Abstract
Histone chaperone Asf1 participates in heterochromatin silencing, DNA repair and regulation of gene expression, and promotes the assembly of DNA into chromatin in vitro. To determine the influence of Asf1 on genetic stability, we have analysed the effect of asf1Δ on homologous recombination. In accordance with a defect in nucleosome assembly, asf1Δ leads to a loss of negative supercoiling in plasmids. Importantly, asf1Δ increases spontaneous recombination between inverted DNA sequences. This increase correlates with an accumulation of double-strand breaks (DSBs) as determined by immunodetection of phosphorylated histone H2A and fluorescent detection of Rad52–YFP foci during S and G2/M phases. In addition, asf1Δ shows high levels of sister chromatid exchange (SCE) and is proficient in DSB-induced SCE as determined by physical analysis. Our results suggest that defective chromatin assembly caused by asf1Δ leads to DSBs that can be repaired by SCE, affecting genetic stability.
Keywords: chromatin assembly, homologous recombination, sister chromatid exchange, DNA replication
Introduction
Nucleosome assembly is tightly connected to DNA-associated processes such as DNA replication and repair, which can influence genome integrity (Kolodner et al, 2002). Newly synthesized DNA is assembled into nucleosomes in a stepwise manner, with the deposition of a histone H3/H4 tetramer followed by the binding of two histone H2A/H2B dimers (Tyler, 2002). Asf1 is a highly conserved histone chaperone that co-purifies with histones H3 and H4 carrying lysine acetylation patterns characteristic of newly synthesized histones (Tyler et al, 1999). Asf1 interacts physically with the chromatin assembly factor CAF-1 (Cac1–Cac3 in Saccharomyces cerevisiae) (Tyler et al, 2001; Krawitz et al, 2002; Mello et al, 2002), with which it cooperates in the assembly of nucleosomes on newly replicated DNA in vitro (Tyler et al, 1999). Consistent with defects in nucleosome assembly during DNA replication, yeast asf1 and cac mutants are delayed in the S and G2/M phases and are defective in heterochromatin silencing mediated by the replication clamp PCNA (Tyler et al, 1999; Sharp et al, 2001). These defects increase synergistically in asf1 cac double mutants, indicating that Asf1 and CAF-1 have redundant chromatin assembly functions (Tyler et al, 1999).
Asf1 also participates in DNA repair in response to DNA damage and replication blocks. Thus, Asf1 cooperates with CAF-1 in the assembly of nucleosomes during nucleotide excision repair in vitro (Mello et al, 2002) and, consistently, asf1Δ cac double mutants are highly sensitive to ultraviolet light compared with either single mutant (Tyler et al, 1999). Also, asf1Δ cells are sensitive to the DNA-damaging agents methylmethanesulphonate (MMS), phleomycine and 4-nitroquinoline-N-oxide (4-NQO), and to the replication inhibitor hydroxyurea (Le et al, 1997; Tyler et al, 1999; Emili et al, 2001), and are defective in the repair of doublestrand breaks (DSBs) induced by the endonuclease (Qin & Parthun, 2002). Asf1 forms with the checkpoint Rad53 protein a dynamic complex that dissociates in response to DNA damage and replication blocks, connecting the role of Asf1 in DNA repair with S-phase replication (Emili et al, 2001). In addition, asf1Δ affects histone gene transcription and is lethal in combination with mutations in transcription elongation factors, suggesting that Asf1 participates in the formation of chromatin structures that influence gene expression (Sutton et al, 2001; Formosa et al, 2002).
The association of Asf1 with a number of processes related to genetic instability, particularly replication and repair, led us to determine its role in homologous recombination. Homologous recombination in vegetatively growing cells participates in the repair of a number of DNA lesions (Kupiec & Steinlauf, 1997; Aguilera et al, 2000). Many of these lesions occur during DNA replication, suggesting that homologous recombination is involved in the rescue of stalled replication forks (Rothstein et al, 2000). If homologous recombination occurs between sister chromatids (sister chromatid exchange, SCE) genome integrity is preserved, but if it takes place between ectopic DNA sequences it can be a source of genetic instability.
