Abstract
The aspartyl protease BACE1 has a pivotal role in the pathogenesis of Alzheimer's disease. Recently, it was shown that in Alzheimer's disease patients, BACE1 levels were elevated although mRNA levels were not changed compared with controls. Here, we demonstrate that the 5′-untranslated region (5′UTR) of BACE1 controls the rate of BACE1 translation. In the presence of the 5′UTR, we observed more than 90% reduction of BACE1 protein levels in HEK293, COS7 and H4 cells, and a similar reduction of BACE1 activity in vitro. mRNA levels were not affected, demonstrating that the 5′UTR repressed the translation but not the transcription of BACE1. The 3′UTR did not affect BACE1 expression. An extensive mutagenesis analysis predicts that the GC-rich region of the 5′UTR forms a constitutive translation barrier, which may prevent the ribosome from efficiently translating the BACE1 mRNA. Our data therefore demonstrate translational repression as a new mechanism controlling BACE1 expression.
Keywords: Alzheimer's disease, neurodegeneration, BACE1, secretases
Introduction
Generation and aggregation of the amyloid β-peptide (Aβ) are assumed to be the first steps in the pathogenesis of Alzheimer's disease (AD) (Selkoe, 2001). Through the action of two protease activities referred to as β- and γsecretase, Aβ is proteolytically generated from the amyloid precursor protein (APP), a type I membrane protein. β-Secretase has been identified as the aspartyl protease BACE1 (Vassar, 2002), whereas γsecretase is a protein complex consisting of presenilin, nicastrin, aph-1 and pen-2 (Haass, 2004). BACE1 cleaves APP at the amino terminus of the Aβ domain and therefore catalyses the rate-limiting step in Aβ generation.
BACE1 is an N-glycosylated, palmitoylated type I membrane protein with a prodomain, which is cleaved by a furin-like protease upon maturation in the secretory pathway (for a review, see Vassar, 2002; Haass, 2004). BACE1 is mainly localized to endosomes, the trans-Golgi network and the plasma membrane, and has been shown to be strictly required for Aβ generation. Mice with a targeted deletion of BACE1 do not produce any Aβ (Vassar, 2002). As such mice do not show any overt phenotype, BACE1 is an ideal drug target. The biological function of BACE1 is not understood. Clearly, BACE1 not only cleaves APP but is also involved in the proteolytic processing of the Pselectin glycoprotein ligand-1 (Lichtenthaler et al, 2003) and the sialyl-transferase ST6Gal I (Kitazume et al, 2001, 2003).
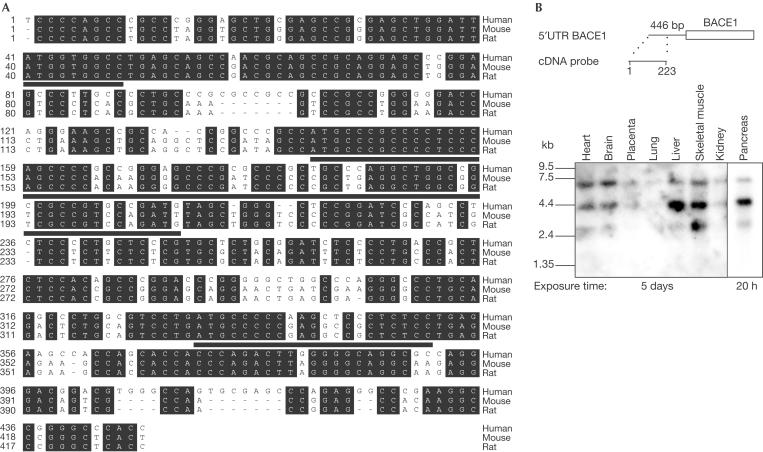
BACE1 is expressed mainly in the brain and pancreas, and at lower levels in most other tissues (Vassar et al, 1999; Yan et al, 1999). Three recent studies showed that BACE1 protein levels were significantly upregulated up to 2.7-fold in the brains of sporadic AD patients compared with non-AD controls (Fukumoto et al, 2002; Holsinger et al, 2002; Yang et al, 2003), whereas mRNA levels were unchanged (Yasojima et al, 2001; Holsinger et al, 2002; Preece et al, 2003). This indicates that post-transcriptional mechanisms, such as translational control or altered BACE1 protein degradation, regulate BACE1 protein levels. Therefore, we analysed the effect of the 5′-untranslated region (5′UTR) of