sulfate (ionic strength = 0.2 M), in the absence (•) and presence (▴) of elemental sulfur. Open circles and triangles indicate killed controls for each group. Concentration of S(0)T was measured by the triphenylphosphine derivatization with high-performance liquid chromatography separation and UV absorbance detection. Error bars represent the standard deviations of measurements.
Comparison of mercury methylation rates in the presence and absence of polysulfides.
Mercury methylation by D. desulfuricans ND132 was measured in cultures equilibrated with cinnabar in the presence and absence of polysulfides. The initial (H2S)T in two separate experiments was 4 or 7.5 mM, which reacted with S8 to generate polysulfides at 0.22 or 0.45 mM S(0)T, respectively. The medium was reduced with 0.1 mM titanium(III)-nitrilotriacetic acid (which does not affect the mercury speciation) so that sulfide would not serve as a reductant. Dissolved methylmercury, total dissolved mercury, (H2S)T, and cell density were measured over time.
In triplicate bottles, initially at 4 mM (H2S)T and pH 7.0, total dissolved mercury at inoculation was eight times greater in the presence of S8. Total dissolved mercury concentration in bottles in the absence of added S8 remained fairly constant, while that in bottles with S8 decreased after inoculation; at the lowest measured concentration, however, the dissolved mercury concentration in the presence of polysulfides was still higher, by a factor of 3, than in the absence of added S8. This temporal pattern of total dissolved mercury is consistent with the transient decrease of polysulfides during cell growth.
At 4 mM initial (H2S)T, growth rates were similar in the absence and presence of S8 (95% confidence intervals of 1.3 ± 0.4 day−1 and 1.2 ± 0.6 day−1, respectively; data not shown), as were final (H2S)T concentrations (11.5 and 12.5 mM). The methylation rate, Kmeth (amol cell−1 day−1), was calculated according to the following equation: Kmeth = [CH3Hg+]tμ/(xt − x0), where μ is the specific growth rate (day−1) and x0 and xt are numbers of cells per milliliter at time zero and t, respectively. Kmeth was not significantly influenced by polysulfides, being 0.021 ± 0.004 and 0.016 ± 0.004 amol of HgCH3+ cell−1 day−1 in their absence and presence, respectively. (The means of replicate methylmercury analyses for each bottle were averaged for each group [absence and presence of S8] and the relative standard deviation of 15% was used in error propagation.)
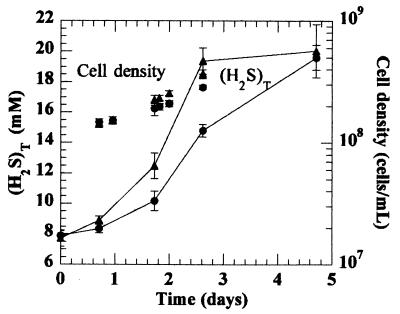
Similar results were obtained at an initial (H2S)T of 7.5 mM, pH 7.3. Growth rates in the absence and presence of S8 were 0.34 ± 0.09 and 0.50 ± 0.05 day−1, respectively (Fig. 3). Total dissolved sulfide (H2S)T was 20 mM for each treatment at the end of the experiment, although it was somewhat higher at intermediate times in the presence of S8 (Fig. 3). In these cultures, methylmercury was measured at the beginning and the end of the experiment. The rates of methylation were 0.027 ± 0.008 and 0.020 ± 0.003 amol of HgCH3+ cell−1 day−1 in the absence and presence of polysulfides, respectively, again not significantly different at the 95% confidence level.
FIG. 3.
Cell density and (H2S)T throughout a methylation experiment with triplicate cultures of D. desulfuricans ND132 (I = 0.2 M) with initial (H2S)T of 7.5 mM in the absence (•) and presence (▴) of elemental sulfur. Open circles and triangles indicate killed controls for each group. Cinnabar was present in all bottles. Error bars represent the standard deviations of measurements.
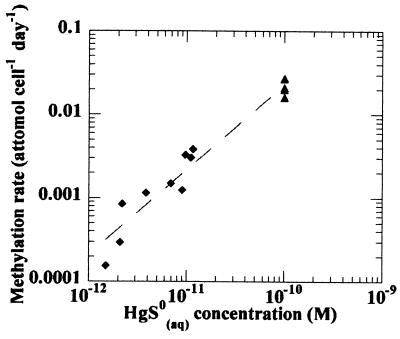
The addition of elemental sulfur in the presence of cinnabar markedly increased the concentration of dissolved mercury in the culture medium, as expected. This was not accompanied, however, by an increase in methylation rate, consistent with the hypothesis that the concentration of membrane-permeative species controls the methylation rate. Methylation rates observed in these systems (at saturation with cinnabar) agree well with data by Benoit et al. (6) relating the concentration of HgS0(aq) to the bacterial methylation rate (Fig. 4).
FIG. 4.
Observed methylation rates as a function of HgS0(aq) concentration. Diamonds and triangles represent data from Benoit et al. (7) and this study, respectively. A formation constant of 10−10 for HgS0(aq) has been used in all calculations for consistency.