In this study, we show that asf1Δ mutants present chromatin assembly defects that are related to an increase in spontaneous recombination. This increase correlates with an accumulation of DSBs and with high levels of SCE. In accordance, we show that asf1Δ cells are proficient in DSB-induced SCE. We propose that asf1Δ accumulates DSBs that can be repaired by SCE, affecting genetic stability.
Results
A role of Asf1 in nucleosome assembly in vivo
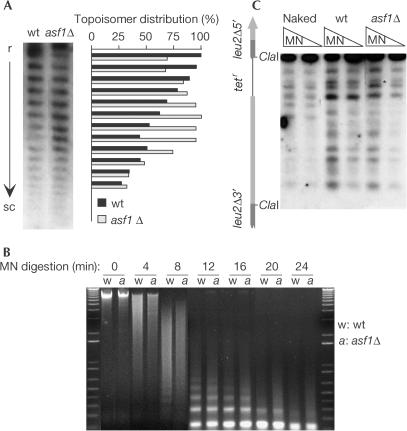
As histone chaperones are redundant in yeast, we considered the determination of the effect of asf1Δ on nucleosome assembly to be important. For this purpose, we analysed plasmid superhelical density and chromatin accessibility to micrococcal nuclease (MNase), which cuts linker DNA between nucleosomes. As one nucleosome introduces one negative superhelical turn in a circular DNA molecule (Wang, 1982), a defect in nucleosome assembly should lead to a loss of negative supercoiling of plasmid pRS316sU, which was used later for recombination analysis (see below). As shown in Fig 1A, the absence of Asf1 shifted the distribution of plasmid topoisomers relative to the wild type, indicating a loss of negative superhelical density.
Figure 1.
Effect of asf1Δ on plasmid topology and chromatin structure in strains BY4741 (wt) and Y11310 (asf1Δ). (A) Topoisomer distribution of plasmid pRS316sU. (B) MNase accessibility of bulk chromatin and (C) nucleosome positioning in the SU recombination system. r and sc, relaxed and supercoiled plasmids, respectively; MN, MNaseI. A scheme of the SU region analysed is shown. The asterisk indicates an MNase cleavage site protected in asf1Δ versus wild type.
To determine the effect of asf1Δ on nucleosome organization, the accessibility of MNase to bulk chromatin and the nucleosome positioning at the SU inverted repeat region were analysed. As can be seen in Fig 1B, bulk chromatin accessibility was not altered, indicating that asf1Δ does not lead to a major loss of nucleosome density. As shown in Fig 1C, only a minor modification in the chromatin structure of the SU system was detected. Altogether, our results are consistent with an impairment of nucleosome assembly in asf1Δ cells that affects DNA topology without important alterations in nucleosome density and positioning.
Asf1Δ increases recombination and accumulates DSBs
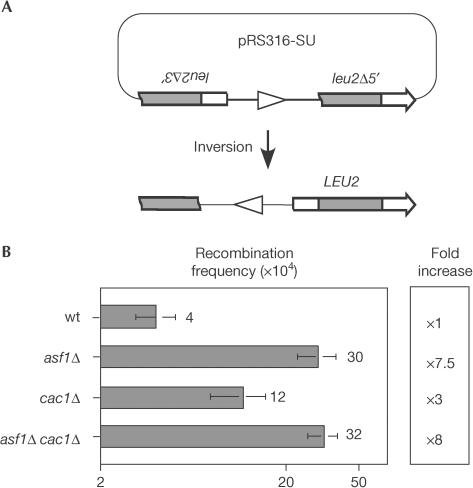
To determine the effect of asf1Δ on spontaneous ectopic recombination, we used the plasmidic inverted repeat system SU, which is based on two truncated leu2 alleles that share a 0.6-kb internal fragment. Homologous recombination between the repeats leads to the inversion of the intervening sequence and the formation of a LEU2 wild-type copy (Prado & Aguilera, 1995) (Fig 2A). As shown in Fig 2B, asf1Δ increased the frequency of inversions eightfold above the wild type. Therefore, the absence of Asf1 leads to genetic instability mediated by ectopic recombination.
