Summary
Workshop on Mechanisms of Nuclear Transport
Keywords: karyopherins, mRNA export, NPC, nuclear transport, Ran
Introduction
During five days in November 2003, new results and exciting hypotheses in the nucleocytoplasmic transport field were discussed intensely at the base of the Etna volcano in the beautiful Sicilian town of Taormina, Italy. The main underlying theme that emerged was that the term 'nucleocytoplasmic transport' is no longer limited to the events that occur in close proximity to the nuclear pore complex (NPC)—the channel through which all macromolecules enter and exit the nucleus (for a review, see Fahrenkrog & Aebi, 2003)—but that it also encompasses the events that occur from the time the cargo molecule is first synthesized until it is delivered to its site of action. The basic mechanisms of protein transport have been known for some time, but the meeting provided important new evidence of how larger and more complex cargoes are transported. An obvious example was the series of talks that followed the fate of messenger RNA (mRNA) from its synthesis in the nucleus until its delivery to the ribosomes in the cytoplasm. Importantly, the close integration of synthesis, processing, packaging and transport is now becoming evident. In addition, the dynamics and exciting regulatory functions of the NPC itself are being revealed through a range of technologically advanced approaches. Multifunctionality is also being recognized for many transport receptors that apparently regulate crucial cellular processes, such as the cell cycle, in addition to chaperoning cargo through the nuclear pore. The field of nucleocytoplasmic transport is therefore expanding to ask important questions about how dynamic compartmentalization regulates crucial cellular processes. Some of the highlights of the work presented at the meeting are described in this report.

This EMBO workshop took place between 1 and 5 November 2003, and was organized by E. Hurt and V. Doye.
The nuclear pore complex
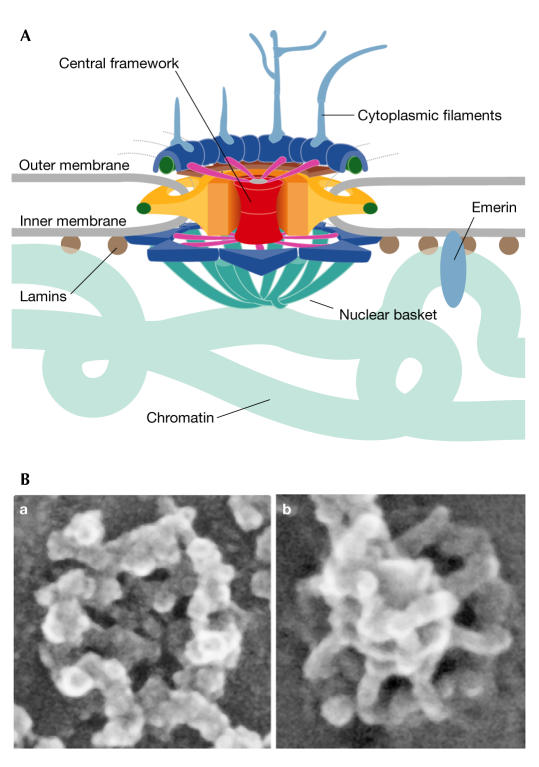
NPCs (Fig 1) serve as conduits between the nucleus and the cytoplasm. The large proteinaceous NPCs are composed of proteins that are referred to as nucleoporins or Nups, many of which contain phenylalanine–glycine (FG) repeats. Although, in the past, NPCs have been thought of as large structural complexes that are static once assembled, evidence is accumulating that these structures are in fact highly dynamic. Inverse fluorescence-recovery after photobleaching (iFRAP) experiments from the J. Ellenberg laboratory (Heidelberg, Germany) systematically analysed the dynamics of green fluorescent protein (GFP)-tagged nucleoporins that were expressed at physiological levels in stable cell lines to reveal that NPC components fall into three classes: the first type remain bound to the NPC for longer than 30 h (for example, Nup107 and Nup133); the second exchange moderately over 2–20 h (for example, Nup62 and Nup98); and the third show only transient interactions with the NPC in the seconds to minutes range (for example, Nup50 and Nup153). Although the iFRAP method can only be applied to GFP-tagged proteins, some of which might behave differently from endogenous proteins, this work clearly stimulated discussion of a dynamic NPC. M. Powers (Atlanta, GA, USA) described the dynamics and NPC associations of Nup98 and Nup153. The mobility of each protein is dependent on ongoing transcription and the domains that are required for this dependence were mapped in each nucleoporin. The mechanistic basis for nucleoporin dynamics is not yet known, but one possibility is that remodelling of the nuclear basket of the pore might occur during transport and that this results in the temporary dissociation of some peripheral nucleoporins.
