Abstract
Our present in-depth knowledge of the physiology and regulatory mechanisms of microorganisms has arisen from our ability to remove them from their natural, complex ecosystems into pure liquid cultures. These cultures are grown under optimized laboratory conditions and allow us to study microorganisms as individuals. However, microorganisms naturally grow in conditions that are far from optimal, which causes them to become organized into multicellular communities that are better protected against the harmful environment. Moreover, this multicellular existence allows individual cells to differentiate and acquire specific properties, such as forming resistant spores, which benefit the whole population. The relocation of natural microorganisms to the laboratory can result in their adaptation to these favourable conditions, which is accompanied by complex changes that include the repression of some protective mechanisms that are essential in nature. Laboratory microorganisms that have been cultured for long periods under optimized conditions might therefore differ markedly from those that exist in natural ecosystems.
Keywords: differentiation, laboratory strains, multicellular structures, natural strains, signalling, yeast colonies
Introduction
Microorganisms are traditionally good models for the investigation of basic cellular processes on a unicellular level. However, these seemingly unicellular organisms naturally form multicellular communities, differentiate into specialized cells that benefit the whole population and synchronize their behaviour under certain conditions. Wild microorganisms are able to modify their properties according to their nutrient supply (for example, phase variation in bacteria) and, in particular, are able to adapt to limited nutrients. This is a common situation in nature and microorganisms are able to decrease their rate of metabolism and survive using rare nutrient sources, including their own waste products (for example, ammonia signalling in yeast colonies). Some of the mechanisms that are apparently important in nature, such as the formation of the extracellular matrix, are switched off when microorganisms are transferred to comfortable laboratory conditions, whereas others that appear to be more general persist. This review discusses the properties of multicellular microorganisms, including their differentiation, interaction and long-range signalling, and considers how these properties differ in the microorganisms that are routinely studied in the laboratory.
Multicellular microorganisms have an advantage in nature
In the laboratory, under conditions of optimal nutrient sources, temperature, humidity and so on, microorganisms are usually treated as individuals and are studied predominantly during the relatively small window of time in which they grow as exponential pure, liquid cultures. However, the natural environment rarely provides microorganisms with conditions that allow them to grow and reproduce at a maximal rate. Microorganisms in the wild need to cope with, for example, nutrient starvation, temperature changes and a lack of moisture, and should be able to survive for extended periods without reproduction. In addition, microorganisms rarely exist naturally as individuals, because one of their survival mechanisms is the ability to organize themselves into multicellular communities and to differentiate into specialized cell variants.
Traditionally studied fruiting bodies of slime moulds and myxobacteria are formed by motile amoebas and bacteria under starvation conditions. The cell differentiation that occurs during the development of these structures results in the formation of highly resistant spores (germinal cells) and fruiting body stalks that finally lyse (terminal phenotype cells). The environment of the fruiting body seems to drive the development of mature spores that are able to survive in extreme environments. During this period, the material released from lysed stalk cells is incorporated into the spore cell wall, which makes the spores fully resistant to harsh conditions (Shimkets, 1990; Kaiser, 1999). These findings reveal the existence of differentiation programmes that are specific to distinct cells and areas of the fruiting body, and which cannot be pursued by individual free-living cells.
In contrast to fruiting bodies, which are formed by the active aggregation and consequent differentiation of non-dividing motile microorganisms, the formation of colonies and biofilms is conditioned by the growth and division of bacteria, yeast and moulds. However, similar to the cells in fruiting bodies, cells in colonies and biofilms also have the ability to differentiate and behave differently in distinct areas of these multicellular communities (see below).