Figure 2.
Frequency of homologous recombination in chromatin assembly mutants. (A) Plasmid pRS316sU carrying the inverted repeat system SU and its recombination product. Grey boxes indicate the inverted repeats. (B) Frequency of Leu+ recombinants in strains BY4741 (wt), Y11310 (asf1Δ), Y05437 (cac1Δ) and BYac1-1B (asf1Δ cac1Δ).
Given the physical and genetic interactions between Asf1 and CAF-1, we analysed the possibility that the effect of asf1Δ on recombination was mediated by CAF-1. We determined the frequency of inversions in a cac1Δ mutant, which lacks the largest subunit of the CAF-1 complex. As shown in Fig 2B, cac1Δ increased inversions threefold above wild-type levels, whereas the asf1Δ cac1Δ double mutant showed the same level of recombination as asf1Δ (eightfold above the wild type). These results indicate that Asf1 and CAF-1 act in the same pathway of nucleosome assembly to prevent hyper-recombinogenicity, with a more prominent role of Asf1.
As DSBs efficiently stimulate homologous recombination in yeast (Paques & Haber, 1999), we wondered whether the increase in spontaneous recombination in asf1Δ cells was related to an accumulation of DSBs. For this purpose, we studied the accumulation of phosphorylated histone H2A and Rad52–yellow fluorescent protein (YFP) foci. It has been previously shown that phosphorylation of Ser 129 of histone H2A (P-H2A) occurs at DSB sites via the Mec1 kinase (Downs et al, 2000) and that Rad52–YFP foci are formed and colocalize in situ with DSBs (Lisby et al, 2003). Rad52–YFP foci appear both spontaneously and in response to DSBs during S and G2/M phases, providing a specific role of the recombination Rad52 protein during DNA replication (Lisby et al, 2001).
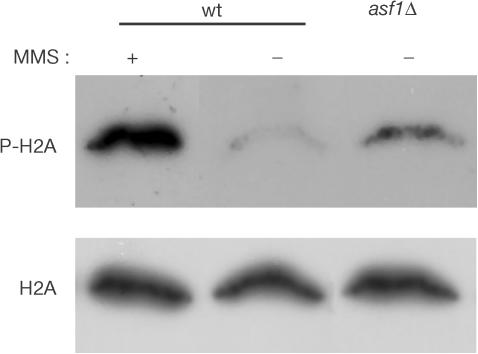
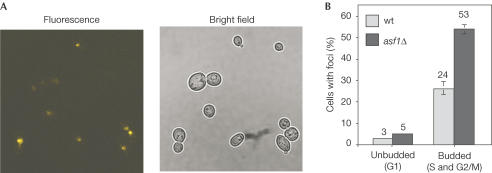
As expected, an accumulation of P-H2A was observed in wild-type cells treated with the DSB inducer agent MMS but not in untreated cells, as determined by western analysis (Fig 3). However, P-H2A was clearly detected in the absence of induced DNA damage in asf1Δ cells. Consistent with previous results, Rad52–YFP foci were detected in 3% of unbudded cells (G1) and in 24% of budded cells (S and G2/M) in the wild type (Fig 4) (Lisby et al, 2001). In asf1Δ, the Rad52–YFP foci occurred at wild-type levels (5%) in unbudded cells, but increased to 53% in budded cells (Fig 4). This increase was observed in both small (S phase) (from 18% in wild type to 30% in asf1Δ) and large budded cells (mostly G2/M) (from 24 to 53%), as expected for events that originated in S phase (Lisby et al, 2003). asf1Δ cells contained on average one Rad52–YFP focus per cell, which has led to the interpretation that each focus represents a centre of recombinational repair of multiple DSBs (Lisby et al, 2001, 2003). The extensive phosphorylation of histone H2A relative to the twofold increase in Rad52–YFP foci is, indeed, consistent with this interpretation. Altogether, these results indicate that the absence of Asf1 leads to an accumulation of DSBs, preferentially during DNA replication.