Figure 1.
Structure of the nuclear pore complex. (A) Schematic diagram of the nuclear pore complex (NPC). The complex has a vertical eightfold symmetry. The nuclear and cytoplasmic sides differ from each other in that the nuclear basket projects into the nucleoplasm and eight filaments extend into the cytoplasm. (B) Field-emission scanning electron microscopy shows the surface structure of the cytoplasmic (a) and nucleoplasmic (b) faces of the NPC. The micrographs were kindly provided by M. Goldberg.
V. Cordes (Stockholm, Sweden) used RNA interference (RNAi) directed against the nuclear basket protein Tpr, and immunoelectron-microscopy localization of Tpr subdomains as well as other nucleoporins, to analyse structurally the nuclear basket of the NPC. Cordes concluded from these and other experiments that Tpr molecules are the central architectural elements of the nuclear basket. By contrast, M. Goldberg (Durham, UK) argued that none of the proteins that he analysed using field-emission scanning microscopy (Nup153, Nup98, lamins, Nup107 and possibly Tpr) seemed to be essential structural components of the NPC basket.
An exciting piece of work that addressed the important issue of how macromolecules are translocated through the NPC was presented by S. Wente (Nashville, TN, USA), who asked which elements are required for a functional NPC. Using a genetic approach in Saccharomyces cerevisiae, her laboratory systematically deleted the FG-repeat region of each of the 11 FG-repeat-containing nucleoporins. This region of the nucleoporins is located in the central aqueous channel of the NPC and extends from the tips of the cytoplasmic filaments to the distal ring of the nuclear basket. The FG repeats have been proposed to either function as initial docking sites for the transport receptors or to create a central meshwork within the NPC to form a permeability barrier that can be overcome by the highly hydrophobic transport receptors (for a review, see Fried & Kutay, 2003). The Wente laboratory created a minimal NPC in living yeast cells with approximately one-third of the normal complement of FG nucleoporins by systematically combining the FG-repeat-region-deleted mutants until it was not possible to delete another domain and still maintain viability. Surprisingly, the FG mass that was deleted did not correlate with either the severity of the analysed transport defects or the effects on cell viability. Equally unexpectedly, the deletion of all FG-repeat domains that were located exclusively on the cytoplasmic or nucleoplasmic sides of the NPC resulted in viable cells with no gross transport defects. By contrast, only specific subsets of the FG-repeat domains that were located on both sides of the NPC could be deleted. This thorough analysis indicates that there are distinct functions for different FG nucleoporins in specific transport pathways.
The nuclear envelope
The nuclear envelope not only accommodates the NPC, but also separates the genetic material from the site of translation. However, during each cell cycle, both nuclear envelope breakdown and reassembly occur. It is therefore of great interest to identify and characterize the components of the nuclear envelope and the integral membrane components of the pore, as they could give us some insight into how these processes are regulated. How proteins are targeted to the inner nuclear membrane was addressed by several genome-wide approaches. A. Hopper's laboratory (Hershey, PA, USA) used the S. cerevisiae gene-deletion collection to analyse the targeting of an inner nuclear envelope protein, Trm1, which was fused to GFP. They found that a heterotrimeric amino-terminal acetyltransferase is essential for Trm1–GFP localization to the inner nuclear membrane. A large-scale genome-wide RNAi approach in Caenorhabditis elegans was performed by I. Mattaj's laboratory (Heidelberg, Germany) to identify the proteins that are involved in nuclear assembly. The screen identified many genes that had already been implicated in this process, the products of which include the Ran system proteins (Fig 2), Nups and nuclear envelope proteins, including emerin. In addition, they identified new molecules, such as Mel28, which is an inner nuclear membrane protein that binds to chromatin. One hypothesis is that Mel28 might have a role in chromatin condensation. Whereas Mattaj focused on nuclear envelope assembly, K. Ullman's group (Salt Lake City, UT, USA) addressed the mechanism of nuclear envelope breakdown using Xenopus egg extracts. They reported that the recruitment of the coat protein 1 (COPI) complex to the nuclear envelope by Nup153 is an important first step to induce vesiculation and nuclear envelope breakdown (Liu et al, 2003).