Wild and laboratory microorganisms differ
There are several indications that environmental conditions have an important influence on the properties of the multicellular communities that are formed by microorganisms. This is particularly evident in biofilms and colonies of bacteria and yeast. Complex social communities of different, coexisting microorganisms are common in nature and these highly organized biofilms can attach to solid surfaces in liquid environments or form a pellicle on the surface of water (for example, see Costerton et al, 1995; Davey & O'Toole, 2000). Indeed, their formation is so efficient that it is an arduous task to keep surfaces free from an invading biofilm. Conversely, in the laboratory, it is difficult to create artificial experimental conditions that simulate natural conditions; in other words, to grow artificial biofilms that have properties similar to those of fully developed natural biofilms. Therefore, despite the fact that there is plenty of information about the occurrence and ultrastructure of biofilms in situ, relatively little is known about the ecology and biology of cells within biofilms and about biofilm-dedicated pathways (Ghigo, 2003).
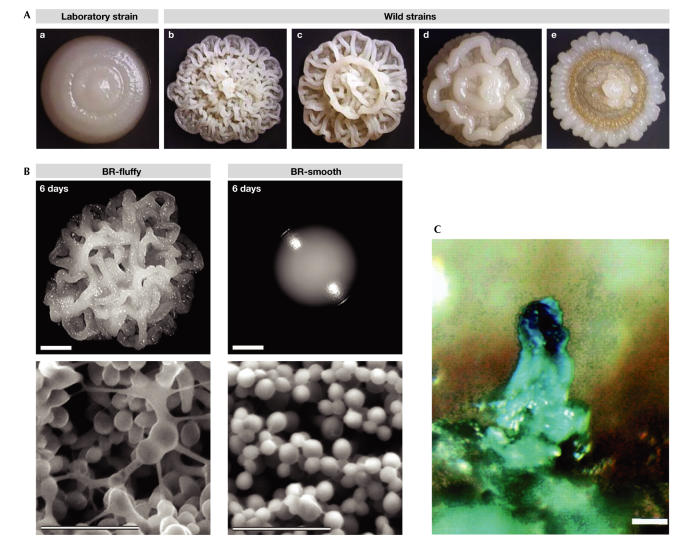
Colonies are another example of multicellular communities of microorganisms that occur under natural conditions. They seem to be less complex and less resistant to the environment than biofilms, and therefore represent an 'intermediate' between unprotected individual cells and fully protected natural biofilms. This, together with the fact that colonies of bacteria, yeast and other microorganisms are commonly grown in laboratories, makes them an interesting subject for studies of, for example, the interactions and signalling between microorganisms within multicellular communities. One intriguing question is whether (and how) colonies in nature differ from those growing under laboratory conditions. Recent findings on the differences between colonies formed by Saccharomyces cerevisiae laboratory strains and those formed by strains freshly isolated from nature have provided some clues (Kuthan et al, 2003). S. cerevisiae laboratory strains usually form smooth colonies with no markedly structured pattern (Fig 1Aa), whereas colonies of wild S. cerevisiae strains form 'fluffy'structured colonies even in the laboratory (Fig 1Ab–e). Ultrastructural native electron microscopy and biochemical analyses revealed that cells within fluffy colonies are connected by an abundant highly glycosylated extracellular matrix that forms a scaffold for the colony (Fig 1B). No such material was observed within the smooth colony. This extracellular matrix is important for the protection of the colony from perilous environmental factors and desiccation. Moreover, similarly to biofilms, it can form small channels for nutrient and water flow, and also chambers and microenvironments for the next generations of cells. This is supported by the observation that fluffy colonies expand and occupy new territory more rapidly than those that are smooth (Fig 1B; Kuthan et al, 2003).
Figure 1.
Differences between wild and laboratory microorganisms. (A) Colonies of laboratory (a) and wild (b–e) strains of Saccharomyces cerevisiae. (B) Colonies and ultrastructure of wild BR-fluffy strain (a) and its domesticated derivative BR-smooth strain. Both colonies are 6 days old (reproduced with permission from Kuthan et al, 2003). (C) Sporulation sites on the surfaces of colonies formed by wild Bacillus subtilis (reproduced with permission from Branda et al, 2001).