Figure 3.
Accumulation of P-H2A in BY4741 (wt) and Y11310 (asf1Δ) strains grown with 0.05% (+) or without (−) MMS for 2 h. Total and Ser 129-phosphorylated histone H2A were detected by western blot as described previously (Downs et al, 2000).
Figure 4.
Accumulation of Rad52–YFP foci in asf1Δ cells. Rad52–YFP foci were visualized by fluorescence microscopy in asynchronous cultures of BY4741 (wt) and Y11310 (asf1Δ) cells transformed with plasmid pWJ1344. (A) Fluorescence and brightfield microscopy of asf1Δ cells expressing the Rad52–YFP fusion protein. (B) Percentage of unbudded (G1) and budded (S and G2/M) wild-type and asf1Δ cells containing Rad52–YFP foci. The total numbers of analysed cells were 55 and 199, and 62 and 396 for unbudded and budded wild-type and asf1Δ cells, respectively.
Spontaneous and DSB-induced Sce in asf1Δ cells
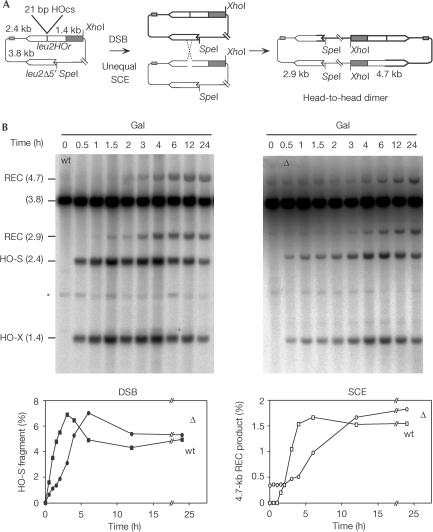
The hyper-recombination phenotype and the accumulation of DSBs in asf1Δ suggest that DSBs are repaired by SCE, a recombination mechanism that occurs during the S and G2 phases. To test this, we used a recently developed recombination system (TINV) for the molecular analysis of SCE (Fig 5A; González-Barrera et al, 2003). In this system, spontaneous unequal SCE between the leu2 inverted repeat copies yields a head-to-head dimer that is detected after SpeI–XhoI digestion by the appearance of two specific recombinant bands (2.9 and 4.7 kb). Southern analysis revealed that spontaneously occurring dimers, identified by the 2.9-kb and 4.7-kb bands, were undetectable in wild-type cells but could be detected in asf1Δ cells, regardless of whether cells were grown in 3% glycerol-lactate (Fig 5B; t=0) or 2% glucose (data not shown). These dimers were unlikely to have been the result of recombination between two copies of the plasmid accumulated by missegregation, because no dimers were detected in cells carrying two plasmids that differed only in the selection marker (data not shown). These results indicate that asf1Δ increases spontaneous SCE. In this sense, it is worth noticing that the proportion of dimers is in the range of spontaneous Leu+ recombinants observed for the TINV system in asf1Δ (2 × 10−3 in asf1Δ versus 3 × 10−4 in the wild type).
Figure 5.
