Abstract
Transcriptional on and off states of HOX genes and other developmental control genes are maintained by antagonistic regulators encoded by trithorax group (trxG) and Polycomb group (PcG) genes. The trxG proteins Ash1 and hTRX and the PcG repressor E(z) are histone methyltransferases (HMTases) that methylate distinct lysine residues in the N-terminal tail of histone H3. trxG proteins are generally thought to function as activators of HOX genes, but how histone methylation by Ash1 and Trx promotes HOX gene transcription is not clear. Here, we show that in ash1 and trx mutants expression of HOX genes is lost within their normal expression domains, but we find that, contrary to expectation, this expression is restored in ash1 and trx mutants that also lack PcG gene function. Moreover, such trxG PcG double mutants show severe misexpression of HOX genes and, hence, ectopic activation of HOX genes caused by the removal of PcG gene function also occurs in the absence of ash1 and trx function. Together, these results suggest that the Ash1 and Trx HMTases are not ‘coactivators' required for transcriptional activation of HOX genes, but function specifically as anti-repressors. We propose that histone methylation by Ash1 and Trx is required continuously throughout development to prevent inappropriate PcG silencing of HOX genes in cells in which they must stay transcriptionally active.
Introduction
Regulation of homeotic gene expression in Drosophila represents a paradigm for understanding how heritable transcriptional states are established and maintained during development. In the early embryo, activators and repressors encoded by segmentation genes determine in which cells homeotic genes are expressed and in which cells these genes must remain inactive. These factors are, however, only transiently present and, during subsequent development, the on and off states of homeotic genes are then heritably maintained by two different groups of regulatory proteins. Polycomb group (PcG) gene products repress homeotic genes in cells in which these genes must stay inactive, whereas trithorax group (trxG) gene products maintain expression of homeotic genes in appropriate cells (reviewed in Francis & Kingston, 2001; Simon & Tamkun, 2002).
Recent progress in understanding the PcG/trxG system has come from the purification and characterization of PcG and trxG protein complexes. Two different PcG protein complexes with distinct biochemical activities have been characterized: Polycomb repressive complex 1 (PRC1) and the Esc–E(z) complex. PRC1 does not contain any enzymatic activity, but in in vitro assays it inhibits chromatin remodelling by SWI–SNF chromatin remodelling complexes (Shao et al, 1999; Francis et al, 2001; Saurin et al, 2001). In contrast, the Esc–E(z) complex functions as a histone methyltransferase (HMTase) that selectively methylates K27 and, to a minor extent, also K9 in the N-terminal tail of histone H3 (Cao et al, 2002; Czermin et al, 2002; Kuzmichev et al, 2002; Müller et al, 2002). Recent biochemical studies on trxG proteins showed that Drosophila Ash1 and mammalian ALL-1/MLL, the homologue of the Drosophila trithorax (Trx) protein, also function as HMTases. Ash1 selectively methylates K4 and K9 on histone H3 and K20 on histone H4 (Beisel et al, 2002; Byrd & Shearn, 2003), whereas ALL-1/MLL selectively methylates K4 in histone H3 (Milne et al, 2002; Nakamura et al, 2002).
What is the role of the Ash1 and Trx methyltransferases in maintaining expression of homeotic genes? The predominant view is that these trxG proteins function as transcriptional coactivators that are needed for expression of homeotic genes (Petruk et al, 2001; Beisel et al, 2002; Poux et al, 2002). Experiments in tissue culture cells appear to support this coactivator model and suggest that transcriptional activation by Ash1 and Trx requires their HMTase activity (Beisel et al, 2002; Milne et al, 2002). More recently, it has been proposed that Ash1 and Trx may not only function as coactivators but could also have a role in preventing the establishment of PcG silencing (Milne et al, 2002; Poux et al, 2002). At present, there is only very little evidence to support this view. One circumstantial argument is provided by the observation that expression of homeotic genes in ash1 mutants (LaJeunesse & Shearn, 1995) or expression of a homeotic reporter gene in trx heterozygotes (Poux et al, 2002) is stochastically lost in some cells but is maintained in others. LaJeunesse & Shearn (1995) and Poux et al (2002) proposed that this variegated all-or-none loss of expression might reflect ectopic repression by PcG proteins, an assumption that was, however, not tested in either of these studies.
Here, we analysed HOX gene expression in different trxG PcG double-mutant combinations. Unexpectedly, these experiments reveal that ash1 and trx are not required for transcriptional activation of HOX genes, but are needed throughout development to prevent the establishment of PcG silencing on HOX genes. We discuss and contrast these results with those obtained in earlier studies that analysed the antagonistic relationship between trxG and PcG proteins.