Recent data on the sporulation of the bacterium Bacillus subtilis also reveal a remarkable divergence between bacteria growing in the laboratory and those growing in the wild. In contrast to the laboratory strains, the sporulation of bacteria that are freshly isolated from nature occurs within small fruiting bodies that are formed at the top of the bacterial community (Fig 1C). These structures resemble the fruiting bodies of myxobacteria and contain regions of cells that differentiate into resistant spores as well as the layer of stalk cells (Branda et al, 2001).
Under heterogeneous environmental conditions, microorganisms can diversify into specific subpopulations that are genotypically and phenotypically different. This 'phase variation' has been described in a range of bacteria (for example, Salmonella spp., Escherichia coli and Neisseria spp.) and is usually associated with changes on the cell surface (membrane antigens, flagella and fimbriae), which result in morphologically altered colonies of the respective strain. This high-frequency switching of phenotype is often the result of genome rearrangements (Henderson et al, 1999) and has been described in, among other examples, Pseudomonas aeruginosa during biofilm formation (Deziel et al, 2001). It is not known whether phase-variation events are mostly random, such that under different stress conditions a mixed bacterial population is generated that is able to colonize different areas of an ecosystem or, alternatively, whether they are determined by a particular environment.
Wild microorganisms are domesticated in the laboratory
The differences in behaviour between freshly isolated microorganisms and those that have been grown in abundant laboratory conditions could be caused either by adaptive mechanisms or by the long-term accumulation of mutations that lead to the gradual selection of laboratory strains. Findings in natural S. cerevisiae strains indicate that, at least in some cases, the laboratory phenotype does not emerge owing to the accumulation of mutations, but seems to be the result of relatively quick and efficient active adaptation (Kuthan et al, 2003). The ability of wild yeast to create structured fluffy colonies is not maintained under laboratory conditions. After passages on complex agar medium, smooth colonies that resemble those formed by laboratory strains start to appear with high frequency (∼2–3%). This 'domestication' is accompanied by the loss of the extracellular matrix (Fig 1B) and extensive changes in gene expression that denote a complex reprogramming of the yeast lifestyle (Kuthan et al, 2003). The fluffy-tosmooth switch includes the repression of the AQY1 gene that encodes the water channel aquaporin (Laize et al, 1999), which indicates differences in the water permeability and the surface properties of cells within the two types of colony. Interestingly, AQY1 seems to be mutated in a broad range of laboratory strains (Bonhivers et al, 1998). For the adaptation to occur, chromatin remodelling might be important, as indicated by transcription changes in genes that are involved in chromatin structure (Kuthan et al, 2003). Also indicated is the involvement of histone deacetylases and the silent information regulator 2 (Sir2) protein in the phenotypic switching of Candida albicans colonies (Perez-Martin et al, 1999; Srikantha et al, 2001; see below).
What might be the reasons for this domestication? The production of the abundant extracellular matrix network in colonies of 'wild' yeast, similar to biofilms, seems to be important for protection against the environment and for territory occupancy. Both of these factors are particularly relevant under hostile conditions. Under laboratory conditions with rich nutrient sources, controlled humidity and sufficient space, there is no longer a need for the extracellular matrix to form, and the massive amounts of energy that this would otherwise consume can be used more profitably. After growing for 1,000 generations in liquid culture at low densities, the bacteria Myxococcus xanthus displayed significantly reduced social behaviour, which was dependent on cell–cell interactions and extracellular matrix production (Velicer et al, 1998). Detailed knowledge of the changes in regulation that occur during the fluffy-tosmooth switch of S. cerevisiae colonies could denote similar mechanisms in more complicated biofilms. In this regard, it is important to define the 'natural' conditions that would allow the reverse (smooth-to-fluffy) switch and the consequent clarification of the mechanisms involved. However, this might be as complicated as growing biofilms in a laboratory.