Physical analysis of spontaneous and HO-induced SCE in WS (wt) and WS-asf1 (asf1Δ) strains. (A) Plasmid pRS316-TINV carrying the inverted repeat system TINV and the dimer resulting from unequal SCE between the inverted repeats. (B) Kinetics of HO-induced SCE. Mid-log-phase cells from SC–3% glycerol/2% lactate plus 5 μg/ml doxycycline were grown for different times in SC–2% galactose (Gal). Spontaneous SCE in the absence of HO breaks is observed as the accumulation of the 2.9-kb and 4.7-kb bands in asf1Δ cells in glycerol-lactate (t=0). The quantification of the HO-induced DSB (HOs 2.4 kb) and the 4.7-kb recombination product (REC 4.7) is shown. The average of two experiments performed with independent transformants is plotted. Variation for each point was <30%. DNA was isolated, digested with XhoI–SpeI and analysed by Southern blot with the ClaI–EcoRV LEU2 internal fragment for spontaneous and HO-induced SCE. Bands resulting from HO cleavage (HO-S and HO-X) and recombination intermediates (REC bands) were quantified in a Fuji FLA3000 and normalized relative to the total plasmid DNA. The asterisk shows unspecific hybridization.
These results suggest that the DSBs accumulated in asf1Δ cells are repaired by SCE. However, Asf1 has recently been shown to be required for efficient repair of DSBs induced by the endonuclease HO during mating-type switching (Qin & Parthun, 2002). Therefore, we decided to determine whether SCE could repair HO-induced DSBs in asf1Δ cells. For this purpose, DSBs were induced at the 21-bp HO site of the leu2HOr allele in the TINV system. As the 21-bp HO site is cut by HO at low efficiency (7%), most HO-induced DSBs are limited to one chromatid and can be repaired using as template any of the leu2 alleles of the intact chromatid (González-Barrera et al, 2003). If the donor copy is the leu2Δ5′ allele located in the sister chromatid, the head-to-head dimer could be detected (Fig 5A). As the HO endonuclease was under the control of the GAL1 promoter, the kinetics of DSB repair can be analysed by Southern analysis after galactose induction. As can be seen in Fig 5B, the kinetics of DSBs accumulation, determined by the accumulation of the 1.4-kb and 2.4-kb bands, was similar in wild-type and asf1Δ cells, although it was delayed 2–3 h in the mutant. Compared with the wild type, the efficiency of SCE, determined by the accumulation of the 2.9-kb and 4.7-kb bands, was not reduced in the absence of Asf1. Although dimer accumulation was also delayed in asf1Δ, it is worth noticing that the maximum level of dimers appeared 3 and 6 h after the peaks of DSBs in wild-type and mutant cells, respectively (Fig 5B).
Discussion
In this study, we provide evidence that the absence of the chromatin assembly factor Asf1 influences genetic stability by the accumulation of DSBs that are repaired via SCE. asf1Δ and cac1Δ increase the frequency of inversions between homologous DNA sequences eightfold and threefold above wild-type levels, respectively. The hyper-recombinogenic effect of asf1Δ is epistatic over cac1Δ, suggesting that Asf1 and CAF-1 participate in the same pathway in the maintenance of genome integrity. Therefore, the absence of chromatin assembly factors, particularly Asf1, causes genetic instability. This is consistent with recent reports showing that a dominant-negative mutant of p150CAF-1 causes DNA breaks in human cells (Ye et al, 2003), and that yeast asf1Δ and cac1Δ mutants strongly increase gross chromosomal rearrangements (GCRs) such as telomere addition, translocations and deletions (Myung et al, 2003). These GCRs involve microhomology or nonhomology breakpoints and increase synergistically in rad52Δ cells, indicating that they do not occur by homologous recombination; instead, homologous recombination prevents their formation (Myung et al, 2003). Our results extend the effect of defective chromatin assembly to rearrangements between DNA homologous sequences. This observation is in accordance with the increase in homologous recombination caused by either partial depletion of histone H4 (F. Prado and A. Aguilera, unpublished data) or a thermosensitive mutation in the histone chaperone Spt6 (Malagon & Aguilera, 2001).
