Abstract
In the genetic code, the UGA codon has a dual function as it encodes selenocysteine (Sec) and serves as a stop signal. However, only the translation terminator function is used in gene annotation programs, resulting in misannotation of selenoprotein genes. Here, we applied two independent bioinformatics approaches to characterize a selenoprotein set in prokaryotic genomes. One method searched for selenoprotein genes by identifying RNA stem–loop structures, selenocysteine insertion sequence elements; the second approach identified Sec/Cys pairs in homologous sequences. These analyses identified all or almost all selenoproteins in completely sequenced bacterial and archaeal genomes and provided a view on the distribution and composition of prokaryotic selenoproteomes. In addition, lineage-specific and core selenoproteins were detected, which provided insights into the mechanisms of selenoprotein evolution. Characterization of selenoproteomes allows interpretation of other UGA codons in completed genomes of prokaryotes as terminators, addressing the UGA dual-function problem.
Keywords: selenocysteine, selenoprotein, TGA codon, genome, annotation
Introduction
In the genetic code, the codon UGA differs from other codons in that it has a dual function: although it most often terminates protein synthesis, it also encodes the 21st amino acid in protein, selenocysteine (Sec) (reviewed in Low & Berry, 1996; Bock, 2000; Hatfield & Gladyshev, 2002). Available gene annotation programs interpret UGA as a stop codon, resulting in misannotation or completely missing selenoprotein genes. This problem is particularly serious for prokaryotic genomes as they currently account for >90% of completely sequenced genomes. Yet, neither which prokaryotes contain selenoproteins nor the number of selenoprotein genes in any of the prokaryotic genomes is known. Recent identification of the 22nd amino acid, pyrrolysine, which is encoded by UAG codon in several methanogenic organisms (Srinivasan et al, 2002), suggests that misannotation of genes due to dual functions of termination signals is not limited to Sec.
To insert Sec at UGA codons, selenoprotein genes evolved an RNA stem–loop structure, designated Sec insertion sequence (SECIS) element. SECIS elements are present immediately downstream of Sec UGA codons in bacteria (Zinoni et al, 1990; Huttenhofer et al, 1996; Liu et al, 1998) and in 3′ untranslated regions (UTRs) in archaea (Wilting et al, 1997) and eukaryotes (Berry et al, 1991; Walczak et al, 1996). The bacterial, archaeal and eukaryotic SECIS elements have no similarities to each other with regard to sequence and structure. The eukaryotic SECIS element consensus has been well characterized, which allowed identification of these structures in sequence databases (Kryukov et al, 1999; Lescure et al, 1999; Castellano et al, 2001; Martin-Romero et al, 2001), including mammalian genomes (Kryukov et al, 2003). However, conservation of bacterial SECIS elements has been insufficient for their computational description, which precluded identification of bacterial selenoprotein genes by searching for the stem–loop structure.
Conversely, SECIS elements in known selenoprotein genes in archaea exhibited significant conservation (Wilting et al, 1997), suggesting that searches for these stem–loop structures might be useful in identifying archaeal selenoprotein genes. Previous homology screens and manual analyses of selected genomic regions identified seven selenoprotein genes in Methanococcus jannaschii (Wilting et al, 1997) and, subsequently, Secspecific translation elongation factors were identified in Methanococcus and Methanopyrus species (Rother et al, 2000, 2001a, 2001b).
In the present work, we applied independent bioinformatics approaches to identify entire selenoprotein sets in completed bacterial and archaeal genomes.