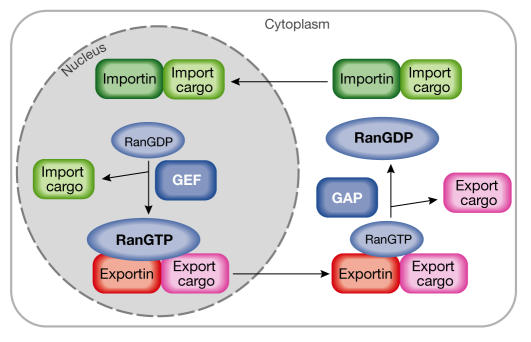
Figure 2.
Cargo binding and release from the karyopherins is controlled by Ran. The asymmetric distribution of the GTPase-activating protein (RanGAP) and the guanine-exchange factor (RanGEF) generate a high RanGTP concentration in the nucleus and a high RanGDP concentration in the cytoplasm, respectively. Import cargoes bind to their receptors in the cytoplasm in the absence of RanGTP. In the nucleus, RanGTP binds to the import complex and triggers the release of the import cargoes. By contrast, export complexes form obligate trimeric complexes that contain receptor, cargo and RanGTP. Export complexes are dissociated in the cytoplasm where they encounter the RanGAP, and the RanGTP is hydrolysed to RanGDP.
The links between the inner nuclear membrane and gene expression emerged as another theme of the meeting owing to talks from several speakers, including R. Goldman (Chicago, IL, USA) and P. Silver (Boston, MA, USA). The lamins that underlie the inner nuclear envelope are important for many cellular events, including DNA replication and transcription (for a review, see Goldman et al, 2002). Goldman reported on several recent studies that link Emery–Dreifuss muscular dystrophy (EDMD) and the premature ageing disease Hutchinson–Gilford progeria syndrome (HGPS) to mutations in lamin A (Eriksson et al, 2003). Fibroblast analysis of EDMD and HGPS patients showed notable alterations in nuclear architecture with respect to the nuclear lamins, the lamina and the nuclear envelope/pore complex. Therefore, it seems that the lamins and their binding partners might act as a complex scaffold for organizing factors that could be responsible for genomic instability and ageing. This functional link lends credence to the idea that the inner nuclear envelope and nuclear pores might be functionally and physically linked to chromatin and, hence, to gene expression. P. Silver's laboratory took this a step further by carrying out a modified genome-wide chromatin immunoprecipitation (ChIP) analysis to examine the interaction of each nucleoporin with specific regions of the genome. Interestingly, they found that distinct subsets of nucleoporins associate with specific regions of the chromatin, which indicates that the nucleus and, in particular, the chromatin are strictly ordered in a eukaryotic cell. For example, they found that a yeast homologue, Mlp1, of the nuclear basket protein Tpr, associates with highly transcribed genes. Indeed, the use of global genomic and proteomic approaches was another exciting underlying theme of the meeting. These approaches should help to unlock the intricacies of the many cellular processes that are linked to nucleocytoplasmic transport.
The Ran cycle and its roles
The Ran GTPase is the driving force for nuclear transport (for a review, see Fried & Kutay, 2003). Whereas the cytoplasmic GTPase-activating protein (RanGAP) maintains a high RanGDP concentration in the cytoplasm, the nuclear guanine-exchange factor (RanGEF) ensures a high RanGTP concentration in the nucleus. The different nucleotide-bound states of Ran regulate cargo binding to a conserved family of transport receptors that are known as importins, exportins or, more generally, karyopherins (Fig 2). Although the general mode of action of Ran in the regulation of cargo/receptor binding has been explained, several intriguing questions remain about Ran function. For example, what happens during mitosis, when the nuclear envelope breaks down and all components can intermingle? Also, are there other transport-independent functions for the components of the Ran cycle? Several researchers approached these questions from different directions and revealed interesting new insights.