Chromatin assembly is tightly associated with DNA synthesis during replication (Shibahara & Stillman, 1999; Sharp et al, 2001). Defective nucleosome assembly in asf1Δ could uncouple this connection and impair DNA synthesis, leading to the accumulation of DNA damage. We provide molecular evidence consistent with defective nucleosome assembly in asf1Δ. Thus, asf1Δ changes the superhelical density of a plasmid without a major loss of nucleosome density or positioning. This could be explained by a change in the path of the DNA around an altered histone octamer in asf1Δ. This possibility is consistent with the proposed role for Asf1 in H3/H4 tetramer deposition on the DNA (Tyler et al, 1999), as the tetramer is a major determinant in the organization of DNA in the nucleosome (Ramakrishnan, 1997). In accordance with impaired DNA synthesis and accumulation of DNA damage, asf1Δ accumulates DSBs during S and G2/M phases of the cell cycle—as determined by Rad52–YFP foci and P-H2A detection (Figs 3 and 4)—and shows high levels of spontaneous SCE (Fig 4), and is delayed in the S and G2/M phases (Tyler et al, 1999; data not shown). Indeed, the high accumulation of Rad52–YFP foci in asf1Δ is similar to that previously observed with mutants affected in the replicative DNA polymerase α complex (Lisby et al, 2001). Consistently, expression of a dominant-negative mutant of p150CAF-1 in human cells causes accumulation of P-H2A and S-phase arrest (Ye et al, 2003). Therefore, a tight regulation of DNA synthesis and histone deposition is essential to prevent genetic instability during DNA replication.
The observation that SCE-mediated dimers accumulate spontaneously at high levels in asf1Δ cells suggests that SCE could be responsible for the increase in homologous recombination observed in asf1Δ. Indeed, asf1Δ cells are proficient in DSB repair by SCE, as determined by physical analysis of HO-induced dimers. This result contrasts with the requirement of Asf1 in the repair of HO-induced DSBs by intramolecular gene conversion in mating-type switching (Qin & Parthun, 2002). Whether or not this is due to the existence of redundant nucleosome assembly functions associated with SCE or due to the formation of DSBs at the 21-bp HO site specifically during the S phase remains to be determined.
In summary, our results support the importance of chromatin assembly in the maintenance of genetic stability, and establish a new link between DNA replication and homologous recombination mediated by chromatin assembly factors. The molecular evidence that SCE is a major mechanism of DSB repair in asf1Δ supports the idea that the hyper-recombinogenicity of asf1Δ cells is linked to replication impairment.
Methods
Yeast strains and plasmids. Yeast strains used in this study were BY4741 (MATa his3Δ0 leu2Δ0 ura3Δ0 met15Δ0) and its isogenic derivatives Y11310 (MATα asf1Δ:KAN), Y05437 (MATα cac1Δ:KAN) (Euroscarf) and BYac1-1B (MATα asf1Δ:KAN cac1Δ:KAN), as well as WS (MATα-inc ade3:GAL-HO leu2Δ:SFA1 ade2-1 can1-100 his3-11,15 trp1-1 ura3-1) (González-Barrera et al, 2003) and its isogenic WS-asf1 (asf1Δ:KAN). Yeast cells were grown at 30°C in synthetic complete (SC) medium as described (Kaiser et al, 1994). pRS316sU—identical to pRS314-SU (Prado & Aguilera, 1995) but in vector pRS316—and pRS316-TINV (González-Barrera et al, 2003) are centromeric plasmids containing the recombination systems used in this study. pWJ1344 is a centromeric plasmid containing the Rad52–YFP construct (by R. Rothstein, Columbia University).
Genetic and physical analysis of recombination. Spontaneous recombination frequencies were obtained by a fluctuation test as the median value of six independent colonies as described previously (Prado & Aguilera, 1995). The average value of three of four independent assays performed with two or three independent transformants is shown. Physical analysis of SCE was performed as described (González-Barrera et al, 2003).