Results And Discussion
To assess the role of the trxG proteins Trx and Ash1 in the maintenance of HOX gene transcription, we first analysed HOX gene expression in clones of cells that lack trx or ash1 function in imaginal discs. Clones of mutant cells were induced by Flp-mediated recombination, and mutant cells were identified by the lack of a GFP-expressing marker gene. We analysed the effect of eliminating ash1 or trx function by monitoring expression of the HOX gene Ultrabithorax (Ubx) in thoracic imaginal discs using an antibody against Ubx protein. In wild-type animals, Ubx is expressed in the haltere and third leg imaginal disc, but is repressed in the wing imaginal disc. We first analysed ash1 and trx mutant clones, and we find that trx mutant clones in the haltere and third leg disc show complete loss of Ubx signal (Fig 1). ash1 mutant clones also show complete loss of Ubx signal, but most haltere and third leg discs also contain a substantial fraction of mutant cells with apparently undiminished expression of Ubx (Fig 2). These results, together with earlier studies on ash1 mutant larvae (LaJeunesse & Shearn, 1995) and studies on trx mutant clones in the adult epidermis (Ingham, 1981), show that both ash1 and trx are critically needed for the maintenance of HOX gene expression.
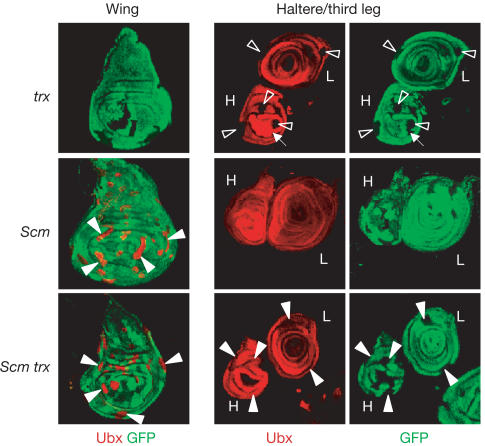
Figure 1.
HOX gene expression in trx and Scm single-mutant clones and in Scm trx double-mutant clones. Wing, haltere (H) and third leg (L) imaginal discs stained with antibodies against GFP (green) and Ubx protein (red); clones of mutant cells (negative for GFP) were induced 96 h before analysis. In wild-type animals, Ubx is expressed in haltere and third leg discs but not in the wing disc. Top: Most trx single-mutant clones in haltere and third leg discs show complete loss of Ubx signal (empty arrowhead); in rare cases, some trx mutant cells within a clone maintain Ubx expression at wild-type levels (small arrow). Middle: Scm mutant clones in wing discs show strong misexpression of Ubx (filled arrowheads); clones in haltere and third leg discs show wild-type expression levels. Bottom: Scm trx double-mutant clones in wing discs show strong misexpression at levels comparable to Scm single-mutant clones. Note that Scm trx double-mutant clones in haltere and third leg discs show wild-type levels of Ubx signal (compare with trx single-mutant clones).
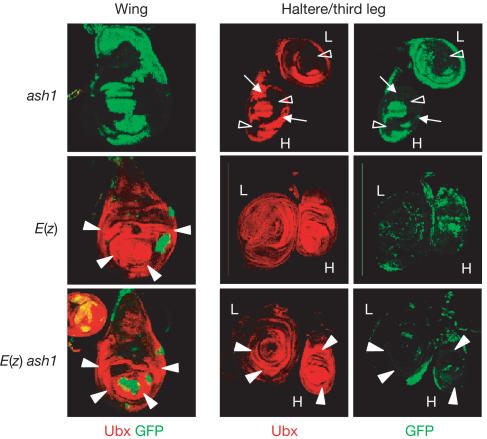
Figure 2.
HOX gene expression in ash1 and E(z) single-mutant clones and in E(z) ash1 double-mutant clones. Wing, haltere (H) and third leg (L) imaginal discs stained with antibodies against GFP (green) and Ubx protein (red); clones of mutant cells (negative for GFP) were induced 96 h before analysis. The Minute technique was used in this experiment, resulting in larger clones. Top: ash1 single-mutant clones in haltere and third leg discs show complete loss of Ubx signal (empty arrowhead), but a significant fraction of mutant cells shows undiminished Ubx expression (small arrows). Middle: E(z) mutant clones in the wing disc show strong misexpression of Ubx (filled arrowheads); clones in the haltere and third leg disc show wild-type expression levels. Bottom: E(z) ash1 double-mutant clones in wing discs show strong misexpression at levels comparable to E(z) single-mutant clones. Note that E(z) ash1 double-mutant clones in haltere and third leg discs all show wild-type levels of Ubx signal (compare with ash1 single-mutant clones).