BACE1 on protein expression. In most vertebrate genes, the 5′UTR is 10–200 nucleotides long, is unstructured, is not very GC-rich and does not contain upstream open reading frames (uORFs). In contrast, fewer than 10% of vertebrate genes, in particular regulatory proteins such as proto-oncogenes, have 5′UTRs longer than 200 nucleotides, which are often GC-rich (70–90%) being indicative of a high degree of secondary structure (Kozak, 1987; Willis, 1999). Additionally, these long 5′UTRs may contain uORFs (for a review, see Clemens & Bommer, 1999). In most cases, the GC-richness and uORFs, especially those in a good context for initiation of translation, are assumed to be involved in inhibition of the translation of the main open reading frame (ORF) (Kozak, 1987; Willis, 1999). The 5′UTR of BACE1 belongs to the class of the long 5′UTRs similar to many regulatory genes. It is 446 nucleotides long, has a GC content of 77% and contains three uORFs (Fig 1A). Furthermore, the 5′UTR is highly conserved in humans, mice and rats (Fig 1A). As the BACE1 5′UTR may thus fulfil the requirements for affecting efficient translation, we investigated whether BACE1 expression is translationally controlled by its 5′UTR.
Figure 1.
The 5′UTR of BACE1 is conserved in different vertebrates and is ubiquitously expressed. (A) The sequence identity between the human and the mouse or the rat sequence is 72.1 and 72.7%, respectively. Residues shown on a black background are conserved among humans, mice and rats. The conserved three uORFs are underlined. (B) A human adult multiple tissue northern blot (Clontech) was hybridized with a human cDNA probe against the first 223 nucleotides of the 5′UTR of BACE1 schematically shown above.
Results And Discussion
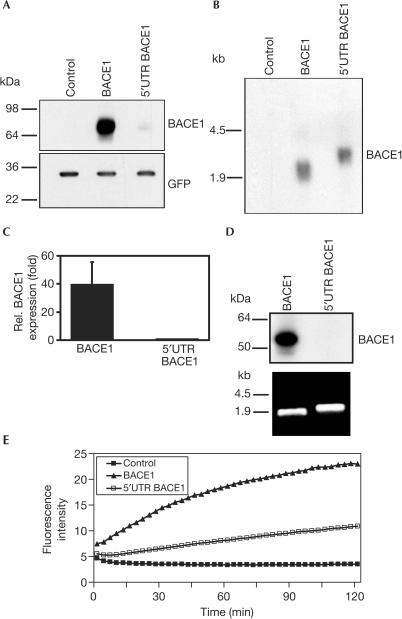
We isolated the BACE1 cDNA containing the 5′UTR from an adult human brain cDNA library and confirmed that it is composed of 446 nucleotides (Fig 1A). To confirm that a significant fraction of the native BACE1 mRNA contains the complete 5′UTR, we performed multiple tissue northern blot analysis using the first 223 nucleotides of the 5′UTR as probe. Indeed this probe detects robust levels of all three previously identified transcripts of 7.0, 4.4 and 2.6 kilobases (kb) (Fig 1B) (Vassar et al, 1999). To determine whether the 5′UTR may affect the expression level of BACE1, expression vectors encoding the ORF of BACE1 with or without the 5′UTR were transiently transfected into human embryonic kidney HEK293 cells. Detection of BACE1 in the cell lysate by immunoblotting revealed that the presence of the 5′UTR strongly reduced BACE1 protein levels (Fig 2A). In contrast, the 3′UTR did not affect BACE1 protein levels (Fig 2A). A similar reduction of BACE1 expression due to the presence of the 5′UTR was observed in African green monkey COS7 cells and human neuroglioma H4 cells (Fig 2B), demonstrating that this effect was not cell-type specific. Moreover, the effect of the 5′UTR did not depend on the promoter used (CMV and EF1α; data not shown). The poor expression of BACE1 in the presence of its 5′UTR may be an explanation of why it is difficult to detect endogenous BACE1 protein levels in many cell lines and a variety of tissues (Basi et al, 2003).