Results And Discussion
SECISearch-based identification of selenoproteins
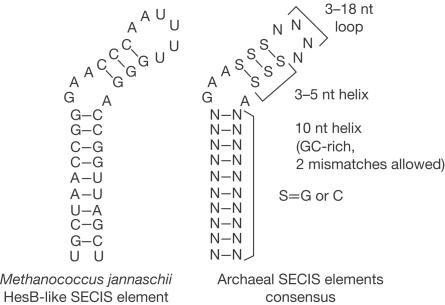
Selenoprotein homologues of known archaeal selenoprotein genes (Wilting et al, 1997) were compiled and their SECIS elements were extracted and structurally aligned (Fig 1). This procedure revealed conserved SECIS regions and resulted in an archaeal SECIS element consensus (Fig 2, right structure). The consensus was very similar to that previously reported (Wilting et al, 1997), although the primary sequence conservation was limited to the unpaired GAA_A region. We identified conserved structural features of these structures, as shown in Fig 2, and developed a computer program, archaeal SECISearch, to recognize an archaeal consensus SECIS element in sequence databases. This threestep program searched for primary sequence, secondary structure and free energy criteria of the predicted stem–loop structures. Additional 'fine structural criteria' were also applied that required the presence of a conserved GAA_A bulge in the predicted structure and removed candidate sequences with predicted Y-shaped structures. A total of 14 completely sequenced archaeal genomes (all that were available at the time of the searches) were analysed with this program (Table 1). Subsequently, sequences flanking the predicted SECIS elements were analysed for the occurrence of open reading frames (ORFs).
Figure 1.
Alignment of SECIS elements in archaeal selenoprotein genes. SECIS elements from eight selenoprotein genes in the M. jannaschii genome and seven selenoprotein genes in the M. kandleri genome were manually aligned on the basis of their primary sequences and secondary structure features. Strictly conserved nucleotides are highlighted.
Figure 2.
Archaeal SECIS element structures. Archaeal SECIS element consensus sequence (right structure) and the SECIS element in M. jannaschii HesB-like gene (left structure) are shown. Conserved structural features in the consensus structure are also indicated.
Table 1.
Selenoprotein genes and SECIS elements identified by SECISearch in completely sequenced archaeal genomes
|
Archaeal SECISearch |
|||||
|---|---|---|---|---|---|
| Primary sequence/secondary structure | Free energy | Fine structure | Selenoprotein genes | False positives | |
| Aeropyrum pernix | 0 | 0 | 0 | 0 | 0 |
| Archaeoglobus fulgidus | 1 | 0 | 0 | 0 | 0 |
| Halobacterium sp. NRC-1 | 7 | 3 | 1 | 0 | 1 |
| Methanobacterium thermoautotrophicum | 0 | 0 | 0 | 0 | 0 |
| Methanococcus jannaschii | 8 | 8 | 8 | 8 | 0 |
| Methanopyrus kandleri | 9 | 7 | 7 | 7 | 0 |
| Pyrobaculum aerophilum | 2 | 0 | 0 | 0 | 0 |
| Pyrococcus abyssi | 2 | 0 | 0 | 0 | 0 |
| Pyrococcus furiosus | 1 | 0 | 0 | 0 | 0 |
| Pyrococcus horikoshii | 0 | 0 | 0 | 0 | 0 |
| Sulfolobus solfataricus | 0 | 0 | 0 | 0 | 0 |
| Sulfolobus tokodaii | 2 | 0 | 0 | 0 | 0 |
| Thermoplasma acidophilum | 1 | 0 | 0 | 0 | 0 |
| Thermoplasma volcanium | 0 | 0 | 0 | 0 | 0 |
See text for details of the searches.
Interestingly, only M. jannaschii and Methanopyrus kandleri had selenoprotein genes (Table 1). Consistent with this finding, known Sec insertion machinery genes, selenophosphate synthetase (SPS) and a Secspecific elongation factor SelB, were detected in M. jannaschii and M. kandleri, but not in other archaeal genomes. The SECIS-based analysis revealed eight selenoprotein genes in M. jannaschii and seven in M. kandleri genomes, with only one false positive and no false negatives in 14 analysed genomes. Of these 15 selenoprotein genes, 13 contained SECIS elements in the 3′-UTRs, and one SECIS element in each organism was found in the 5′-UTR, the location not observed in eukaryotic and bacterial selenoprotein genes. Although incorrectly annotated in genome databases, 14 archaeal selenoproteins were homologues of previously identified selenoproteins (Wilting et al, 1997). The 15th archaeal selenoprotein was a new selenoprotein with distant homology to HesB protein (Figs 2, 3). This ORF was entirely missing in the M. jannaschii genome annotation.
Figure 3.
Amino-acid sequence alignment of archaeal and bacterial HesB-like selenoproteins and their Cys-containing homologues. U is Sec.