M. Dasso (Bethesda, MD, USA) addressed the role of Ran and Ran-binding proteins during mitosis. Both RanGAP and RanBP2, which is a Ran-binding protein that is part of the NPC in interphase and a scaffold for the GAP, bind to kinetochores during mitosis (Joseph et al, 2002). M. Dasso's laboratory asked how these proteins are recruited to kinetochores. They found that two proteins, Hec1 and Nuf2, are required for proper kinetochore localization of RanGAP and RanBP2. Furthermore, using RNAi experiments, they found that RanBP2 is essential for the kinetochore targeting of the mitotic checkpoint proteins Mad1 and Mad2. They suggest a model in which the attachment of microtubules to the kinetochores is unstable in the absence of the RanBP2–RanGAP complex. In another intriguing potential link between Ran and the cell cycle, F. Melchior (Munich, Germany) reported cyclin B/Cdk1-dependent phosphorylation of RanGAP before nuclear envelope breakdown. This phosphorylation persists throughout mitosis. Although functional analysis has not yet revealed a cellular role for this modification, it might clearly regulate the function of the RanGAP.
An area of increasing interest is the direct or indirect involvement of nucleoporins in mitosis. V. Doye (Paris, France) described studies that analysed the role of the Nup107-complex constituents in Schizosaccharomyces pombe. In this yeast, deletion of Nup120 impairs mRNA export and NPC distribution, and also leads to septation defects, chromosome missegregation and abnormal spindles. Whereas in human cells a fraction of the entire Nup107–160 complex, including Nup96 and Sec13, is targeted to kinetochores during mitosis, a similar localization for the homologous proteins in S. pombe could not be demonstrated. In this yeast, however, Nup120 deletion also leads to an altered nuclear accumulation of Ran, which might underlie the reported aberrations in cell-cycle-dependent processes. It will be interesting to determine how these phenotypes can be functionally explained.
Nuclear transport
The separation of the nucleus and the cytoplasm necessitates selective macromolecular transport, which is mediated by the Ran system and the nuclear transport receptors or karyopherins (Fig 2). There are 14 karyopherins in S. cerevisiae and more than 20 in humans (for a review, see Fried & Kutay, 2003). An interesting question that was addressed by several researchers concerned the specificity of these receptor-mediated transport pathways.
Multiple cargoes have been identified for several karyopherins. At the meeting, G. Schlenstedt (Homburg, Germany) reported on the identification of a cargo for Kap120, which was previously the only yeast karyopherin for which no cargo had been described. He showed that Kap120 is the importer for Rpf1—an essential protein that is involved in ribosome biogenesis. Interestingly, two other karyopherins, Nmd5 and Kap114, can participate in the nuclear import of Rpf1, revealing another import pathway by which several nonessential karyopherins cooperate to assure the import of an essential nuclear protein. I. Macara (Charlottesville, VA, USA) provided evidence for another interesting import pathway. His group found that importin 11, which is the human homologue of Kap120, specifically imports ubiquitin-loaded UbcM2, which is a class II ubiquitin E2 ligase. Therefore, in this context, ubiquitin could function as a new type of nuclear localization signal (NLS) that is active only when conjugated to UbcM2. Such a mechanism would ensure that only the ubiquitylated charged version of the ubiquitin ligase would be imported into the nucleus, therefore ensuring the efficient and preferential modification of substrates within the nucleus.
A new export pathway was examined by D. Görlich (Heidelberg, Germany). His group identified exportin 6 (Exp6) as a new karyopherin family member from higher eukaryotes and showed that it mediates the nuclear export of profilin–actin complexes. Profilin therefore functions not only as the nucleotide-exchange factor for actin, but also as a cofactor for actin export. Calculations of the actin concentration in the nucleus resulted in an estimate of approximately 1% of the total nuclear protein concentration. Therefore, the Görlich laboratory postulated that nuclear export of actin is an essential mechanism for cells to prevent toxic concentrations of the protein from accumulating in the nucleus (Stuven et al, 2003).