We next analysed Ubx expression in ash1 and trx mutant clones that also lack PcG function. Using null mutations in the PcG genes Sex combs on midleg (Scm) and Enhancer of zeste (E(z)), we generated Scm trx and E(z) ash1 double-mutant clones and compared them with the corresponding single-mutant clones. As reported previously (Beuchle et al, 2001; Müller et al, 2002), Scm and E(z) single-mutant clones in the wing disc show severe misexpression of Ubx, whereas mutant clones in the haltere and third leg disc show wild-type levels of Ubx expression (Figs 1 and 2). Strikingly, Scm trx and E(z) ash1 double-mutant clones show the same phenotype as Scm and E(z) single-mutant clones (Figs 1 and 2). First, Scm trx and E(z) ash1 double-mutant clones in the wing disc show strong misexpression of Ubx, comparable to Scm and E(z) single-mutant clones in wing discs. Second, clones in the haltere and third leg disc show wild-type levels of Ubx expression. Ubx expression is thus restored in trx and ash1 mutant cells when PcG gene function is removed.
The strong misexpression of HOX genes in these PcG trxG double-mutant clones was unexpected, as a study by Ingham (1983) reported that the morphological defects observed in embryos mutant for the PcG gene extra sex combs (esc) are suppressed in esc trx double-mutant embryos. On the basis of the cuticle phenotype of such esc trx double mutants, Ingham (1983) concluded that HOX genes would be differentially expressed in the absence of PcG and trxG gene function. This notion could not be tested at that time owing to the lack of molecular probes. We therefore analysed HOX gene expression in esc trx and Scm trx double-mutant embryos and in the corresponding single mutants.
Embryos lacking trx function show a reduction of Ubx and Abd-B expression (Fig 3 and data not shown; see also Mazo et al, 1990; Breen & Harte, 1993), and Scm or esc mutant embryos show severe misexpression of both Ubx and Abd-B (Fig 3A and data not shown; see also Struhl & Akam, 1985; McKeon & Brock, 1991; Simon et al, 1992). Strikingly, Scm trx double-mutant embryos show the same strong misexpression of Ubx and Abd-B as Scm single-mutant embryos and the same severe PcG phenotype in the embryonic cuticle (Fig 3A). Thus, in embryos, as in imaginal discs, the Scm mutant phenotype is not suppressed in the absence of trx function. In the case of esc trx double-mutant embryos, the severe PcG phenotype that is observed in the cuticle of esc mutant embryos is partially suppressed, although most segments still show some posteriorly directed homeotic transformations (Fig 3A). At the level of HOX gene expression, we find that esc trx double-mutant embryos show the same widespread misexpression as esc single mutants. However, we find that the onset of misexpression in these esc trx double mutants is delayed and that the level of misexpression is lower compared to esc single mutants (Fig 3A and data not shown); this might explain the partial suppression of the PcG phenotype in the embryonic cuticle. Note that Scm trx double mutants show no such reduction of HOX gene expression compared with Scm single mutants (Fig 3A). Taken together, the widespread misexpression of HOX genes in Scm trx and esc trx double-mutant embryos shows that ectopic activation occurs in the absence of trx function. Nevertheless, it appears that trx function is needed for maximal levels of HOX gene transcription in esc mutant embryos. Surprisingly, the level of Abd-B expression in parasegments (ps) 13 and 14 of esc trx double mutants is even lower than in trx single mutants (Fig 3A); the reason for this is not clear.