Figure 2.
The 5′UTR of BACE1 is responsible for reduced BACE1 expression. (A) HEK293 cells were transiently transfected with BACE1, or BACE1 constructs containing the 5′UTR or 3′UTR of BACE1 as shown. Protein levels were corrected for secretory alkaline phosphatase activity. (B) Reduction of BACE1 expression is not cell-type specific. H4 and COS7 cells were transfected with BACE1 or 5′UTR BACE1 and analysed by western blotting (upper panels). GFP was used as a loading control (lower panels). (C) Influence of the BACE1 5′UTR on the expression of a chimeric luciferase (Luc) construct. HEK293 cells were transfected with luciferase containing the 5′UTR of BACE1 or with luciferase as control. The signal for luciferase without 5′UTR was set to 100%. Luciferase activity was corrected for secretory alkaline phosphatase activity. Results were expressed as the mean and standard deviation of three independent experiments made in duplicate.
We next investigated whether the BACE1 5′UTR could inhibit the expression of a heterologous downstream ORF different from BACE1. For this purpose, vectors encoding luciferase with or without the BACE1 5′UTR or empty control vector were expressed in HEK293 cells and the luciferase activity was measured in cell lysates. A strong reduction in luciferase activity was observed in cells transfected with the construct containing the 5′UTR of BACE1 compared with cells transfected with luciferase control vector (Fig 2C).
To prove that the 5′UTR lowered BACE1 protein levels by selectively reducing the translation of BACE1, vectors encoding BACE1 with or without the 5′UTR or empty control vector were expressed in HEK293 cells stably expressing wild-type APP695 (HEK293-APP) (Haass et al, 1992). BACE1 protein levels in cell lysates were determined by immunoblotting and mRNA levels by northern blotting (Fig 3A,B). The presence of the 5′UTR had no significant effect on mRNA levels (Fig 3B), ruling out the possibility that the 5′UTR altered the transcription of BACE1. In contrast, the presence of the 5′UTR lowered BACE1 protein levels ∼40-fold (Fig 3C), demonstrating that the 5′UTR represses the expression of BACE1 at the translational level. To prove this further, equal amounts of in vitro-transcribed BACE1 mRNA with and without the 5′UTR (Fig 3D, lower panel) were translated in a nuclease-treated rabbit reticulocyte lysate. BACE1 protein was detected when the BACE1 mRNA without the 5′UTR was added to the translation mix. In contrast, no BACE1 protein was observed when the BACE1 mRNA including the 5′UTR was used for in vitro translation (Fig 3D, upper panel). This result clearly demonstrates that the 5′UTR of BACE1 is responsible for translational repression of BACE1. In agreement with this finding, membranes of HEK293-APP cells transiently transfected with BACE1 containing the 5′UTR showed a reduced BACE1 activity in an in vitro assay compared with cells expressing BACE1 without the 5′UTR (Fig 3E).
Figure 3.
BACE1 expression is regulated by translational control mechanisms. (A) HEK293-APP cells were transfected with BACE1, 5′UTR BACE1 or empty vector and GFP as transfection control. Representative western blots of BACE1 (upper panel) and GFP expression (lower panel) are shown. (B) Northern blot analysis of cells shown in (A). Blots were probed for BACE1 mRNA. (C) Quantification of BACE1 expression. The signal for 5′UTR BACE1 was set to one. Results are expressed as the mean and standard deviation of at least three different experiments. (D) In vitro translation of in vitro-transcribed mRNAs with and without the 5′UTR of BACE1 (upper panel). For translation, 4.2 pmol of the in vitro-transcribed mRNAs was used (lower panel). (E) Membranes from BACE1-transfected (filled triangles), 5′UTR BACE1-transfected (open squares) and untransfected (filled squares) HEK293-APP cells were solubilized and analysed in vitro for proteolytic activity.