Most strains of C. albicans and related species are able to switch between two or more colony morphotypes. Switching regulates several phenotypic characteristics, including virulence, and occurs at higher frequencies in virulent pathogenic strains than in commensal ones (Hellstein et al, 1993). It could therefore be related to the efficient adaptation of pathogenic Candida strains in response to factors in the host environment, including drug therapy and the immune response of the host (Odds, 1997). As mentioned above, the high-frequency, reversible 'opaque/white' switch (Slutsky et al, 1987) seems to be associated with reversible chromatin remodelling, as two histone deacetylases (Hda1 and Rpd3) and Sir2—which is important for silencing at specific regions of chromosomes—control the switch (Perez-Martin et al, 1999; Srikantha et al, 2001).
Differentiation in yeast colonies and stalks
A fundamental property of multicellular tissues and organisms is the ability of cells to differentiate and form specialized variants, which have specific roles that benefit the whole tissue or organism. Cellular differentiation is usually elicited by signals that are either on the cell surface and influence their immediate vicinity, or that are released by cells into their surroundings and affect other cells, even those at a distance. During the past years, various studies have indicated that this property can be extrapolated even to multicellular structures that are formed by microorganisms.
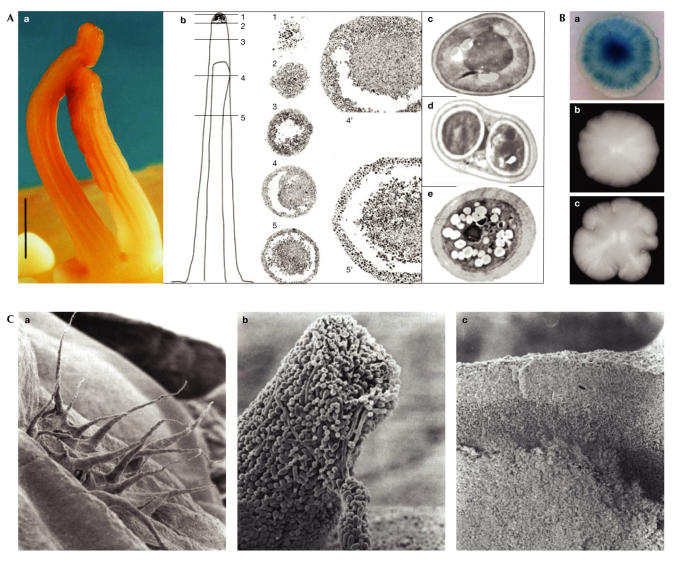
Marked differentiation has been described within multicellular stalks (Fig 2Aa) that are formed by yeast and bacteria (Engelberg et al, 1998). The ultrastructural analysis of thin sections of yeast multicellular stalks (Fig 2Ab; Scherz et al, 2001) revealed at least two distinct layers: a central core containing 'normal' yeast cells (Fig 2Ac), and yeast spores (Fig 2Ad) covered by a surface layer of vacuolized cells with thick cell walls (Fig 2Ae). These cells are probably dying or already dead and seem to form a 'skin' for the stalk, which is likely to protect it against drying and other environmental effects.
Figure 2.
Differentiation in yeast multicellullar communities. (A) Multicellular yeast stalk (a; reproduced with permission from Engelberg et al, 1998) and its anatomical structure (b; reproduced with permission from Scherz et al, 2001). Central core formed by normal cells (c), yeastspores (d) and dying cells (e). (B) Saccharomyces cerevisiae colony expressing the CCR4 promoter–LacZ fusion (a). Colonies of the ccr4-deletion mutant (c) and the isogenic parental strain (b) (reproduced with permission from Minarikova et al, 2001). (C) Ultrastructure of colonies of Candida albicans (reproduced with permission from Radford et al, 1994) showing aerial hyphae (a and b) and three layers of different cells within a colony (c).