Two groups reported that exportin 5 (Exp5), which is another member of the karyopherin family, mediates the nuclear export of short microRNA (miRNA) precursors (pre-miRNAs). miRNAs are 22–24-nucleotide-long RNAs that function as regulators of eukaryotic gene expression. They are generated by sequential processing of longer precursor RNAs in the nucleus and the cytoplasm. Using small interfering RNA (siRNA) or oligonucleotides directed against Exp5, B. Cullen's laboratory (Durham, NC, USA) showed that loss of Exp5 function causes the nuclear accumulation of miRNA (Yi et al, 2003). In a collaboration, the groups of E. Lund & J. Dahlberg (Madison, WI, USA) and U. Kutay (Zurich, Switzerland) showed that Exp5 binds directly and specifically to correctly processed pre-miRNAs in a RanGTP-dependent manner (Lund et al, 2004). Therefore, Exp5, in addition to being an export factor, is central to miRNA biogenesis and might help to coordinate nuclear and cytoplasmic processing steps, which again indicates complex regulatory roles for the karyopherins.
mRNA export
Bulk mRNA export is independent of both the Ran system and the karyopherins (for a review, see Fried & Kutay, 2003). Perhaps because of the numerous processing events that must occur before the export of a mature message, mRNA export seems to be intimately linked to transcription, processing and translation. Several talks touched on this linkage of early and late events in mRNA biogenesis (Fig 3). C. N. Cole (Hanover, NH, USA) discussed his recent results on the RNA helicase Dbp5, which could link its previously known function in the late steps of mRNA export to early steps in mRNA biogenesis. He described genetic interactions between DBP5 and factors that are involved in early transcription events, such as the TFIIH complex (Estruch & Cole, 2003).
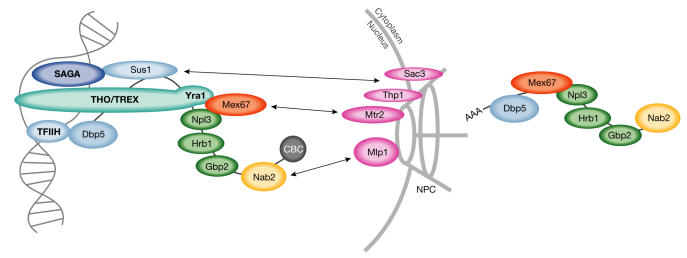
Figure 3.
Recent developments in our understanding of messenger RNA export in Saccharomyces cerevisiae. During transcription, mRNA export factors are recruited to the precursor mRNA. The helicase Dbp5 interacts with components of the transcription factor complex IIH (TFIIH). Sus1 tethers some mRNAs to the nuclear pore complex (NPC) with Sac3 and Thp1 during transcription, through its association with the Spt/Ada/Gcn5/acetyltransferase (SAGA) complex. The transcription and export (TREX) complex consists of Sub2, Yra1 and the THO complex, which functions in transcription and elongation. Yra1 and Npl3 recruit Mex67, which docks the mRNA through Mtr2 to the NPC. Nab2 interacts with Mlp proteins, which act as an mRNA quality control checkpoint before the translocation of the messenger ribonucleoprotein complexes. Apart from Npl3, two novel SR-type mRNA-binding proteins, Gbp2 and Hrb1 are part of the exported mesenger ribonucleoprotein (mRNP) complex. CBC, cap-binding complex.
The transcription and export (TREX) complex is a conserved multisubunit factor containing proteins that are required for mRNA export (Sub2/UAP56 and Yra1/Aly) and transcription elongation (the THO complex). TREX might function to recruit the mRNA-export machinery to the mRNA by a transcription-coupled mechanism. Mutations in TREX-complex components in S. cerevisiae cause pleiotropic phenotypes, such as impaired transcription elongation, mRNA-export defects and increased transcription-associated recombination. A. Aguilera (Seville, Spain) presented data that indicated an important function for Hpr1, which is a component of the THO complex, in preventing the formation of DNA–RNA hybrids during transcriptional elongation (Huertas & Aguilera, 2003). Further compelling evidence for the coupling of early transcriptional events to mRNA export was provided by E. Hurt's laboratory (Heidelberg, Germany). In a synthetic lethal screen with a yra1 mutant, they identified a mutant of sus1. Sus1 is associated with the Spt/Ada/Gcn5/acetyltransferase (SAGA) complex, which functions in the transcriptional regulation of a subset of yeast genes. Sus1 also interacts with two NPC-associated proteins: Thp1 and Sac3. Interestingly, Sus1, which is localized to the nucleus and the nuclear rim, remains in the nuclear interior in sac3-deletion mutants, indicating that Sus1 is involved in tethering some genes to the NPC to specifically enhance the export of their messages (Rodriguez-Navarro et al, 2004).