The actual rates of mercury methylation in our cultures, however, were approximately 10−4 of the calculated maximum possible diffusive rate of uptake of HgS0(aq) through a lipid bilayer membrane. Possibly the vast majority of mercury passing into the cell becomes bound (such as to sulfhydryl groups in proteins), in which case transport limitation per se could limit the intracellular concentration of the substrate for methylation (4). However, this result suggests an alternative mechanism by which the concentration of neutral species in the culture medium may control mercury availability for methylation. Without a large nonmethylation sink within the cell, HgS0(aq) may equilibrate across the cell membrane. In this case, the extracellular concentration of HgS0(aq) would thus determine the intracellular concentration of HgS0(aq), and equilibria within the cell would then fix the mercury species (e.g., Hg2+) that serves as a substrate for methylation.
Regardless of the mechanism by which the extracellular concentration of membrane-permeative species controls methylation rate, these results lead directly to quantitative predictions about the effect of polysulfides on environmental mercury methylation in systems not at equilibrium with cinnabar. In the absence of polysulfides, HgS0(aq) is the dominant species at low sulfide concentrations. At higher sulfide levels, HgS2H− becomes the dominant complex, resulting in a marked decrease in HgS0(aq) and a consequent decrease in the calculated maximum uptake rate for mercury. This model has been employed to explain trends in methylmercury concentrations in the Florida Everglades and the Patuxent River Estuary (3, 7) as well as experimental mercury methylation results (6).
At saturation with elemental sulfur, the dominant species are HgSxOH− at low sulfide levels and Hg(Sx)22− as sulfide levels increase. The HgS0(aq) concentration is expected to be lower than in the purely sulfidic case due to the dominance of these charged complexes, resulting in a greatly reduced estimated ability to permeate the membrane. Other models for the speciation of mercury in porewater (that consider, for example, sorption to surface thiols [3]) would also show a proportionate decrease in the concentration of HgS0(aq) with the addition of polysulfides. These considerations imply that the formation of polysulfides in natural waters would be expected to decrease methylation rates, except when cinnabar is present.
Acknowledgments
We thank Janina Benoit for her helpful insights and advice and Ron Jones and John MacFarlane for their generous method assistance. Also, we are grateful to Dan Brabander, Nicole Keon, Dianne Newman, Martin Polz, David Senn, and anonymous reviewers for their comments on the manuscript.
This work was supported by the NIEHS Superfund Basic Research Fund (grant P42-ES04675-08) and a GE Graduate Fellowship to J.A.J.
REFERENCES
- 1.Andersson, J. T., and U. Holwitt. 1994. An advantageous reagent for the removal of elemental sulfur from environmental samples. J. Anal. Chem. 350:474-480. [Google Scholar]
- 2.Barkay, T., M. Gillman, and R. Turner. 1997. Effects of dissolved organic carbon and salinity on bioavailability of mercury. Appl. Environ. Microbiol. 63:4267-4271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benoit, J., C. Gilmour, R. Mason, and A. Heyes. 1999. Sulfide controls on mercury speciation and bioavailability to methylating bacteria in sediment pore waters. Environ. Sci. Technol. 33:951-957. [Google Scholar]
- 4.Benoit, J., C. Gilmour, A. Heyes, R. P. Mason, and C. L. Miller. Geochemical and biological controls over methylmercury production and degradation in aquatic ecosystems. In Biogeochemistry of environmentally important trace metals. In Y. Chai (ed.), Am. Chem. Soc. Symp. Ser., in press.
- 5.Benoit, J. M., C. C. Gilmour, and R. P. Mason. 2001. Aspects of bioavailability of mercury for methylation in pure cultures of Desulfobulbus propionicus (1pr3). Appl. Environ. Microbiol. 67:51-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Benoit, J. M., C. C. Gilmour, and R. P. Mason. 2001. The influence of sulfide on solid-phase mercury availability for methylation by pure cultures of Desulfobulbus propionicus (1pr3). Environ. Sci. Technol. 35:127-132. [DOI] [PubMed] [Google Scholar]
- 7.Benoit, J. M., C. C. Gilmour, R. P. Mason, G. S. Riedel, and G. F. Riedel. 1998. Behavior of mercury in the Patuxent River estuary. Biogeochemistry 40:249-265. [Google Scholar]
- 8.Benoit, J. M., R. Mason, and C. Gilmour. 1999. Estimation of mercury-sulfide speciation in sediment pore waters using octanol-water partitioning and implications for availability to methylating bacteria. Environ. Toxicol. Chem. 18:2138-2141. [DOI] [PubMed] [Google Scholar]
- 9.Bloom, N., and W. Fitzgerald. 1988. Determination of volatile mercury species at the picogram level by low temperature gas chromatography with cold vapor atomic fluorescence detection. Anal. Chim. Acta 208:151-161. [Google Scholar]
- 10.Bondi, A. 1964. van der Waals volumes and radii. J. Phys. Chem. 68:441-451. [Google Scholar]
- 11.Borchardt, L. G., and D. B. Easty. 1984. Gas chromatographic determination of elemental and polysulfide sulfur in kraft pulping liquors. J. Chromatogr. 299:471-476. [Google Scholar]
- 12.Boulegue, J., and G. Michard. 1978. Constantes de formation des ions polysulfures S62−, S52− et S42− en phase aqueous. J. Fr. Hydrol. 9:27-34. [Google Scholar]
- 13.Compeau, C., and R. Bartha. 1985. Sulfate-reducing bacteria: principal methylators of mercury in anoxic estuarine sediment. Appl. Environ. Microbiol. 50:498-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dyrssen, D. 1989. Biogenic sulfur in two different marine environments. Mar. Chem. 28:241-249. [Google Scholar]
- 15.Dyrssen, D., and M. Wedborg. 1991. The sulfur-mercury (II) system in natural waters. Water Air Soil Pollut. 56:507-519. [Google Scholar]
- 16.Gerhardt, P., R. Murray, W. Wood, and N. Krieg. (ed.). 1994. Methods for general and molecular bacteriology. American Society for Microbiology, Washington, D.C.