Candida spp. are budding yeasts that can form a range of polarized and elongated cell shapes, from pseudohyphae to true unconstricted hyphae, which usually have remarkably structured colonies (Fig 2C). Scanning electron microscopy revealed that C. albicans colonies are composed of various layers of cells that exhibit different morphologies, some of which are connected by fibrillar structures (Whittaker & Drucker, 1970; Radford et al, 1994). The reasons for, and the molecular regulation of, this differentiation are not yet known.
As discussed previously, colonies of S. cerevisiae laboratory strains are usually smooth with no structural pattern. However, the use of a lacZ promoter library (Minarikova et al, 2001) allowed the identification and isolation of different genes that are specifically expressed in distinct areas of the colony (Fig 2Ba). A strain that is defective in one of the identified genes that encodes the Ccr4 transcription factor forms colonies that exhibit a rambling morphology (Fig 2Bc) compared with the parental strain (Fig 2Bb). These results indicate that differentiated gene expression in distinct areas of smooth colonies of S. cerevisiae might be important for the formation of colony structure.
Long-range signalling
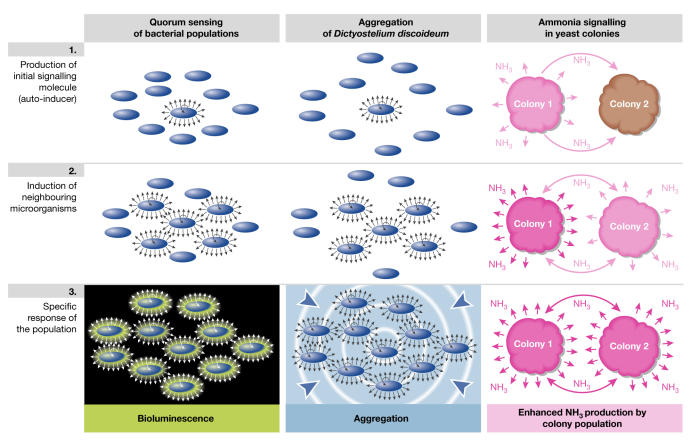
Extracellular receptors and matrix material might participate in direct short-range cell–cell communication between microorganisms and presumably in the differentiation of cells within multicellular communities. However, microorganisms also have the ability to emit and to receive various long-range signals that can mutually influence their behaviour. Such signals can coordinate the behaviour of either individual microorganisms or multicellular communities. Again, this seems to be more important under natural conditions and might help microorganisms to respond better to environmental changes, such as limited nutrient sources or increasing cell density. Long-range signals are often related to different aspects of the social life of microorganisms. The most studied example of long-range signalling is quorum sensing, which has been described in various bacteria (for a review, see Miller & Bassler, 2001; Bassler, 2002). Quorum sensing allows microorganisms to monitor their density and consequently leads to a specific response by the whole bacterial population. It is based on the principle of autoinduction by diffusible signal molecules (known as 'autoinducers' or 'pheromones'), which are produced by bacterial populations (Fig 3). The system of autoinduction allows the amount of a signalling compound to increase more efficiently than could be achieved by a simple increase in the number of non-induced cells. Therefore, the population can change its behaviour before reaching a critical density. Quorumsensing signals are usually specific for particular microorganisms; for example, N-acyl-homoserine lactones in Gram-negative bacteria and post-translationally modified peptides in Gram-positive bacteria. However, Chen and colleagues have described the existence of a universal signalling molecule, furanosyl-borate diester (AI-2), which is used for interspecies bacterial communication (Chen et al, 2002). Quorum sensing regulates diverse physiological processes, including bioluminescence, swarming, antibiotic synthesis, the production of virulence determinants in pathogenic bacteria and biofilm formation.
Figure 3.
Autoinduction: general signalling mechanism in communities of microorganisms. Autoinducers (arrows) induce their own production in the same cell and/or in surrounding cells (colonies).
Autoinduction also has an important role in the formation of multicellular fruiting bodies by Dictyostelium discoideum. During this process, starving vegetative amoebae start to produce the signalling molecule cAMP (Garrod & Malkinson, 1973), which diffuses into the surroundings and induces its own production by other cells (Fig 3). In this way, the signal spreads throughout the neighbouring population and an orientated concentration gradient is established, which determines the direction of cell movement towards an aggregation centre where an unorganized aggregate is formed (Gross, 1994).