Eukaryotic cells have developed complex surveillance mechanisms to ensure that only properly processed messages are exported from the nucleus and only correct mRNAs are translated into proteins. The nonsense-mediated decay (NMD) system recognizes and degrades mRNAs that prematurely terminate translation during a 'pioneer' round of translation (Maquat, 2002). R. Reed (Boston, MA, USA) reported on the identification of a homologue of the translation initiation factor eIF4A, which associates with nuclear mRNAs and co-immunoprecipitates with the cap-binding protein CBP80; this factor might therefore participate in the pioneer round of translation and, hence, in NMD. A. Corbett (Atlanta, GA, USA) presented data obtained by her group that implicate two nuclear basket proteins, Mlp1 and Mlp2, which are yeast homologues of human Tpr, in mRNA surveillance. They identified a functional interaction between the Mlp proteins and the mRNA-binding protein Nab2 (Green et al, 2003). Using a LacZ reporter, they went on to show that unprocessed pre-mRNA can exit the nucleus in mlp mutants. In their model, Mlp proteins act as a checkpoint at the NPC by preferentially concentrating export-competent ribonucleoproteins (RNPs) at the pore. A similar model (Galy et al, 2004) was mentioned by U. Nehrbass (Paris, France), who then went on to discuss his work linking the Mlp proteins to chromatin and, possibly, to the control of gene expression. F. Stutz (Geneva, Switzerland) reported on her group's observation that mutations in YRA1 result in decreased mRNA levels. Deletion of Mlp proteins in a yra1 mutant restores mRNA levels, but increases mRNA leakage into the cytoplasm. They propose that Mlp proteins might contribute to mRNP surveillance by retaining improperly assembled mRNP particles at the nuclear periphery. mRNP retention might, in turn, negatively affect mRNA synthesis or stability. In fact, RNAi studies from the Cordes laboratory also revealed that mammalian Tpr is not essential for NLS-mediated import or nuclear export signal (NES)-dependent export. However, it did have an impact on mRNA export, with the loss of Tpr apparently resulting in facilitated export of both intron-less and intron-containing mRNAs. Together, the quality-control mechanisms at the NPC by means of Mlp/Tpr seem to represent a new checkpoint along the mRNA journey from biogenesis to translation.
Many laboratories presented evidence for the intimate linkage between events that occur before mRNA export from the nucleus, but there was also evidence for crosstalk between later events: mRNA export and translation. H. Krebber (Marburg, Germany) described her finding that significant amounts of the serine/arginine (SR)-type shuttling mRNA-binding proteins, Npl3, Gbp2 and Hrb1 (Hacker & Krebber, 2004), are associated with ribosomes during translation and might deliver the exported mRNAs to the translation machinery. P. Prekker from C. Guthrie's laboratory (San Francisco, CA, USA) discussed the antagonistic phosphatase for the SRspecific protein kinase Sky1, which the Guthrie laboratory identified as being protein phosphatase 1-type, termed Glc7. Their data indicate a model in which Glc7 dephosphorylates RNA-bound Npl3 in the nucleus, which subsequently allows Mex67 to bind Npl3 and promote the export of the mRNP to the cytoplasm (Gilbert & Guthrie, 2004). Another link between nuclear export and translation was presented by M.-L. Hammarskjold (Charlottesville, VA, USA). She showed that the heterotrimeric mRNA-export receptor composed of the TAP and p15/NXT proteins enhances the translation of proteins from the exported constitutive-transport element (CTE) RNA. Pulse-chase experiments showed that TAP and NXT significantly increase the rate of protein synthesis. Furthermore, TAP, but not NXT, is detected in polyribosomes, which indicates a role for these mRNA-export factors in translation (Jin et al, 2003).