SECIS-independent identification of selenoproteins
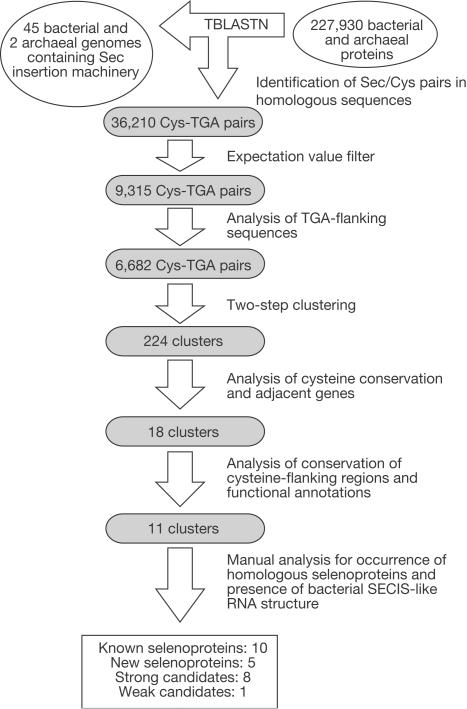
We also applied a SECIS-independent method for identification of selenoprotein genes that searched for Sec/Cys pairs in homologous sequences. It took advantage of the fact that all known prokaryotic selenoproteins except one (selenoprotein A) had homologues in NCBI non-redundant and/or microbial databases that inserted Cys in the place of Sec. This method was applied to both bacterial and archaeal genomes that could contain selenoprotein genes as follows (Fig 4). A database of all available prokaryotic genomes that coded for selenoprotein genes was developed, which was composed of 12 completely and 33 incompletely sequenced bacterial genomes and two completely sequenced archaeal genomes (M. jannaschii and M. kandleri) that possessed components of the Sec insertion machinery (archaeal and bacterial searches were combined in this experiment because sets of known selenoproteins in these organisms showed significant overlap). We also constructed a prokaryotic protein database, which included 227,930 protein sequences from all available annotated prokaryotic genomes. This protein database was searched against the selenoprotein genome database with tblastn to identify local alignments, in which TGA in a genomic sequence corresponded to cysteine in a protein query and the corresponding Sec/Cys flanking sequences showed significant homology. The identified TGA-containing sequences were subjected to computational filters (tests for the presence of upstream in-frame stop signals preceding translation initiation signals and superior blastx and rpsblast hits in different frames than the TGA codon; see Methods for details) and clustered, pseudogenes were removed, and the resulting candidate selenoprotein genes were screened for homologues against NCBI microbial and NR databases. To decrease the possibility that some of the hits were due to sequence errors that replaced TGC or TGT codons with TGA, an ORF containing an in-frame TGA was considered a selenoprotein gene if at least two different sequences in two distant genomes were found that conserved this TGA and in addition at least two corresponding Cys-containing homologues were detected. Protein families in which all homologues conserve Sec would be missed by this approach, but such families appear to be extremely rare (currently, only one family (the selenoprotein A family) conserves Sec in 100% of sequences).
Figure 4.
Identification of prokaryotic selenoproteins by searching for Sec/Cys pairs in homologous sequences (SECIS-independent method). For details of the searches, see Methods.
Analysis of the archaeal subset of identified sequences revealed the same set of seven and eight archaeal selenoprotein genes in the M. kandleri and M. jannaschii genomes (Table 2), with no other archaeal selenoprotein candidates or false-positive sequences. Thus, both SECIS-based and Sec/Cys homology approaches, being independent methods, were efficient in identifying selenoprotein genes in archaeal genomes. Analysis of the bacterial hits revealed ten previously known and five new selenoprotein families (Fig 4, Table 2 and supplementary Figs S1–S5 online). In addition, candidate selenoproteins, each of which was found only in one bacterial genome, were identified (Fig 4, Table 2 and supplementary Figs S6–S13 online). Potential bacterial SECIS structures were identified downstream of TGA in known, new and candidate selenoprotein genes (supplementary Fig S14 online). Only one previously known selenoprotein, selenoprotein A of the glycine reductase complex, was not detected, because no Cys-containing homologues were found for this protein. Thus, our ability to complete the analyses of genomic sequences with an excellent true-positive rate suggested that we identified all or almost all selenoprotein genes in completely sequenced prokaryotic genomes.