Another example of autoinduction through a simple signalling molecule is the 'ping-pong' mechanism of ammonia induction in yeast colonies (Fig 3; Palkova & Forstova, 2000). Volatile, unprotonated ammonia has been identified as a signalling molecule that is spread through the air by colonies of various unrelated yeasts and is important for colony development and survival (Palkova et al, 1997; Palkova & Vachova, 2003). Yeast colonies periodically change the pH of their surroundings from acidic to alkaline and vice versa. Colonies grow in the acidic phase, whereas they produce ammonia and their growth is transiently inhibited in the alkaline phase. Ammonia induces colonies of different yeast species to produce their own ammonia, regardless of their developmental phase. Therefore, all colonies in a respective area simultaneously switch to producing ammonia after the oldest colony starts to release ammonia molecules (Palkova & Forstova, 2000). Changes occurring within colonies of S. cerevisiae during their transition to ammonia production indicate that their metabolism alters to use previously excreted waste products and that there is a parallel decrease in stress (Palkova et al, 2002). This simple volatile compound might therefore universally function as an alarm signal that is produced by the colony that first senses a stress, such as limited nutrients or toxic levels of a compound. Released ammonia then induces its own production in surrounding colonies and, consequently, the whole 'colony population' switches on adaptive mechanisms. Moreover, the observed transient growth inhibition in the direction of the ammonia gradient, together with the universal nature of this signalling mechanism, might help yeast to orientate growth towards unoccupied areas and away from those where competition for nutrients is high.
As indicated, long-range signalling is usually based on the production of a chemical compound that is spread through liquid or air. However, an intriguing case of non-chemical signalling between bacterial colonies has been described. Experiments performed by Matsuhashi et al (1995) implied the existence of ultrasound signalling between bacterial colonies. Using the example of the bacteria Bacillus carbophilus growing dependently on activated charcoal, this study showed that bacteria growing around the carbon material transmitted colony-forming potential to cells and spores in distant locations. Moreover, transmission was not interrupted by plastic or glass barriers, which points to the existence of a physical signal. The authors showed that signal transmission still occurred even in the absence of graphite and when B. carbophilus was substituted with Bacillus subtilis or E. coli (Matsuhashi et al, 1995).
Most of the described signalling mechanisms allow individual microorganisms or populations to respond to environmental changes that are associated with nutrient depletion. Such conditions are common in nature but, even in the laboratory, microorganisms need to retain properties that allow them to survive under non-optimal growth conditions. It is not yet known whether (and how) these signalling systems differ between natural strains and their laboratory counterparts. The importance of extracellular signalling molecules has also been highlighted by findings in 'viable-but-non-culturable' (VNC) bacteria. Studies on the density and variability of microorganisms that exist in different environments indicate that they represent one-half of all cellular carbon, but that 99% of them are unknown and cannot be grown under standard laboratory conditions (Whitman et al, 1998); in other words, only a small proportion (1%) of microorganisms that occur in nature can be grown in a laboratory. At least in some cases, VNC bacteria need specific factors that are produced by other cells for their growth. For example, after long-term (several months) starvation, Micrococcus luteus bacteria convert to dormant cells that are able to survive for long periods of time but have lost the ability to grow on agar plates. This ability is restored by adding a picomolar concentration of the specific resuscitation factor Rpf, which is the peptide that is produced by a growing M. luteus culture. Rpf homologues have been identified in other bacteria, including those of Mycobacterium spp. (Kell & Young, 2000). Therefore, in nature, a large proportion of bacteria probably survive long periods of starvation and other stresses as dormant cells, and only continue growth after receiving a signal from growing individuals that indicates a possible favourable change in the environment.