In summary, the expanding nuclear transport field is bringing together topics and expertise from a wide range of research areas. This multidisciplinary approach is contributing to an enhanced understanding of a range of crucial cellular processes. It is also providing insight into how transport into and out of the nucleus contributes to the regulation of numerous cellular functions, including gene expression, protein translation, cell division and nuclear dynamics. The meeting in Sicily left its participants confident that the field of nucleocytoplasmic transport will continue to expand and be a source of exciting ideas that will help to “read the instruction manual for the cell”.

Acknowledgments
The meeting was sponsored by EMBO and Merck Biosciences. We thank the speakers for agreeing to the included citations. Special thanks go to E. Hurt and V. Doye for organization of the meeting. There were, of course, many other exciting talks that are not mentioned here owing to space limitations, and we sincerely apologize to our colleagues whose work was not mentioned. A.H.C. is supported by grants from the National Institutes of Health and H.K. is supported by the Deutsche Forschungsgemeinschaft (SFB593).
References
- Eriksson M et al. (2003) Recurrent de novo point mutations in lamin A cause Hutchinson–Gilford progeria syndrome. Nature 423: 293–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estruch F, Cole CN (2003) An early function during transcription for the yeast mRNA export factor Dbp5p/Rat8p suggested by its genetic and physical interactions with transcription factor IIH components. Mol Biol Cell 14: 1664–1676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrenkrog B, Aebi U (2003) The nuclear pore complex: nucleocytoplasmic transport and beyond. Nat Rev Mol Cell Biol 4: 757–766 [DOI] [PubMed] [Google Scholar]
- Fried H, Kutay U (2003) Nucleocytoplasmic transport: taking an inventory. Cell Mol Life Sci 60: 1659–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U (2004) Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell 116: 63–73 [DOI] [PubMed] [Google Scholar]
- Gilbert W, Guthrie C (2004) The glc7p nuclear phosphatase promotes mRNA export by facilitating association of mex67p with mRNA. Mol Cell 13: 201–212 [DOI] [PubMed] [Google Scholar]
- Goldman RD, Gruenbaum Y, Moir RD, Shumaker DK, Spann TP (2002) Nuclear lamins: building blocks of nuclear architecture. Genes Dev 16: 533–547 [DOI] [PubMed] [Google Scholar]
- Green DM, Johnson CP, Hagan H, Corbett AH (2003) The C-terminal domain of myosin-like protein 1 (Mlp1p) is a docking site for heterogeneous nuclear ribonucleoproteins that are required for mRNA export. Proc Natl Acad Sci USA 100: 1010–1015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacker S, Krebber H (2004) Differential export requirements for shuttling SR-type mRNA binding proteins. J Biol Chem 279: 5049–5052 [DOI] [PubMed] [Google Scholar]
- Huertas P, Aguilera A (2003) Cotranscriptionally formed DNA:RNA hybrids mediate transcription elongation impairment and transcription-associated recombination. Mol Cell 12: 711–721 [DOI] [PubMed] [Google Scholar]
- Jin L, Guzik BW, Bor YC, Rekosh D, Hammarskjold ML (2003) Tap and NXT promote translation of unspliced mRNA. Genes Dev 17: 3075–3086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph J, Tan SH, Karpova TS, McNally JG, Dasso M (2002) SUMO-1 targets RanGAP1 to kinetochores and mitotic spindles. J Cell Biol 156: 595–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Prunuske AJ, Fager AM, Ullman KS (2003) The COPI complex functions in nuclear envelope breakdown and is recruited by the nucleoporin Nup153. Dev Cell 5: 487–498 [DOI] [PubMed] [Google Scholar]
- Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U (2004) Nuclear export of microRNA precursors. Science 303: 95–98 [DOI] [PubMed] [Google Scholar]
- Maquat LE (2002) Nonsense-mediated mRNA decay. Curr Biol 12: R196–R197 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro S, Fischer T, Luo MJ, Antunez O, Brettschneider S, Lechner J, Perez-Ortin JE, Reed R, Hurt E (2004) Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell 116: 75–86 [DOI] [PubMed] [Google Scholar]
- Stuven T, Hartmann E, Gorlich D (2003) Exportin 6: a novel nuclear export receptor that is specific for profilin–actin complexes. EMBO J 22: 5928–5940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Qin Y, Macara IG, Cullen BR (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev 17: 3011–3016 [DOI] [PMC free article] [PubMed] [Google Scholar]