Chromatin analysis by MNase digestion. Mapping of MNase cleavage sites was performed as previously published (Chavez et al, 1995). For bulk chromatin accessibility to MNase, chromatin was digested with 400 mU of MNase and DNA samples were taken at different times and resolved in 1% agarose gel. For nucleosome positioning by indirect end labelling, chromatin was digested with different amounts of MNase during 20 min. DNA was extracted and restricted with ClaI, resolved in 1.5% agarose gel and probed with the 221-bp fragment located immediately downstream of the LEU2 ClaI site.
Plasmid supercoiling. Total DNA from mid-log-phase cultures was extracted and resolved in chloroquine gels according to Brill & Sternglanz (1988). Topoisomer distribution of plasmid pRS316sU was detected with the ClaI–EcoRV LEU2 probe, quantified in a Fuji FLA3000 and normalized relative to the most abundant topoisomer.
Detection of Rad52–YFP. Rad52–YFP foci from mid-log-phase cells transformed with plasmid pWJ1344 were visualized by a confocal microscope Leica TCS SL (excitation 514 nm; emission 535 nm), according to Lisby et al (2001).
Acknowledgments
We thank S.P. Jackson for providing anti-P-H2A antibodies, R. Rothstein for the Rad52–YFP construct, M. Lisby for technical tips on the analysis of Rad52–YFP foci, M. Fidalgo for help with confocal microscopy, M. Nieto for technical assistance, S. González-Barrera and R.E. Wellinger for critical reading of the manuscript, and Diane Haun for style supervision. The research was funded by the Spanish Ministry of Science and Technology (BMC2000-0439 and SAF2003-00204 grants). F.C.-L.is recipient of a predoctoral training grant from the Spanish Ministry of Health.
References
- Aguilera A, Chavez S, Malagon F (2000) Mitotic recombination in yeast: elements controlling its incidence. Yeast 16: 731–754 [DOI] [PubMed] [Google Scholar]
- Brill SJ, Sternglanz R (1988) Transcription-dependent DNA supercoiling in yeast DNA topoisomerase mutants. Cell 54: 403–411 [DOI] [PubMed] [Google Scholar]
- Chavez S, Candau R, Truss M, Beato M (1995) Constitutive repression and nuclear factor I-dependent hormone activation of the mouse mammary tumor virus promoter in Saccharomyces cerevisiae. Mol Cell Biol 15: 6987–6998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downs JA, Lowndes NF, Jackson SP (2000) A role for Saccharomyces cerevisiae histone H2A in DNA repair. Nature 408: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Emili A, Schieltz DM, Yates JR Jr, Hartwell LH (2001) Dynamic interaction of DNA damage checkpoint protein Rad53 with chromatin assembly factor Asf1. Mol Cell 7: 13–20 [DOI] [PubMed] [Google Scholar]
- Formosa T, Ruone S, Adams MD, Olsen AE, Eriksson P, Yu Y, Rhoades AR, Kaufman PD, Stillman DJ (2002) Defects in SPT16 or POB3 (yFACT) in Saccharomyces cerevisiae cause dependence on the Hir/Hpc pathway: polymerase passage may degrade chromatin structure. Genetics 162: 1557–1571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Barrera S, Cortés-Ledesma F, Wellinger RE, Aguilera A (2003) Equal sister chromatid exchange is a major mechanism of doublestrand break repair in yeast. Mol Cell 11: 1661–1671 [DOI] [PubMed] [Google Scholar]
- Kaiser C, Michaelis M, Mitchel A (1994) Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press [Google Scholar]
- Kolodner RD, Putnam CD, Myung K (2002) Maintenance of genome stability in Saccharomyces cerevisiae. Science 297: 552–557 [DOI] [PubMed] [Google Scholar]
- Krawitz DC, Kama T, Kaufman PD (2002) Chromatin assembly factor I mutants defective for PCNA binding require Asf1/Hir proteins for silencing. Mol Cell Biol 22: 614–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kupiec M, Steinlauf R (1997) Damage-induced ectopic recombination in the yeast Saccharomyces cerevisiae. Mutat Res 384: 33–44 [DOI] [PubMed] [Google Scholar]