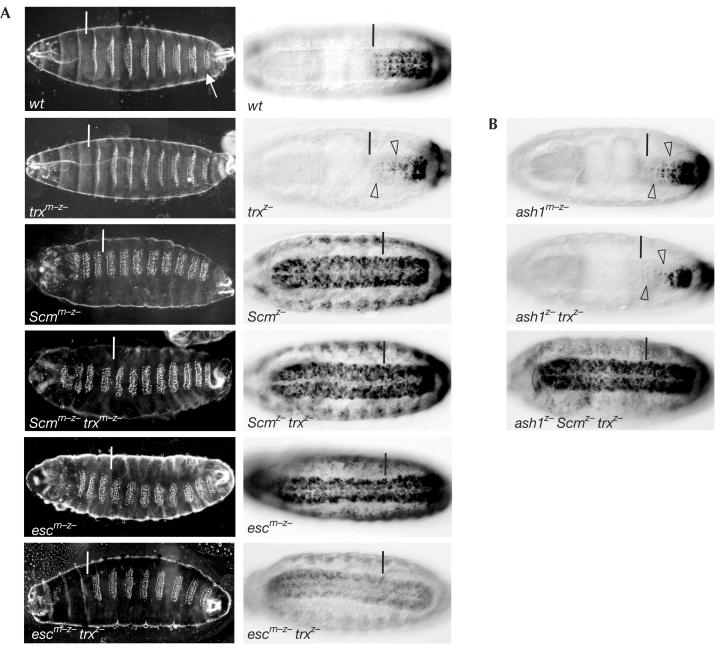
Figure 3.
HOX genes are misexpressed in PcG trxG double-mutant embryos. Ventral views of embryonic cuticles (A, left column) and stage-16 embryos stained with antibody against Abd-B protein (A (right column), B). In cuticle images, the white bar marks the boundary between thoracic and abdominal segments, and the arrow in wt marks the eighth abdominal segment. In stained embryos, the anterior margin of parasegment 10 is marked by a black bar. Genotypes are as indicated; ‘m–z–' lack maternal and zygotic product, ‘z–' lack zygotic product. (A) In wt embryos, denticle belts in each segment have a characteristic shape (left) and Abd-B is expressed from ps 10 to ps 14 (right). Cuticles of trxm–z– and trxz– embryos are indistinguishable and show only mild homeotic transformations (for details, see Ingham, 1983); Abd-B expression is reduced in the CNS (empty arrowhead) and epidermis (not visible in this focal plane). Cuticles of Scmm–z–, Scmm–z– trxm–z– and escm–z– embryos are indistinguishable; they show complete homeotic transformation of all segments into copies of the eighth abdominal segment and strong misexpression of Abd-B in all segments. escm–z– trxz– embryos show partial suppression of homeotic transformations in the cuticle (i.e. thoracic denticle belts are comparable to those in wt embryos), but the head is abnormal and abdominal segments are still partly transformed towards the eighth abdominal segment; Abd-B is misexpressed in all segments, but expression levels are reduced compared with escm–z embryos. For unknown reasons, Abd-B expression in ps 13 and 14 of escm–z– trxz– mutant embryos is lower than in wt, trxz– or escm–z single-mutant embryos. (B) ash1m–z embryos show reduction of Abd-B expression similar to trxz– mutant embryos (empty arrowheads). ash1z– trxz– mutant embryos show slightly more extensive reduction of Abd-B expression (empty arrowheads). ash1z– Scmz– trxz– triple-mutant embryos show severe misexpression of Abd-B comparable to Scmz– or Scmz– trxz– mutant embryos, but note that expression levels in the epidermis are slightly reduced.
Human Trx methylates K4 in the N-terminal tail of histone H3 (Milne et al, 2002; Nakamura et al, 2002), whereas Ash1 methylates K4 and K9 in H3 and K20 in H4 (Beisel et al, 2002; Byrd & Shearn, 2003). The loss of HOX gene expression in either trx or ash1 mutant clones suggests that, despite this overlap in enzymatic specificity, both HMTases are required for maintaining HOX gene expression. To test a possible functional redundancy between Trx and Ash1, we next analysed ash1 trx double mutants and ash1 Scm trx triple mutants. ash1 homozygous embryos that are derived from heterozygous parents show virtually normal HOX gene expression (data not shown), but ash1 homozygous embryos that are derived from ash1 mutant germ cells show reduction of HOX gene expression similar to trx homozygotes (Fig 3B). ash1 trx double homozygotes show slightly more extensive reduction of HOX gene expression than either trx or ash1 single mutants, suggesting that these two HMTases synergize only to a minor extent to maintain HOX gene expression (Fig 3B and data not shown). Nevertheless, we find that ash1 Scm trx triple-mutant embryos show severe misexpression like Scm trx or Scm mutant embryos (Fig 3B). Thus, even the removal of both H3 K4specific HMTases does not result in suppression of the PcG phenotype.