Table 2.
Prokaryotic selenoproteins
|
Example |
||||
|---|---|---|---|---|
| Protein name | Bacterial genomes | Archaeal genomes | Genome | Sec location (protein length) |
| Known selenoproteins | ||||
| Formate dehydrogenase α-chain | 37 | 3 | Methanococcus jannaschii | 144 (686) |
| Formylmethanofuran dehydrogenase, subunit B | 0 | 2 | Methanococcus jannaschii | 121 (435) |
| Coenzyme F420-reducing hydrogenase, α-subunit | 0 | 3 | Methanococcus jannaschii | 389 (413) |
| Methylviologen-reducing hydrogenase, α-subunit | 3 | 2 | Methanococcus jannaschii | 441 (464) |
| Coenzyme F420-reducing hydrogenase, δ-subunita | 2 | 3 | Methanococcus jannaschii | 13,64 (134) |
| Heterodisulphide reductase, subunit A | 1 | 2 | Methanococcus jannaschii | 207 (668) |
| Selenophosphate synthetase | 17 | 2 | Methanococcus jannaschii | 19 (349) |
| Peroxiredoxin (Prx) | 2 | 0 | Eubacterium acidaminophilum | 47 (203) |
| Glycine reductase complex, selenoprotein Ab | 9 | 0 | Clostridium difficile | 45 (157) |
| Glycine reductase complex, selenoprotein B | 8 | 0 | Clostridium difficile | 350 (436) |
| Proline reductase | 3 | 0 | Clostridium difficile | 151 (241) |
| New selenoproteins | ||||
| HesB-like protein | 5 | 1 | Methanococcus jannaschii | 35 (95) |
| Thioredoxin (Trx) | 4 | 0 | Treponema denticola | 32 (107) |
| Prx-like thiol:disulphide oxidoreductase | 3 | 0 | Geobacter sulfurreducens | 66 (209) |
| SelW-like protein | 3 | 0 | Campylobacter jejuni | 12 (81) |
| Glutathione peroxidase | 1 | 0 | Treponema denticola | 35 (155) |
| Candidate selenoproteins | ||||
| Glutaredoxin (Grx) | 1 | 0 | Geobacter sulfurreducens | 49 (122) |
| Protein similar to the N-terminal domain of Prx reductase AhpF | 1 | 0 | Carboxydothermus hydrogenoformans | 147 (218) |
| Thiol:disulphide interchange protein | 1 | 0 | Geobacter metallireducens | 33 (190) |
| DsbG-like protein | 1 | 0 | Chloroflexus aurantiacus | 104 (253) |
| Fe–S oxidoreductase | 1 | 0 | Desulfovibrio vulgaris | 244 (432) |
| DsrE-like protein | 1 | 0 | Desulfovibrio vulgaris | 70 (107) |
| NADH oxidase | 1 | 0 | Geobacter metallireducens | 45 (451) |
| Distant homologue of peroxidase/peroxynitrite reductase system component AhpD | 1 | 0 | Geobacter sulfurreducens | 50 (98) |
A total of 11 previously known prokaryotic selenoproteins are shown, along with five new selenoproteins (supported by occurrences in multiple genomes and the presence of potential SECIS elements) and eight candidate selenoproteins (supported by occurrences in single genomes and the presence of potential SECIS elements). The number of bacterial and archaeal genomes in which indicated selenoproteins were found is shown, followed by names of representative genomes (full list is in supplementary Fig S1 online) and Sec locations and lengths of representative selenoproteins.
a Contains two Sec residues.
b Protein was not detected in genomic searches.