Conclusions
The increasing number of studies that have specifically investigated biofilms and quorum sensing clearly indicate that the interest of microbiologists in natural microbial life has intensified during the past few years. Shapiro wrote in 'Bacteria as Multicellular Organisms' that bacteria should be viewed “as sensitive, communicative, decisive organisms integrating information from their environment and from their neighbours in order to carry out the complex tasks of reproduction and survival in organised multi-cellular populations” (Shapiro, 1997). However, this view has not yet been completely accepted, and much more effort is needed to, at least partly, understand 'multicellular microorganisms' and their implications for microbiological research.

Zdena Palková is the recipient of an EMBO Young Investigator Award
Acknowledgments
I thank L. Váchová and V. Závada for comments and for help in improving the manuscript. This work is supported by grants from GACR 204/02/0650, GAUK 141/2001/B-BIO/PrF, the Ministry of Education, Youth and Sports (MSMT) of the Czech Republic projects LA141 and J13/98:113100003, and the EMBO Young Investigator Programme.
References
- Bassler BL (2002) Small talk. Cell-to-cell communication in bacteria. Cell 109: 421–424 [DOI] [PubMed] [Google Scholar]
- Bonhivers M, Carbrey JM, Gould SJ, Agre P (1998) Aquaporins in Saccharomyces. Genetic and functional distinctions between laboratory and wild-type strains. J Biol Chem 273: 27565–27572 [DOI] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R (2001) Fruiting body formation by Bacillus subtilis. Proc Natl Acad Sci USA 98: 11621–11626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Schauder S, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM (2002) Structural identification of a bacterial quorumsensing signal containing boron. Nature 415: 545–549 [DOI] [PubMed] [Google Scholar]
- Costerton JW, Lewandowski Z, Caldwell DE, Korber DR, Lappinscott HM (1995) Microbial biofilms. Annu Rev Microbiol 49: 711–745 [DOI] [PubMed] [Google Scholar]
- Davey ME, O'Toole GA (2000) Microbial biofilms: from ecology to molecular genetics. Microbiol Mol Biol Rev 64: 847–867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deziel E, Comeau Y, Villemur R (2001) Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J Bacteriol 183: 1195–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelberg D, Mimran A, Martinetto H, Otto J, Simchen G, Karin M, Fink GR (1998) Multicellular stalk-like structures in Saccharomyces cerevisiae. J Bacteriol 180: 3992–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrod DR, Malkinson AM (1973) Cyclic AMP, pattern formation and movement in the slime mould, Dictyostelium discoideum. Exp Cell Res 81: 492–495 [DOI] [PubMed] [Google Scholar]
- Ghigo JM (2003) Are there biofilmspecific physiological pathways beyond a reasonable doubt? Res Microbiol 154: 1–8 [DOI] [PubMed] [Google Scholar]
- Gross JD (1994) Developmental decisions in Dictyostelium discoideum. Microbiol Rev 58: 330–351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellstein J, Vawter-Hugart H, Fotos P, Schmid J, Soll DR (1993) Genetic similarity and phenotypic diversity of commensal and pathogenic strains of Candida albicans isolated from the oral cavity. J Clin Microbiol 31: 3190–3199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson IR, Owen P, Nataro JP (1999) Molecular switches-the ON and OFF of bacterial phase variation. Mol Microbiol 33: 919–932 [DOI] [PubMed] [Google Scholar]
- Kaiser D (1999) Cell fate and organogenesis in bacteria. Trends Genet 15: 273–277 [DOI] [PubMed] [Google Scholar]
- Kell DB, Young M (2000) Bacterial dormancy and culturability: the role of autocrine growth factors. Curr Opin Microbiol 3: 238–243 [DOI] [PubMed] [Google Scholar]