The translation-inhibitory effect may be mediated by three short uORFs in the BACE1 5′UTR (Fig 1A), encoding putative peptides of 3 residues (uORF1), 23 residues (uORF2) or 7 residues (uORF3). From these uORFs, uORF1 and uORF2 have a fairly good context for translation initiation with a purine at position −3 and in the case of uORF1 with a purine at position +4. However, the authentic start codon of BACE1 occurs in an excellent context for initiation. All three uORFs are highly conserved in the mouse and rat orthologues (Fig 1A), suggesting a biological function. The role of uORFs in translational control is not fully understood. They are assumed to cause the ribosome to stall during its scanning for the right start codon and may or may not be translated into short peptides (Morris & Geballe, 2000). For example, the uORFs in the 5′UTR of huntingtin and embryonic proinsulin (Lee et al, 2002; Hernandezsanchez et al, 2003) inhibit the translation of the downstream cistrons. However, in other cases, such as the platelet-derived growth factor (Horvath et al, 1995) or the NR2A subunit of the N-methyl-D-aspartate receptor (Wood et al, 1996), the uORFs within the 5′UTR were not required for the translational repression. Instead, owing to the high GC content, the 5′UTR was assumed to form a stable mRNA stem–loop secondary structure preventing an efficient scanning of the ribosome (Wood et al, 1996; Clemens & Bommer, 1999).
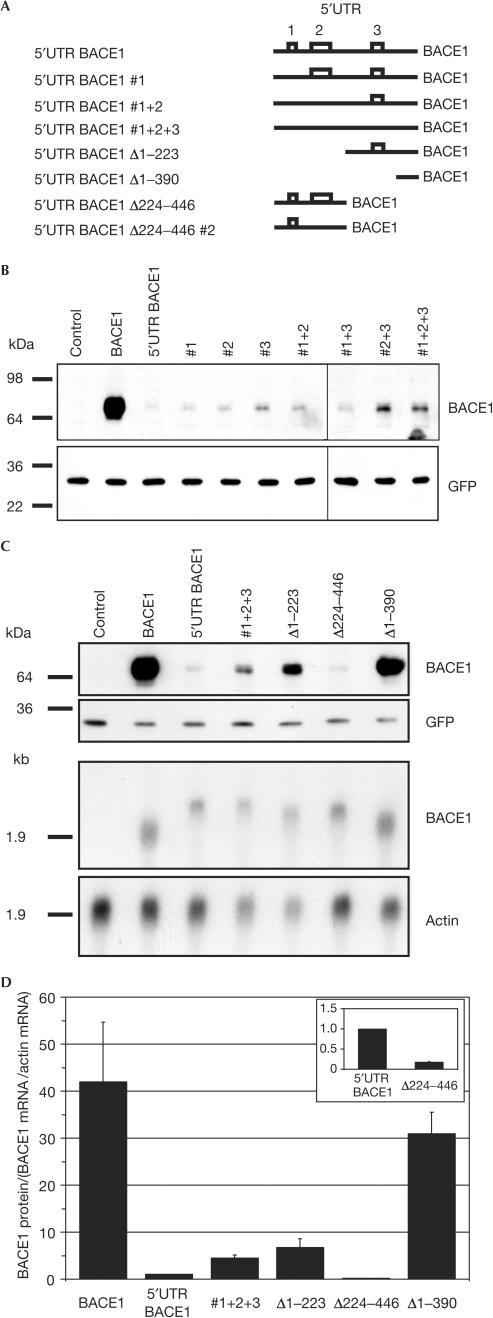
To determine whether the high GC content of the long 5′UTR is sufficient for repressing BACE1 expression or whether the uORFs and their encoded putative short peptides are required, the start codon of the three uORFs was mutated from ATG to ATA (Fig 4A). The mutated BACE1 plasmids were transfected into HEK293-APP cells and BACE1 protein levels were determined in the cell lysate by immunoblotting. Mutation of uORF1 (#1) had no significant effect on BACE1 expression, whereas mutation of uORF2 (#2) and uORF3 (#3) very slightly increased the expression of BACE1 (Fig 4B). Combined mutations of the upstream ATGs, for example #1+3, #2+3 or #1+2+3, showed a slight but significant increase of BACE1 expression compared with the single mutations (Fig 4B). These experiments reveal that the uORFs account only to a minor extent for the repressed BACE1 expression mediated by its 5′UTR. This suggests instead that the GC-rich BACE1 5′UTR forms a tightly folded secondary structure, which may be the major element determining the repression of BACE1 expression. Indeed, an mRNA structure prediction for the BACE1 5′UTR suggests a stable secondary structure with several stem–loops. Its free energy of −215.3 kcal/mol (as calculated by the MFOLD program) indicates a very stable structure, as a free energy of −30 to −70 kcal/mol is known to be sufficient for inhibiting translation in vitro (Gray & Hentze, 1994; Willis, 1999). Therefore, we investigated the effect of several deletion mutants of the 5′UTR of BACE1 (Fig 4A,C,D). The deletion mutant Δ1–223, lacking the first 223 nucleotides of the 5′UTR, slightly increased BACE1 expression compared with the full-length 5′UTR BACE1 (Fig 4C,D), whereas the deletion construct