Prokaryotic selenoproteomes
Our data allowed a view on entire selenoproteomes in completed bacterial and archaeal genomes. Although selenoproteins were present in all three major domains of life, only ∼20% of completed bacterial and ∼14% of completed archaeal genomes had them. In addition, the number of selenoproteins in a genome varied from one to more than ten, with Carboxydothermus hydrogenoformans, Eubacterium acidaminophilum, Geobacter metallireducens, Geobacter sulfurreducens and M. jannaschii having the largest numbers of selenoproteins among partially and completely sequenced genomes. Analysis of the composition of selenoproteomes revealed that most selenoproteins were redox proteins, which used Sec either to coordinate a redox-active metal (molybdenum, nickel or tungsten) or in Sec: thiol redox catalysis. Interestingly, in contrast to mammals, where new selenoproteins identified through SECIS elements represent a significant fraction of selenoproteomes (Kryukov et al, 2003), the data suggested that in many prokaryotes, entire selenoprotein sets were already known before our study. For example, seven out of eight selenoproteins in M. jannaschii (Wilting et al, 1997) and two out of two selenoproteins in Haemophilus influenzae (Wilting et al, 1998) were previously identified.
The presence of selenoproteins that were found in a small number of genomes (mostly homologues of thiol-dependent oxidoreductases) (Table 2) contrasted with the broad occurrence of their Cys homologues. This observation suggested that these selenoproteins evolved recently, probably from Cys-containing proteins. Conversely, formate dehydrogenases, which were present in most genomes with functional Sec insertion systems (supplementary Fig S1 online), appeared to be of ancient origin. In addition, bacterial formate dehydrogenase genes often clustered with Sec insertion machinery genes (data not shown), suggesting correlated expression. Thus, both a lineage-specific expansion (recent origin) and the presence of core selenoproteins (ancient origin) contribute to the composition of selenoproteomes. Our data suggest that as new genomic sequences become available, additional examples of lineage-specific selenoproteins are likely to emerge.
In conclusion, we identified genes encoding selenocysteine-containing proteins in completely sequenced genomes of archaea and bacteria. This essentially solved the UGA dual-function gene prediction problem in prokaryotes, as other in-frame UGA codons may now be assigned a terminator function. Further systematic characterization of selenoprotein functions should reveal a full set of biological processes that are dependent on the trace element selenium.
Methods
Databases and resources. Completely and incompletely sequenced genomes of archaea and bacteria were obtained from the NCBI data repository (ftp://ftp.ncbi.nlm.nih.gov/genomes) (Wheeler et al, 2003) and TIGR, and blast programs were obtained from NCBI (http://www.ncbi.nlm.nih.gov/BLAST) (Altschul et al, 1990). The searches were performed on a Prairiefire 256-processor Beowulf cluster supercomputer at the Research Computing Facility of the University of Nebraska-Lincoln.
SECIS-based identification of selenoprotein genes in archaeal genomes. Archaeal SECISearch contains three modules. The first module is based on the PatScan program (http://www-unix.mcs.anl.go v/compbio/PatScan/HTML/patscan .html) (Dsouza et al, 1997) and searches for RNA structures that match the archaeal SECIS element primary sequence and the secondary structure consensus. The second module, based on the RNAfold program from Vienna RNA package (http://www.tbi.univie.ac.at/~i vo/RNA) (Hofacker et al, 1994), predicts secondary structure and calculates free energy for the entire putative SECIS element. The predicted bulge-forming GAA_A nucleotides are constrained to be unpaired. Predicted RNA structures, the calculated free energies of which are above the −16 kcal/mol threshold determined from the analysis of known archaeal SECIS elements, are excluded from further analysis. The third module of archaeal SECISearch filters out structures that are either Yshaped or do not have a pronounced GAA_A bulge (nucleotide pairs exactly 'above' and at most one nucleotide 'below' the GAA_A bulge are required to be present in the SECIS element stem).
Identification of selenoprotein genes by searching for Sec/Cys pars in homologous sequences (SECIS-independent method) in prokaryotic genomes. Identification of Sec/Cys pairs in homologous sequences: The protein database was constructed by automated extraction of bacterial and archaeal protein sequences from the NCBI genome data repository. This database had 227,930 unique ORFs, which at the time of analysis represented nearly all predicted protein sequences in completely sequenced prokaryotic genomes. To identify prokaryotic genomes that encode Sec-containing proteins, the NCBI microbial genome database was searched for homologues of known protein components of Sec insertion machinery (Sec synthase, SPS and Sec-specific elongation factor). We identified 12 completely and 33 partly sequenced bacterial and two completely sequenced archaeal genomes, in which at least one Sec insertion machinery gene was detected. These 47 genomes were extracted and combined to generate a selenoprotein genome database.