Two main conclusions can be drawn from the results reported here. First, E(z) ash1 and Scm trx double mutants show misexpression of HOX genes outside and wild-type levels of HOX gene expression within HOX expression domains. This suggests that the Trx and Ash1 HMTases are not generally required for transcriptional activation of HOX genes. From our data set, the reduced levels of HOX gene misexpression in esc trx double-mutant embryos compared to esc single-mutant embryos provide the only indication that Trx may, under some circumstances, be required for transcriptional activation. The second main conclusion comes from the observation that removal of PcG gene function in trx and ash1 mutant cells in imaginal discs restores HOX gene expression. This implies that in the absence of Trx or Ash1, PcG proteins repress HOX genes also within their normal expression domains. Taken together, our findings therefore suggest that Trx and Ash1 are not transcriptional ‘coactivators', but function primarily as ‘anti-repressors' that prevent PcG silencing of HOX genes within their normal expression domains. We note that other trxG regulators in Drosophila such as the SWI/SNF complex components Brahma (Brm), Osa and Moira (Mor) may have a much more direct role in the transcriptional activation process (e.g. Kal et al, 2000). The role of SWI/SNF proteins is therefore distinct from the anti-repressor function of Ash1 and Trx that we propose here. Unfortunately, the pleiotrophic phenotype of SWI/SNF mutants did not allow us to analyse their specific role in HOX gene regulation. Finally, it is interesting to note that in mice that are double mutant for the trx homologue Mll and the PcG gene bmi-1, the reduction of HOX c8 expression observed in Mll single mutants is at least partly rescued (Hanson, 1999). This observation would be consistent with the interpretation that, in mice, Mll could exert a similar anti-repressor function to prevent HOX gene silencing by PcG proteins.
How do Ash1 and Trx counteract PcG repression? One possible scenario is that binding and/or histone methylation by Ash1 and Trx prevents the binding of PcG proteins to HOX genes within normal HOX expression domains. For example, methylation of the H3/H4 tails by Ash1 and/or Trx could prevent methylation of K27 in the H3 tail by the E(z) HMTase (Cao et al, 2002; Czermin et al, 2002; Kuzmichev et al, 2002; Müller et al, 2002) or prevent the binding of PcG protein complexes such as PRC1 (Shao et al, 1999; Fischle et al, 2003; Min et al, 2003). It is also possible that methylation by Trx and Ash1 does not prevent the binding of PcG proteins but interferes at a subsequent step by blocking their silencing function.
Methylation of histone tails appears to be a modification that is able to persist, at least for some time, even after dissociation of the HMTase (Ng et al, 2003). We previously proposed that H3 K27 methylation by E(z) may serve as a molecular mark of the silenced state of HOX genes (Müller et al, 2002). Similarly, histone methylation by Trx and Ash1 may provide a mark that allows the propagation of the active state of HOX genes through replication and mitosis. In particular, the methylation mark created by Trx and Ash1 may be critical to prevent PcG silencing during stages of the cell cycle when no transcription is taking place.
Methods
The following fly strains were used in this study: w;FRT82B trxe2/TM6C, w;FRT82B ScmD1/TM6C, w;FRT82B ScmD1trxe2/TM6C, w;ash122FRT2A/TM6C,w;E(z)731FRT2A/TM6C, w;E(z)731ash122FRT2A/TM6C, esc6/CyO;FRT82B trxe2/TM2,w;ash122FRT2A trxe2/TM6C, w;ash122FRT2A ScmD1trxe2/TM6C.
The PcG and trxG mutant alleles used in this study are all genetically or molecularly defined as null alleles (http://flybase.bio.indiana.edu/). Marked clones in imaginal discs were induced and stained as described (Beuchle et al, 2001). Germ-line clone analysis, cuticle preparations and embryo stainings were carried out following standard protocols; balancer chromosomes carrying GFP marker genes were used to genotype mutant embryos.
Acknowledgments
We thank Allen Shearn for fly strains. Suggestions from Andreas Ladurner, Elisa Izaurralde and Gary Struhl helped to improve the manuscript. T.K. is supported by a grant from the DFG.