- Kuthan M, Devaux F, Janderova B, Slaninova I, Jacq C, Palkova Z (2003) Domestication of wild Saccharomyces cerevisiae is accompanied by changes in gene expression and colony morphology. Mol Microbiol 47: 745–754 [DOI] [PubMed] [Google Scholar]
- Laize V, Gobin R, Rousselet G, Badier C, Hohmann S, Ripoche P, Tacnet F (1999) Molecular and functional study of AQY1 from Saccharomyces cerevisiae: role of the C-terminal domain. Biochem Biophys Res Commun 257: 139–144 [DOI] [PubMed] [Google Scholar]
- Matsuhashi M, Pankrushina AN, Endoh K, Watanabe H, Mano Y, Hyodo M, Fujita T, Kunugita K, Kaneko T, Otani S (1995) Studies on carbon material requirements for bacterial proliferation and spore germination under stress conditions: a new mechanism involving transmission of physical signals. J Bacteriol 177: 688–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller MB, Bassler BL (2001) Quorum sensing in bacteria. Annu Rev Microbiol 55: 165–199 [DOI] [PubMed] [Google Scholar]
- Minarikova L, Kuthan M, Ricicova M, Forstova J, Palkova Z (2001) Differentiated gene expression in cells within yeast colonies. Exp Cell Res 271: 296–304 [DOI] [PubMed] [Google Scholar]
- Odds EC (1997) Switch of phenotype as an escape mechanism of the intruder. Mycoses 40: 9–12 [DOI] [PubMed] [Google Scholar]
- Palkova Z, Forstova J (2000) Yeast colonies synchronise their growth and development. J Cell Sci 113: 1923–1928 [DOI] [PubMed] [Google Scholar]
- Palkova Z, Vachova L (2003) Ammonia signaling in yeast colony formation. Int Rev Cytol 225: 229–272 [DOI] [PubMed] [Google Scholar]
- Palkova Z, Janderova B, Gabriel J, Zikanova B, Pospisek M, Forstova J (1997) Ammonia mediates communication between yeast colonies. Nature 390: 532–536 [DOI] [PubMed] [Google Scholar]
- Palkova Z, Devaux F, Icicova M, Minarikova L, Le Crom S, Jacq C (2002) Ammonia pulses and metabolic oscillations guide yeast colony development. Mol Biol Cell 13: 3901–3914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martin J, Uria JA, Johnson AD (1999) Phenotypic switching in Candida albicans is controlled by a SIR2 gene. EMBO J 18: 2580–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radford DR, Challacombe SJ, Walter JD (1994) A scanning electronmicroscopy investigation of the structure of colonies of different morphologies produced by phenotypic switching of Candida albicans. J Med Microbiol 40: 416–423 [DOI] [PubMed] [Google Scholar]
- Scherz R, Shinder V, Engelberg D (2001) Anatomical analysis of Saccharomyces cerevisiae stalk-like structures reveals spatial organization and cell specialization. J Bacteriol 183: 5402–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro J (1997) Multicellularity: the Rule, not the Exception. Lessons from Escherichia coli Colonies. Oxford Univ Press, New York, USA [Google Scholar]
- Shimkets LJ (1990) Social and developmental biology of the myxobacteria. Microbiol Rev 54: 473–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller M, Soll DR (1987) “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol 169: 189–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikantha T, Tsai L, Daniels K, Klar AJ, Soll DR (2001) The histone deacetylase genes HDA1 and RPD3 play distinct roles in regulation of high-frequency phenotypic switching in Candida albicans. J Bacteriol 183: 4614–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velicer GJ, Kroos L, Lenski RE (1998) Loss of social behaviors by Myxococcus xanthus during evolution in an unstructured habitat. Proc Natl Acad Sci USA 95: 12376–12380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitman WB, Coleman DC, Wiebe WJ (1998) Prokaryotes: the unseen majority. Proc Natl Acad Sci USA 95: 6578–6583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker DK, Drucker DB (1970) Scanning electron microscopy of intact colonies of microorganisms. J Bacteriol 104: 902–909 [DOI] [PMC free article] [PubMed] [Google Scholar]