The protein database was searched against a selenoprotein genome database (47 genomes, ∼200 Mb) with the NCBI-tblastn program. In this search, the threshold for extending hits was set to 11 and the expectation value was cut off to 10. The resulting data set was analysed for the presence of local alignments, in which cysteine in a protein query from a protein database corresponded to TGA in a translated nucleotide sequence from the selenoprotein genome database. If at least one such alignment with an expectation value below 10−3 or at least two alignments with expectation values below 1 were identified for a particular TGA codon, the corresponding TGA-containing sequence was chosen for further analysis. We identified 9,315 such local alignments.
Analysis of TGA-flanking sequences: In each of the TGA-containing sequences from 9,315 alignments, a region upstream of the TGA was analysed for the presence of in-frame start and stop codons. If a stop codon (TGA, TAA or TAG) occurred closer to the TGA codon than an appropriate start codon (ATG or GTG), such sequences were discarded. For each remaining sequence, a 1 kb TGA-flanking region (500 nt–TGA–500 nt) was searched against the protein database with NCBI-blastx program. If the best hit that covered the TGA codon with at least a seven-nucleotide overlap was in a different frame from the TGA, the corresponding sequence was filtered out. The 1 kb TGA-flanking regions (500 nt–TGA–500 nt) were then translated in all three possible ORFs and searched using rpsblast against an NCBI collection of known conserved domains (ftp://ftp.ncbi.nih.gov/pub/mmd b/cdd). If the best hit that covered the TGA codon with at least six amino-acid residue overlap was not in the same frame as the TGA codon, the sequence was removed from further analysis. In addition, if additional stop codons were found within the predicted conserved domain that covered TGA in the correct frame, such a sequence was considered a pseudogene and was also discarded. A total of 6,682 local alignments remained after application of these filters.
Clustering: The set of 2,071 unique proteins that corresponded to the remaining 6,682 TGA-containing sequences was extracted. These protein sequences were compared pairwise by the NCBI-bl2seq program. If two proteins produced a local alignment that had an expectation value below 10−5 and was at least 20 amino-acid residues long, they were assigned to the same cluster. Clusters containing proteins that were initially aligned with the same TGA-containing region were joined into larger clusters. This analysis resulted in 244 clusters.
Analyses of cysteine conservation and adjacent genes: Cysteines that corresponded to the TGA codons in the local alignments were analysed for conservation in other homologous proteins. If a cysteine was not conserved, the hit was discarded. In addition, ORFs that contained TGA-flanking regions were analysed for domain conservation in homologues. The 1 kb TGA-flanking regions were searched with the NCBI-blastx program against the NCBI microbial and NR protein databases. If a TGA codon was located on the edge of the homology region (on either side of the sequence), the candidate hit was removed from further analyses. The remaining sequences were manually analysed for the occurrence of homologous selenoproteins and corresponding Cys-containing proteins in the NCBI microbial and NR databases and for the presence of potential SECIS elements immediately downstream of the TGA codon using mfold.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v5/n5/7400126s1.pdf).