References
- Beisel C, Imhof A, Greene J, Kremmer E, Sauer F (2002) Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419: 857–862 [DOI] [PubMed] [Google Scholar]
- Beuchle D, Struhl G, Müller J (2001) Polycomb group proteins and heritable silencing of Drosophila Hox genes. Development 128: 993–1004 [DOI] [PubMed] [Google Scholar]
- Breen TR, Harte PJ (1993) Trithorax regulates multiple homeotic genes in the bithorax and Antennapedia complexes and exerts different tissuespecific, parasegment-specific and promoter-specific effects on each. Development 117: 119–134 [DOI] [PubMed] [Google Scholar]
- Byrd KN, Shearn A (2003) ASH1, a Drosophila trithorax group protein, is required for methylation of lysine 4 residues on histone H3. Proc Natl Acad Sci USA 100: 11535–11540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R et al. (2002) Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Czermin B, Melfi R, McCabe D, Seitz V, Imhof A, Pirrotta V (2002) Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 111: 185–196 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S (2003) Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17: 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis NJ, Kingston RE (2001) Mechanisms of transcriptional memory. Nat Rev Mol Cell Biol 2: 409–421 [DOI] [PubMed] [Google Scholar]
- Francis NJ, Saurin AJ, Shao Z, Kingston RE (2001) Reconstitution of a functional core polycomb repressive complex. Mol Cell 8: 545–556 [DOI] [PubMed] [Google Scholar]
- Hanson RD et al. (1999) Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc Natl Acad Sci USA 96: 14372–14377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingham PW (1981) Trithorax: a new homoeotic mutation of Drosophila melanogaster. Wilhelm Roux's Arch 190: 365–369 [DOI] [PubMed] [Google Scholar]
- Ingham PW (1983) Differential expression of bithorax complex genes in the absence of the extra sex combs and trithorax genes. Nature 306: 591–593 [DOI] [PubMed] [Google Scholar]
- Kal AJ, Mahmoudi T, Zak NB, Verrijzer CP (2000) The Drosophila brahma complex is an essential coactivator for the trithorax group protein zeste. Genes Dev 14: 1058–1071 [PMC free article] [PubMed] [Google Scholar]
- Kuzmichev A, Nishioka K, Erdjument-Bromage H, Tempst P, Reinberg D (2002) Histone methyltransferase activity associated with a human multiprotein complex containing the Enhancer of Zeste protein. Genes Dev 16: 2893–2905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaJeunesse D, Shearn A (1995) Trans-regulation of thoracic homeotic selector genes of the Antennapedia and bithorax complexes by the trithorax group genes: absent, small, and homeotic discs 1 and 2. Mech Dev 53: 123–139 [DOI] [PubMed] [Google Scholar]
- Mazo AM, Huang DH, Mozer BA, Dawid IB (1990) The trithorax gene, a trans-acting regulator of the bithorax complex in Drosophila, encodes a protein with zinc-binding domains. Proc Natl Acad Sci USA 87: 2112–2116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKeon J, Brock HW (1991) Interactions of the Polycomb group of genes with homeotic loci of Drosophila. Roux's Arch Dev Biol 199: 387–396 [DOI] [PubMed] [Google Scholar]
- Milne TA et al. (2002) MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol Cell 10: 1107–1117 [DOI] [PubMed] [Google Scholar]
- Min J, Zhang Y, Xu RM (2003) Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev 17: 1823–1828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J et al. (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Nakamura T et al. (2002) ALL-1 is a histone methyltransferase that assembles a supercomplex of proteins involved in transcriptional regulation. Mol Cell 10: 1119–1128 [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K (2003) Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11: 709–719 [DOI] [PubMed] [Google Scholar]
- Petruk S et al. (2001) Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science 294: 1331–1334 [DOI] [PubMed] [Google Scholar]
- Poux S, Horard B, Sigrist CJ, Pirrotta V (2002) The Drosophila trithorax protein is a coactivator required to prevent re-establishment of polycomb silencing. Development 129: 2483–2493 [DOI] [PubMed] [Google Scholar]
- Saurin AJ, Shao Z, Erdjument-Bromage H, Tempst P, Kingston RE (2001) A Drosophila Polycomb group complex includes Zeste and dTAFII proteins. Nature 412: 655–660 [DOI] [PubMed] [Google Scholar]
- Shao Z et al. (1999) Stabilization of chromatin structure by PRC1, a Polycomb complex. Cell 98: 37–46 [DOI] [PubMed] [Google Scholar]
- Simon J, Chiang A, Bender W (1992) Ten different Polycomb group genes are required for spatial control of the abdA and AbdB homeotic products. Development 114: 493–505 [DOI] [PubMed] [Google Scholar]
- Simon JA, Tamkun JW (2002) Programming off and on states in chromatin: mechanisms of Polycomb and trithorax group complexes. Curr Opin Genet Dev 12: 210–218 [DOI] [PubMed] [Google Scholar]
- Struhl G, Akam M (1985) Altered distributions of Ultrabithorax transcripts in extra sex combs mutant embryos of Drosophila. EMBO J 4: 3259–3264 [DOI] [PMC free article] [PubMed] [Google Scholar]