- 17.Giggenbach, W. 1972. Optical spectra and equilibrium distribution of polysulfide ions in aqueous solution at 20 degrees C. Inorg. Chem. 11:1201-1207. [Google Scholar]
- 18.Gill, G., and W. Fitzgerald. 1987. Picomolar mercury measurements in seawater and other materials using stannous chloride reduction and two-stage gold amalgamation with gas phase detection. Mar. Chem. 20:227-243. [Google Scholar]
- 19.Gilmour, C., E. Henry, and R. Mitchell. 1992. Sulfate stimulation of mercury methylation in fresh-water sediments. Environ. Sci. Technol. 26:2281-2287. [Google Scholar]
- 20.Gilmour, C., J. Tuttle, and J. Means. 1985. Tin methylation in sulfide bearing sediments, p. 239-258. In A. Sigleo and A. Hattori (ed.), Marine and estuarine geochemistry. Lewis Publishers, Inc., Chelsea, Mich.
- 21.Greenberg, A. E., L. S. Clesceri, and A. D. Eaton (ed.). 1985. Standard methods for the examination of water and wastewater, 16th ed. American Public Health Association, Washington, D.C.
- 22.Gutknekt, J. 1981. Inorganic mercury (Hg2+) transport through lipid bilayer membranes. J. Membr. Biol. 61:61-66. [Google Scholar]
- 23.Hedderich, R., O. Klimmek, A. Kroger, R. Dirmeier, M. Keller, and K. Stetter. 1999. Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol. Rev. 22:353-381. [Google Scholar]
- 24.Jay, J. A. 1999. Ph.D. dissertation. Massachusetts Institute of Technology, Cambridge.
- 25.Jay, J. A., F. M. M. Morel, and H. F. Hemond. 2000. Mercury speciation in the presence of polysulfides. Environ. Sci. Technol. 34:2196-2200. [Google Scholar]
- 26.Jones, R., M. Jacobson, R. Jaffe, J. West-Thomas, C. Arfstrom, and A. Alli. 1995. Method development and sample processing of water, soil, and tissue for the analysis of total and organic mercury by cold vapor atomic fluorescence spectrometry. Water Air Soil Pollut. 80:1285-1294. [Google Scholar]
- 27.Mason, R., J. Reinfelder, and F. Morel. 1995. Bioaccumulation of mercury and methylmercury. Water Air Soil Pollut. 80:915-921. [Google Scholar]
- 28.Mason, R., J. Reinfelder, and F. Morel. 1996. Uptake, toxicity, and trophic transfer of mercury in a coastal diatom. Environ. Sci. Technol. 30:1835-1845. [Google Scholar]
- 29.Newman, D., T. Beveridge, and F. Morel. 1997. Precipitation of arsenic trisulfide by Desulfotomaculum auripigmentum. Appl. Environ. Microbiol. 63:2022-2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paquette, K. E., and G. R. Helz. 1997. Inorganic speciation of mercury in sulfidic waters: the importance of zero-valent sulfur. Environ. Sci. Technol. 31:2148-2153. [Google Scholar]
- 31.Schwarzenbach, G., and M. Widmer. 1963. Solubility of metal sulfides. I. Black metal sulfide. Helv. Chim. Acta 46:2613-2628. [Google Scholar]
- 32.Stein, W., and W. Lieb. 1986. Transport and diffusion across cell membranes. Academic Press, Inc., New York, N.Y.
- 33.Taylor, B. F., T. A. Hood, and L. A. Pope. 1989. Assay of sulfur as TPPS by high-performance liquid chromatography: applications to studies of sulfur bioproduction and sulfur in marine sediments. J. Microbiol. Methods 9:221-231. [Google Scholar]
- 34.Teder, A. 1971. The equilibrium between elementary sulfur and aqueous polysulfide solutions. Acta Chem. Scand. 25:1722-1728. [Google Scholar]