Supplementary Material
Supplementary Figures
Acknowledgments
We thank the Research Computing Facility of the University of Nebraska-Lincoln for the use of Prairiefire 256-processor Beowulf cluster supercomputer. This work was supported by NIH grant GM61603 (to V.N.G.).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR (1991) Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 353: 273–276 [DOI] [PubMed] [Google Scholar]
- Böck A (2000) Biosynthesis of selenoproteins—an overview. Biofactors 11: 77–78 [DOI] [PubMed] [Google Scholar]
- Castellano S, Morozova N, Morey M, Berry MJ, Serras F, Corominas M, Guigo R (2001) In silico identification of novel selenoproteins in the Drosophila melanogaster genome. EMBO Rep 2: 697–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dsouza M, Larsen N, Overbeek R (1997) Searching for patterns in genomic data. Trends Genet 13: 497–498 [DOI] [PubMed] [Google Scholar]
- Hatfield DL, Gladyshev VN (2002) How selenium has altered our understanding of the genetic code. Mol Cell Biol 22: 3565–3576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofacker IL, Fontana W, Stadler PF, Bonhoeffer S, Tacker M, Schuster P (1994) Fast folding and comparison of RNA secondary structures. Monatsh Chem 125: 167–188 [Google Scholar]
- Huttenhofer A, Westhof E, Böck A (1996) Solution structure of mRNA hairpins promoting selenocysteine incorporation in Escherichia coli and their basespecific interaction with special elongation factor SELB. RNA 2: 354–366 [PMC free article] [PubMed] [Google Scholar]
- Kryukov GV, Kryukov VM, Gladyshev VN (1999) New mammalian selenocysteine-containing proteins identified with an algorithm that searches for selenocysteine insertion sequence elements. J Biol Chem 274: 33888–33897 [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Castellano S, Novoselov SV, Lobanov A, Zehtab O, Guigo R, Gladyshev VN (2003) Characterization of mammalian selenoproteomes. Science 300: 1439–1443 [DOI] [PubMed] [Google Scholar]
- Lescure A, Gautheret D, Carbon P, Krol A (1999) Novel selenoproteins identified in silico and in vivo by using a conserved RNA structural motif. J Biol Chem 274: 38147–38154 [DOI] [PubMed] [Google Scholar]
- Liu Z, Reches M, Groisman I, Engelberg-Kulka H (1998) The nature of the minimal 'selenocysteine insertion sequence' (SECIS) in Escherichia coli. Nucleic Acids Res 26: 896–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low SC, Berry MJ (1996) Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem Sci 21: 203–208 [PubMed] [Google Scholar]
- Martin-Romero FJ, Kryukov GV, Lobanov AV, Carlson BA, Lee BJ, Gladyshev VN, Hatfield DL (2001) Selenium metabolism in Drosophila: selenoproteins, selenoprotein mRNA expression, fertility, and mortality. J Biol Chem 276: 29798–29804 [DOI] [PubMed] [Google Scholar]
- Rother M, Wilting R, Commans S, Bock A (2000) Identification and characterisation of the selenocysteinespecific translation factor SelB from the archaeon Methanococcus jannaschii. J Mol Biol 299: 351–358 [DOI] [PubMed] [Google Scholar]
- Rother M, Resch A, Wilting R, Bock A (2001a) Selenoprotein synthesis in archaea. Biofactors 14: 75–83 [DOI] [PubMed] [Google Scholar]
- Rother M, Resch A, Gardner WL, Whitman WB, Bock A (2001b) Heterologous expression of archaeal selenoprotein genes directed by the SECIS element located in the 3′ non-translated region. Mol Microbiol 40: 900–908 [DOI] [PubMed] [Google Scholar]
- Srinivasan G, James CM, Krzycki JA (2002) Pyrrolysine encoded by UAG in Archaea: charging of a UAG-decoding specialized tRNA. Science 296: 1459–1462 [DOI] [PubMed] [Google Scholar]
- Walczak R, Westhof E, Carbon P, Krol A (1996) A novel RNA structural motif in the selenocysteine insertion element of eukaryotic selenoprotein mRNAs. RNA 2: 367–379 [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL et al. (2003) Database resources of the National Center for Biotechnology. Nucleic Acids Res 31: 28–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilting R, Schorling S, Persson BC, Böck A (1997) Selenoprotein synthesis in archaea: identification of an mRNA element of Methanococcus jannaschii probably directing selenocysteine insertion. J Mol Biol 266: 637–641 [DOI] [PubMed] [Google Scholar]
- Wilting R, Vamvakidou K, Bock A (1998) Functional expression in Escherichia coli of the Haemophilus influenzae gene coding for selenocysteine-containing selenophosphate synthetase. Arch Microbiol 169: 71–75 [DOI] [PubMed] [Google Scholar]
- Zinoni F, Heider J, Bock A (1990) Features of the formate dehydrogenase mRNA necessary for decoding of the UGA codon as selenocysteine. Proc Natl Acad Sci USA 87: 4660–4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures