Abstract
Histone H3 lysine 9 methylation is associated with long-term transcriptional repression through recruitment of heterochromatin protein 1 (HP1) proteins. These proteins are believed to promote the formation of dense chromatin structures interfering with DNA accessibility. During the G2 phase of the cell cycle, HP1 proteins are delocalized from foci of pericentromeric heterochromatin, while a wave of H3 serine 10 phosphorylation is initiated within these regions. Here, we show that in vivo phosphorylation of serine 10 in G2 can occur on histone tails methylated on lysine 9. Unexpectedly, this modification favours rather than prevents HP1 binding to chromatin. Dissociation of HP1 from the methylated histone H3 tails is observed only after a third modification by acetylation of lysine 14, which occurs in prophase. We propose that phosphoacetylation of histone H3 could be a general mechanism allowing the cell to overcome HP1-mediated transcriptional repression.
Keywords: kinetic, microinjection, mitosis, surface plasmon resonance
Introduction
Heterochromatin protein 1 (HP1) proteins preferentially localize within condensed inactive heterochromatin. However, several recent observations suggest that this protein can also function as a co-repressor of genes located within active, transcribed chromatin (for a review, see Li et al, 2002). Recruitment of HP1 to chromatin is mediated by methylation of the histone H3 tail on the lysine at position 9 (H3 metK9). Methylation is a very stable modification and, to date, no histone demethylase has been identified. Yet, the cyclin E gene, for example, is targeted by HP1 upon Rb-induced repression (Nielsen et al, 2001) and nevertheless is re-activated later in the cell cycle. This observation clearly indicates that repression by HP1 can be reversed. Global delocalization of HP1 from chromatin has been observed at the G2/M transition, resulting in disappearance of pericentromeric accumulation of the protein and its increased extractability (Murzina et al, 1999). This HP1 delocalization coincides with the wave of mitotic histone H3 serine 10 phosphorylation (H3 pS10) that initiates within the pericentromeric heterochromatin in late G2 and then spreads to the rest of the chromatin as M phase approaches. This observation suggested that H3 S10 phosphorylation may displace HP1 from methylated H3 tails (Fischle et al, 2003a). However, another report claimed that in vitro methylation of H3 K9 prevented phosphorylation of H3 S10 (Rea et al, 2000), suggesting that the effect of H3 pS10 on HP1 binding was more likely to be indirect. Here, we provide evidence that in vivo H3 metK9 and pS10 modifications can in fact occur concomitantly on the same histone tail. Surprisingly, this double modification does not prevent HP1 binding, and a third modification, namely acetylation of H3 lysine 14 (H3 acK14), is required for delocalization of the protein.
Results
HP1 delocalization and H3 phosphorylation in G2
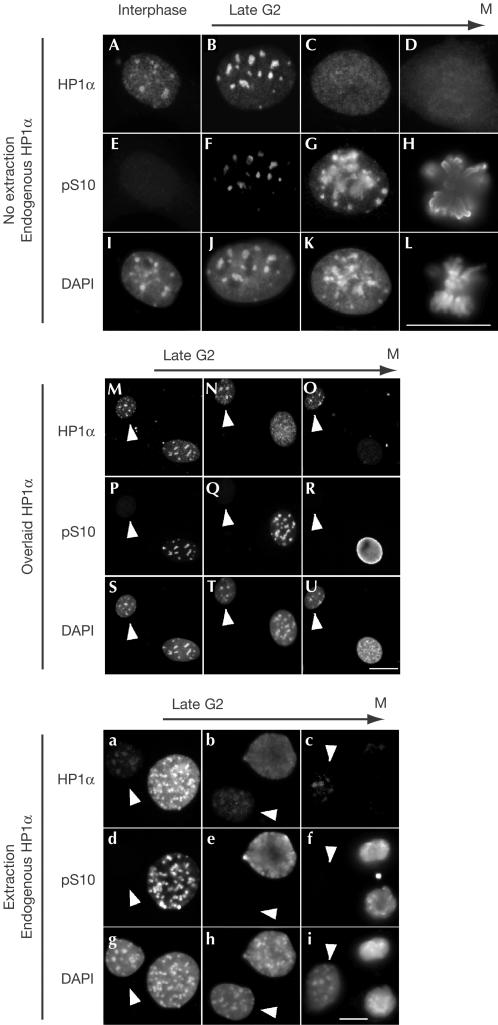
To dissect the mechanism leading to HP1 delocalization from the pericentromeric heterochromatin at the G2/M transition, we co-stained fixed NIH3T3 cells with anti-HP1α and anti-H3 pS10 antibodies. Using the pattern of anti-H3 pS10 staining as a criterion, we identified cells fixed at different steps in late G2 and early M (Fig 1A–L). As previously described (Murzina et al, 1999), HP1 was delocalized from pericentromeric foci as cells entered M phase (Fig 1, compare A and C). However, careful observation revealed that early phases of H3 S10 phosphorylation were correlated with more concentrated rather than diffuse distribution of HP1 (Fig 1B,F). We also tested binding of a glutathione S-transferase (GST)–HP1α fusion protein in overlay assays. Fixed cells were incubated in the presence of recombinant GST–HP1α, which in turn was detected using anti-GST antibodies (Muchardt et al, 2002). Similar to the endogenous HP1α, GST–HP1α associated well with the chromatin in late G2 after initiation of H3 S10 phosphorylation (Fig 1M,N,P,Q,S,T). Decreased binding of GST–HP1α (compared with interphasic cells) was observed only at later stages when the cells had entered prophase (Fig 1O,R,U). These observations suggested that delocalization of HP1 at the G2/M transition was likely to follow a more complex mechanism than simple inhibition of HP1 binding by phosphorylation of the H3 tails. As GST–HP1α was produced in Escherichia coli, they also showed that decreased mitotic chromatin binding of HP1 did not require any post-translational modification of the protein itself.
Figure 1.
Mitotic phosphorylation of histone H3 is associated first with concentration, then with delocalization of HP1 from pericentromeric heterochromatin. (A–L) Fixed NIH3T3 cells were labelled with the indicated antibodies and DAPI. (M–U) NIH3T3 cells were extracted with Triton X-100, fixed, incubated with recombinant GST–HP1α and then labelled with anti-GST (M–O), anti-H3 pS10 (P–R) and DAPI (S–U). (a–i) NIH3T3 cells were extracted with Triton X-100, fixed and labelled with the indicated antibodies and DAPI. Scale bars, 10 μm. The presented nuclei were ordered according to the levels of H3 S10 phosphorylation and chromatin compaction. The arrows indicate interphasic cells.
H3 S10 phosphorylation has also been correlated with increased HP1 extractability (Murzina et al, 1999). To re-investigate this issue, we used cells that were treated with the non-ionic detergent Triton X-100 before fixation. Labelling with anti-HP1α and anti-H3 pS10 antibodies surprisingly showed that in early stages of the G2/M transition, HP1α was less, rather than more, extractible than in interphasic cells (Fig 1a,d,g; compare HP1α signals in the interphasic (arrow) and pS10-positive cells). High extractability was observed in cells selected at later stages, and extractability appeared to increase gradually as M phase progressed (Fig 1a–i; see also Fig 3 for more intermediate steps). We also noted that in prophase the diffusely distributed HP1α is not fully extractable (Fig 1b,e,h), suggesting that HP1α nuclear redistribution and its release from chromatin are two uncoupled events.
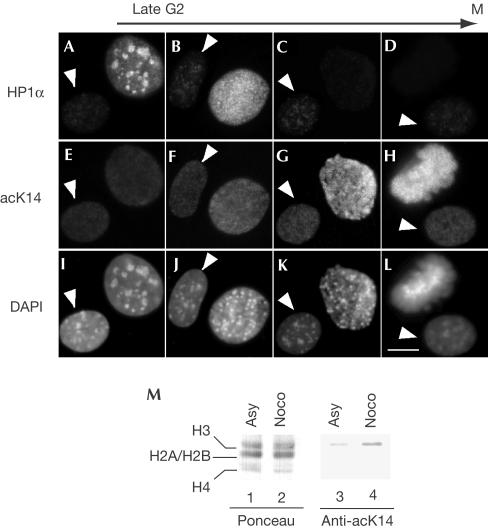
Figure 3.
HP1 extractability in M phase is correlated with increased H3 K14 acetylation. (A–L) NIH3T3 cells were extracted with Triton X-100, fixed and labelled with the indicated antibodies and DAPI. The presented nuclei were ordered according to the level of HP1 extractability and chromatin compaction. The arrows indicate a cell in interphase. Scale bar, 10 μm. (M) Total extracts from HeLa cells either asynchronous (lanes 1 and 3) or arrested in M phase with nocodazole (lanes 2 and 4) were resolved by 18% SDS–polyacrylamide gel electrophoresis, and levels of H3 K14 acetylation were estimated by western blot.
Taken together, these experiments show that mitotic phosphorylation of H3 S10 cannot be simply correlated either with diffused HP1α distribution or increased extractability.
Concomitant methylation and phosphorylation
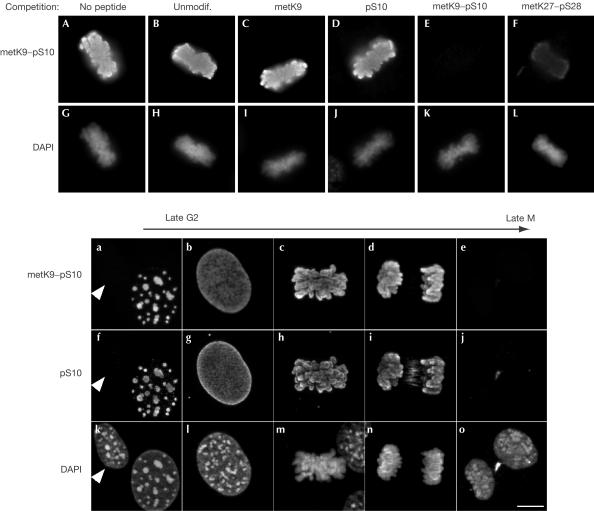
To determine whether mitotic phosphorylation of histone H3 on S10 can occur on histone tails already methylated on K9, we generated antibodies directed against the double H3 metK9–pS10 modification. Competition experiments using synthetic peptides mimicking modified H3 histone tails showed that these antibodies specifically recognized the double modification and not unmodified or singly modified histone tails carrying either the metK9 or the pS10 modifications (Fig 2A–E,G–K). We noted some crossreactivity with a peptide carrying a metK27–pS28 modification (Fig 2F,L). Competition with this peptide was, however, significantly less efficient than with the metK9–pS10 peptide (Fig 2, compare E and F). Double staining with anti-H3 metK9–pS10 and anti-H3 pS10 showed simultaneous appearance and disappearance of the two signals during G2 and M phases (Fig 2a–o). We also doublestained cells with anti-H3 metK9–pS10 and anti-H3 pS28 antibodies. Whereas anti-H3 metK9–pS10 signal was detected in late G2, anti-pS28 signal was observed only as starting in prophase (supplementary Fig 1 online). This is consistent with earlier observations showing that phosphorylation occurs later on H3 S28 than on H3 S10 (Goto et al, 1999). From these experiments, we concluded that although the anti-H3 metK9–pS10 antibodies potentially crossreacted with H3 metK27–pS28, histone H3 tails carrying the double metK9–pS10 modification were detected with certainty in late G2. Besides, the phosphorylation of methylated H3 tails appeared to follow a pattern similar to that of nonmethylated tails. It must be noted here that the anti-H3 pS10 antibodies used did not recognize the double modification (supplementary Fig 2 online).
Figure 2.
H3 histone tails can be phosphorylated on S10 and methylated on K9 in vivo. (A–L) Fixed NIH3T3 cells were labelled with antibodies raised against an H3 peptide carrying a dimethylation on K9 and a phosphorylation on S10. To test specificity, the antibodies were challenged with an excess of the indicated peptides. Panels show mitotic cells strongly labelled by the antibodies. (a–o) Fixed NIH3T3 cells were labelled with the indicated antibodies and DAPI. The presented nuclei were ordered according to the level of H3 S10 phosphorylation and chromatin compaction. The arrows point to an interphasic cell. Scale bars, 10 μm.
Acetylation and HP1 extractability in mitosis
The histone deacetylase inhibitor trichostatin A can induce HP1 delocalization from pericentromeric heterochromatin (Taddei et al, 2001; Cimini et al, 2003) and reduces HP1-mediated transcriptional repression (Nielsen et al, 1999). We therefore examined H3 acetylation during mitosis with a focus on K14, which is located in close proximity to the methylated K9. Cells were extracted with Triton X-100 before fixation and then stained with anti-HP1α and anti-H3 AcK14 antibodies. Cells fixed at different steps in late G2 and M were identified based on HP1α extractability and degree of chromatin condensation observed by 4,6-diamidino-2-phenylindole (DAPI) staining of the DNA. AcK14 staining was detected in all cells including interphasic cells (Fig 3, arrows). It was unchanged during the early phases of the G2/M transition, then increased in M phase (Fig 3F–H). Interestingly, this increase was well correlated with augmented HP1α extractability (Fig 3B–D). As a confirmation, we also detected increased mitotic H3 K14 acetylation by western blot on extracts from HeLa cells blocked in prophase with nocodazole (Fig 3M, lanes 3 and 4).
Acetylation inhibits HP1 binding
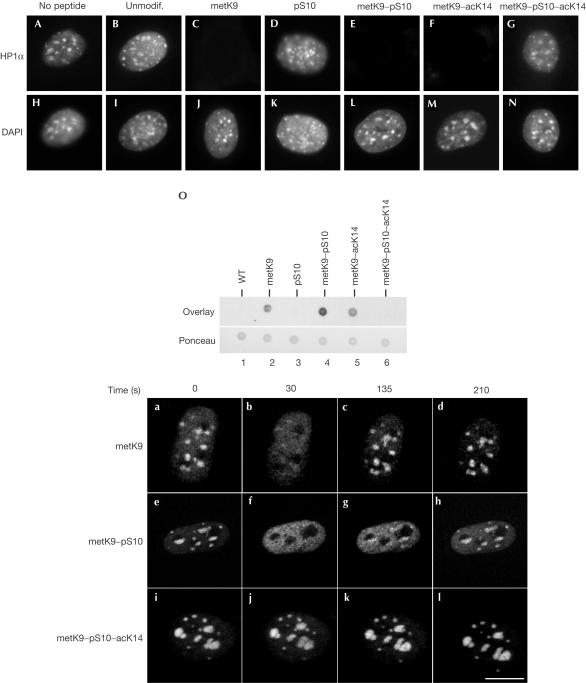
To characterize the effect of H3 S10 phosphorylation and K14 acetylation on HP1 binding to H3 metK9, we performed overlay assays in the presence of excess of various H3 peptides. In addition to H3 metK9, HP1 binding to chromatin was competed for by H3 metK9–pS10 and H3 metK9–acK14 peptides, indicating that neither S10 phosphorylation nor K14 acetylation could prevent HP1 from associating with an H3 peptide also methylated on K9 (Fig 4C,E,F,J,L,M). As expected, no competition was observed with either unmodified or H3 pS10 peptide (Fig 4B,D,I,K). These observations suggested that a triple modification of the H3 histone tail might be necessary for inhibition of HP1 binding. We therefore tested an H3 peptide carrying a metK9–pS10–acK14 modification. This peptide failed to compete for HP1 binding (Fig 4G,N), suggesting that it is the combined effect of phosphorylation and acetylation of the histone H3 tails that interferes with HP1 binding. Overlays on peptides spotted on nitrocellulose membrane confirmed that HP1 could bind metK9, metK9–pS10 and metK9–acK14, but not metK9–pS10–acK14 peptide (Fig 4O). Interestingly, in this assay, we noted some preference of HP1 for the metK9–pS10 peptide (Fig 4O, compare lane 4 with lanes 2 and 5). To examine HP1 peptide binding in vivo, we microinjected the various peptides into the nucleus of an NIH3T3-derived cell line expressing a green fluorescent protein (GFP)–HP1β fusion protein. Injection of H3 metK9 peptide led to rapid delocalization of GFP–HP1β (Fig 4a,b). Most probably due to rapid diffusion of the injected peptide out of the nucleus, this delocalization was only transient and relocalization of GFP–HP1β to pericentromeric heterochromatin was observed within 135 s, with images captured every 15 s (Fig 4c,d). A similar delocalization was observed after injection of the H3 metK9–pS10 peptide, confirming that this peptide can compete for chromatin binding of HP1 in vivo (Fig 4e,f). Interestingly, relocalization of HP1 was significantly slower than with H3 metK9 peptide, suggesting that HP1 associates more stably with the H3 metK9–pS10 than the H3 metK9 peptide (Fig 4g,h). No delocalization of GFP–HP1β was observed when using either unmodified (data not shown) or metK9–pS10–acK14 peptide (Fig 4i–l), confirming that the triply modified peptide was unable to bind HP1.
Figure 4.
HP1α is displaced from its chromatin targets by metK9, metK9–pS10 and metK9–acK14, but not by metK9–pS10–acK14 peptide. (A–N) Fixed NIH3T3 cells were incubated with recombinant GST–HP1α in the presence of the indicated peptides. Cells were then labelled with anti-GST (A–G) and DAPI (H–N). (O) 1 μg of the indicated peptides was spotted on a nitrocellulose membrane and stained with Ponceau (bottom panel). The membrane was then blocked with bovine serum albumin and incubated first with GST–HP1α and then with anti-GST antibodies (top panel). (a–l) Indicated peptides were microinjected into the nuclei of an NIH3T3-derived cell line expressing GFP–HP1β. Injected cells were then photographed every 15 s. Scale bars, 10 μm.
Effects of phosphorylation on the H3–HP1 complex
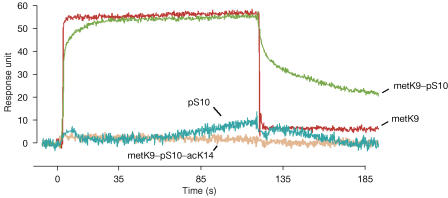
To evaluate the affinity of HP1 for the H3 metK9 and H3 metK9–pS10 peptides, we performed surface plasmon resonance (SPR) assays with recombinant 6 × His–HP1α bound to the sensor-chip surface (Fig 5). The equilibrium constant (Kd) of the interaction with the H3 metK9 peptide was 4.5±0.8 μM. This value is compatible with earlier measurements performed using other techniques (Nielsen et al, 2002; Fischle et al, 2003b). The affinity of 6 × His–HP1α for H3 metK9–pS10 peptide was in the same range as that for H3 metK9 (Kd=16.2±4.2 μM). However, the kinetic parameters of both interactions were significantly different. Indeed, the dissociation rate of the HP1α/H3metK9–pS10 complex (koff=(4.6±0.6)× 10−3 s−1) was more than 20-fold lower than that of the HP1α/H3metK9 complex (koff>0.1 s−1), showing that the phosphorylation of S10 increases the stability and the half-life of the association between H3 and HP1. Once again, we observed no or little binding of the unmodified (data not shown), the H3 pS10 and H3 metK9–pS10–acK14 peptides to the immobilized 6 × His–HP1α fusion protein.
Figure 5.
6 × His–HP1α binds in vitro to the metK9 and metK9–pS10 peptides, but not to the triple-modified metK9–pS10–acK14 peptide. A typical SPR measurement is shown for the different peptides at 100 μM. Note the slower dissociation phase observed for metK9–pS10 compared with metK9 peptide.
Discussion
In the present paper, we have followed HP1 localization and chromatin binding in late G2 and M phases, and we provide several observations suggesting that the mitotic redistribution of this protein is a multistep mechanism. First, we realized that extractability of HP1 is decreased after the onset of S10 phosphorylation in late G2 or early M. This can be paralleled with the slower dissociation of HP1 from an H3 metK9–pS10 peptide (compared to an H3 ‘metK9-only' peptide) observed in both microinjection and SPR experiments. Taken together, these observations indicate that association of HP1 with phosphomethylated H3 tails is more stable than association with only methylated tails. Interestingly, the distribution of H3 methylated on K9 does not coincide with the distribution of HP1 (Muchardt et al, 2002), suggesting that a large fraction of the histone H3 tails methylated on K9 are potential binding sites not associated with an HP1 protein. HP1 proteins are known to be highly mobile and are able to recolonize quickly areas subject to laser-induced photobleaching (Cheutin et al, 2003). We therefore suggest that an early effect of mitotic S10 phosphorylation is to ‘trap' HP1 on methylated histone H3 tails outside the heterochromatin foci, leading to diffuse HP1 localization. This model is compatible with the poor extractability of the diffused HP1 in late G2 (see, for example, Fig 3B).
Acetylation of H3 K14 gradually increases as M phase progresses and this increase is correlated with increased HP1 extractability. In vitro, we also find that an H3 peptide carrying a triple metK9–pS10–AcK14 modification no longer binds HP1. These observations strongly suggest that the combined effect of S10 phosphorylation and K14 acetylation is responsible for the delocalization of HP1 from chromatin in mitosis. Acetylation of K14 alone did not affect binding of HP1 to H3 metK9, indicating that some histone H3 tails bound by HP1 may be acetylated before phosphorylation. However, the chronology that we observe suggests that K14 acetylation occurs after S10 phosphorylation.
As mentioned above, S10 phosphorylation, in the absence of K14 acetylation, surprisingly stabilizes the HP1–H3 complex. This observation is compatible with an earlier study showing that phosphorylation of H3 S10 does not prevent binding of plant HP1γ in GST pull-down assays (Fass et al, 2002). In the structure derived from co-crystals of the HP1 chromodomain and a methylated histone H3 tail (Jacobs & Khorasanizadeh, 2002; Nielsen et al, 2002), S10 appears to point outwards from the HP1–H3 interface. This positioning of S10 would sterically allow the accommodation of an additional phosphate group. The crystal structure also shows that HP1 undergoes a conformational change when binding to methylated H3 tails. As suggested by the decreased off-rate of HP1 bound to H3 metK9–pS10 peptide in the SPR experiments, further conformational changes may occur upon binding to phosphomethylated H3 tails. Possibly, the presence of an acetyl group on H3 K14 may prevent HP1 from performing a proper conformational shift upon contact with histone H3. The mitotic H3 S10 kinase Ipl1/Aurora, when assayed in vitro, poorly phosphorylates an H3 peptide methylated on K9 (Rea et al, 2000). However, using the anti-H3 metK9–pS10 antibodies that we developed, we find that S10 phosphorylation of nonmethylated and K9-methylated histone tails initiates simultaneously during the late G2 phase of the cell cycle. This observation shows that in vivo methylation of K9 is not an obstacle for the phosphorylation of the neighbouring residue. It is possible that, in vivo, S10 is displayed in a conformation that is more favourable for phosphorylation by Ipl1/Aurora. Alternatively, other kinases, such as the NIMA-related kinases, may phosphorylate methylated histone H3 tails in vivo (Roig et al, 2002).
The data presented here have been collected from cells at the G2/M transition. Nevertheless, the mechanism of HP1 delocalization that we have uncovered may apply in other phases of the cell cycle. In particular, the transition from growth arrest (G0) to G1 is a time of extensive re-expression of transiently silenced genes. This transition, when induced by mitogenic stimulation, is followed by a wave of H3 S10 phosphorylation and K14 acetylation (Cheung et al, 2000; Clayton & Mahadevan, 2003). At the same time, we also observe a wave of phosphomethylation using the anti-H3 metK9–pS10 antibodies (data not shown). It is therefore tempting to speculate that in addition to recruitment of chromatin remodelling and transcription factors, these modifications serve as ways to overcome HP1-mediated repression. Redistribution of HP1 has recently been observed upon dedifferentiation in plant cells (Williams et al, 2003). Further studies will be required to determine whether re-entry into the cell cycle also affects HP1 targeting in mammalian cells.
Methods
Antibodies and peptides. Anti-H3 metK9–pS10 antibodies were produced in rabbits using a peptide coupled to KLH with the following sequence: AR(di-metK)(pS)TGGKAPRKQLC. These antibodies were purified by protein A-affinity columns and labelled with Fluorescein-EX (from Molecular Probes). Rabbit anti-H3 pS10 and anti-H3 acK14 antibodies were purchased from Upstate. Anti-H3 acK14 can recognize dimetK9–pS10–acK14 peptide. Monoclonal mouse anti-HP1α clone 1H5 was purchased from Euromedex. Rat anti-H3 pS28 (H9908) was purchased from Sigma. DNA was labelled with DAPI at 150 ng/ml. Competition experiments were performed with the following peptides either unmodified or with dimetK9, pS10, dimetK9–pS10, dimetK9–acK14 or dimetK9–pS10–acK14 modifications: ARTKQTARKSTGGKAPRC. The sequence of the dimetK27–pS28 peptide was as follows: QLATKAARKSAPATGGVC. Peptides were carefully quantified by amino-acid analysis and the presence of the modifications was confirmed by mass spectrometry.
Immunocytochemistry and microinjection. Immunofluorescent labelling, Triton extractions and overlay assays were performed using NIH3T3 cells as previously described (Muchardt et al, 2002). For microinjection, a GFP–HP1β expression construct (gift from C. Gazin) was stably integrated into NIH3T3 cells. Nuclear microinjection was performed using a Transjector 3246 (Eppendorf). All peptides were resuspended at 90 μM in water supplemented with Texas red dextran (Molecular Probes). Imaging was performed on an Axiovert 200M microscope (Zeiss) coupled with an RS2000 Nipkow-disk confocal system (Perkin Elmer).
In vitro binding studies by SPR. The assays were performed on a Biacore 2000 instrument equilibrated at 25°C with PBS+0.005% Tween 20 at a flow rate of 20 μl/min. The Penta-His monoclonal antibody (Qiagen) was covalently immobilized, using the Amine Coupling Kit (Biacore AB), on the carboxymethylated surface of flow cells 1 and 2 of a CM5 sensor chip. 6 × His–HP1α was captured on flow cell 1, and ten different concentrations of the peptides (0.2–150 μM) were then injected simultaneously on flow channels 1 and 2 for 2 min. The dissociation of the complexes was followed for 100 s. Flow cell 2 served as an anti-6 × His reference surface devoid of 6 × His–HP1α. The residual signals obtained for the unmodified peptide were used as negative controls, and subtracted from the signals obtained for the modified peptides. Between binding cycles, the coated surfaces were regenerated by two injections of 10 mM HCl. The association and dissociation profiles were analysed with a nonlinear least-squares algorithm implemented in the Biaevaluation 3.0 software (Biacore AB).
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v5/n5/extref/7400139s1.jpeg, http://www.nature.com/embor/journal/v5/n5t/extref/7400139s2.jpeg).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Acknowledgments
We thank C. Gazin, M. Guillemé and J. Seeler for the gift of plasmids and reagents. We thank the ‘Plate-Forme d'Imagerie Dynamique, Institut Pasteur' for assistance and use of the equipment, and R. Vazquez-Martinez for microinjection experiments. We also thank B. Bourachot, F. Baleux, E. Batché, M. Lavigne and J.-Y. Lallemand for valuable discussions. This work was supported by grants from the ‘Human Frontier Science Program', ‘L'Association pour la Recherche sur le Cancer' and ‘La Ligue contre le Cancer, Ile-de-France'.
References
- Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD (2000) Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell 5: 905–915 [DOI] [PubMed] [Google Scholar]
- Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T (2003) Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299: 721–725 [DOI] [PubMed] [Google Scholar]
- Cimini D, Mattiuzzo M, Torosantucci L, Degrassi F (2003) Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects. Mol Biol Cell 14: 3821–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton AL, Mahadevan LC (2003) MAP kinase-mediated phosphoacetylation of histone H3 and inducible gene regulation. FEBS Lett 546: 51–58 [DOI] [PubMed] [Google Scholar]
- Fass E, Shahar S, Zhao J, Zemach A, Avivi Y, Grafi G (2002) Phosphorylation of histone H3 at serine 10 cannot account directly for the detachment of human heterochromatin protein 1γ from mitotic chromosomes in plant cells. J Biol Chem 277: 30921–30927 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Allis CD (2003a) Binary switches and modification cassettes in histone biology and beyond. Nature 425: 475–479 [DOI] [PubMed] [Google Scholar]
- Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S (2003b) Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev 17: 1870–1881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto H et al. (1999) Identification of a novel phosphorylation site on histone H3 coupled with mitotic chromosome condensation. J Biol Chem 274: 25543–25549 [DOI] [PubMed] [Google Scholar]
- Jacobs SA, Khorasanizadeh S (2002) Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science 295: 2080–2083 [DOI] [PubMed] [Google Scholar]
- Li Y, Kirschmann DA, Wallrath LL (2002) Does heterochromatin protein 1 always follow code? Proc Natl Acad Sci USA 99(Suppl 4): 16462–16469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Guilleme M, Seeler JS, Trouche D, Dejean A, Yaniv M (2002) Coordinated methyl and RNA binding is required for heterochromatin localization of mammalian HP1α. EMBO Rep 3: 975–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina N, Verreault A, Laue E, Stillman B (1999) Heterochromatin dynamics in mouse cells: interaction between chromatin assembly factor 1 and HP1 proteins. Mol Cell 4: 529–540 [DOI] [PubMed] [Google Scholar]
- Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R (1999) Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J 18: 6385–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen PR, Nietlispach D, Mott HR, Callaghan J, Bannister A, Kouzarides T, Murzin AG, Murzina NV, Laue ED (2002) Structure of the HP1 chromodomain bound to histone H3 methylated at lysine 9. Nature 416: 103–107 [DOI] [PubMed] [Google Scholar]
- Nielsen SJ et al. (2001) Rb targets histone H3 methylation and HP1 to promoters. Nature 412: 561–565 [DOI] [PubMed] [Google Scholar]
- Rea S et al. (2000) Regulation of chromatin structure by sitespecific histone H3 methyltransferases. Nature 406: 593–599 [DOI] [PubMed] [Google Scholar]
- Roig J, Mikhailov A, Belham C, Avruch J (2002) Nercc1, a mammalian NIMA-family kinase, binds the Ran GTPase and regulates mitotic progression. Genes Dev 16: 1640–1658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taddei A, Maison C, Roche D, Almouzni G (2001) Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol 3: 114–120 [DOI] [PubMed] [Google Scholar]
- Williams L, Zhao J, Morozova N, Li Y, Avivi Y, Grafi G (2003) Chromatin reorganization accompanying cellular dedifferentiation is associated with modifications of histone H3, redistribution of HP1, and activation of E2F-target genes. Dev Dyn 228: 113–120 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2