- Le S, Davis C, Konopka JB, Sternglanz R (1997) Two new S-phasespecific genes from Saccharomyces cerevisiae. Yeast 13: 1029–1042 [DOI] [PubMed] [Google Scholar]
- Lisby M, Rothstein R, Mortensen UH (2001) Rad52 forms DNA repair and recombination centers during S phase. Proc Natl Acad Sci USA 98: 8276–8282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisby M, Mortensen UH, Rothstein R (2003) Colocalization of multiple DNA doublestrand breaks at a single Rad52 repair centre. Nat Cell Biol 5: 572–577 [DOI] [PubMed] [Google Scholar]
- Malagon F, Aguilera A (2001) Yeast spt6-140 mutation, affecting chromatin and transcription, preferentially increases recombination in which Rad51p-mediated strand exchange is dispensable. Genetics 158: 597–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello JA, Sillje HH, Roche DM, Kirschner DB, Nigg EA, Almouzni G (2002) Human Asf1 and CAF-1 interact and synergize in a repair-coupled nucleosome assembly pathway. EMBO Rep 3: 329–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myung K, Pennaneach V, Kats ES, Kolodner RD (2003) Saccharomyces cerevisiae chromatin-assembly factors that act during DNA replication function in the maintenance of genome stability. Proc Natl Acad Sci USA 100: 6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paques F, Haber JE (1999) Multiple pathways of recombination induced by doublestrand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63: 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado F, Aguilera A (1995) Role of reciprocal exchange, one-ended invasion crossover and singlestrand annealing on inverted and direct repeat recombination in yeast: different requirements for the RAD1, RAD10, and RAD52 genes. Genetics 139: 109–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin S, Parthun MR (2002) Histone H3 and the histone acetyltransferase Hat1p contribute to DNA doublestrand break repair. Mol Cell Biol 22: 8353–8365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan V (1997) Histone structure and the organization of the nucleosome. Annu Rev Biophys Biomol Struct 26: 83–112 [DOI] [PubMed] [Google Scholar]
- Rothstein R, Michel B, Gangloff S (2000) Replication fork pausing and recombination or ‘gimme a break'. Genes Dev 14: 1–10 [PubMed] [Google Scholar]
- Sharp JA, Fouts ET, Krawitz DC, Kaufman PD (2001) Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol 11: 463–473 [DOI] [PubMed] [Google Scholar]
- Shibahara K, Stillman B (1999) Replication-dependent marking of DNA by PCNA facilitates CAF-1-coupled inheritance of chromatin. Cell 96: 575–585 [DOI] [PubMed] [Google Scholar]
- Sutton A, Bucaria J, Osley MA, Sternglanz R (2001) Yeast Asf1 protein is required for cell cycle regulation of histone gene transcription. Genetics 158: 587–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler JK (2002) Chromatin assembly. Cooperation between histone chaperones and ATP-dependent nucleosome remodeling machines. Eur J Biochem 269: 2268–2274 [DOI] [PubMed] [Google Scholar]
- Tyler JK, Adams CR, Chen SR, Kobayashi R, Kamakaka RT, Kadonaga JT (1999) The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature 402: 555–560 [DOI] [PubMed] [Google Scholar]
- Tyler JK, Collins KA, Prasadsinha J, Amiott E, Bulger M, Harte PJ, Kobayashi R, Kadonaga JT (2001) Interaction between the Drosophila CAF-1 and ASF1 chromatin assembly factors. Mol Cell Biol 21: 6574–6584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JC (1982) The path of DNA in the nucleosome. Cell 29: 724–726 [DOI] [PubMed] [Google Scholar]
- Ye X, Franco AA, Santos H, Nelson DM, Kaufman PD, Adams PD (2003) Defective S phase chromatin assembly causes DNA damage, activation of the S phase checkpoint, and S phase arrest. Mol Cell 11: 341–351 [DOI] [PubMed] [Google Scholar]