Δ224–446 even showed a slightly stronger translational inhibition than the full-length 5′UTR (Fig 4C and inset of Fig 4D). To address whether this inhibition is due to the presence of uORF2 in this construct, we mutated the potential start codon of uORF2 to ATA (Fig 4A). However, protein and mRNA levels did not significantly alter from construct Δ224–446 (data not shown). This shows that both the 5′- and the 3′-half of the UTR have a strong inhibitory effect, with the effect being more pronounced for the 5′-half. Thus, no single motif within either half is uniquely responsible for inhibiting BACE1 translation. To prove this assumption further, we analysed an additional deletion construct Δ1–390, lacking 87% of the 5′UTR, including the highly GC-rich parts of the 5′UTR. Transient expression of this construct led to similar levels of BACE1 protein compared with the construct without 5′UTR (Fig 4C). Furthermore, northern blot analysis showed that similar mRNA levels were detected for the ATG mutation construct #1+2+3 and all deletion constructs (Fig 4C), ruling out any effects on transcription or mRNA stability. Taken together, our data thus demonstrate translational repression as a new mechanism controlling BACE1 expression.
Figure 4.
Effect of uORF mutations and deletion constructs on the expression levels of BACE1. (A) Schematic representation of the analysed constructs. Open boxes represent the three uORFs in the 5′UTR, which are indicated by numbers. Mutation of the potential start codons from ATG to ATA are indicated by #, whereas deletion constructs are indicated by Δ. (B) HEK293-APP cells were transiently transfected with BACE1 plasmids bearing the indicated uORF mutations, or (C) deletion mutants. Western blots were probed for BACE1 and GFP (upper panels). Representative northern blots (lower panels) showed similar mRNA levels for all analysed constructs compared with actin levels. (D) Quantification of the results in (C) from two independent experiments. BACE1 protein levels were corrected for BACE1 and actin mRNA levels. The columns show the changes in BACE1 translation for the indicated constructs relative to the BACE1 construct containing the full-length, wild-type 5′UTR (set to one). The inset shows an enlargement of the effect of the Δ224–446 mutant compared with the 5′UTR BACE1 construct.
Methods
Antibodies
The monoclonal anti-green fluorescent protein (GFP) antibody was obtained from Clontech. Anti-BACE-1 (EE-17) was purchased from Sigma.
Plasmid construction
Two cDNA clones of BACE1 in the peak8 vector were obtained from an adult human brain cDNA library in an expression cloning screen for genes stimulating the ectodomain shedding of APP (S.F. Lichtenthaler, unpublished data). Both BACE1 cDNAs contained the published 446 nucleotides of the 5′UTR (GenBank accession numbers NM_012104 and AF201468), the BACE1 ORF of 1503 nucleotides, but differed in the length of their 3′UTR (1759 and 551 nucleotides, respectively). BACE1 without 5′UTR was cloned into pcDNA3.1Zeo(+) (Invitrogen) using EcoRI and XhoI maintaining the natural Kozak sequence of BACE1 (nucleotides 438–446, according to the numbering in Fig 1A). The 5′UTR of BACE1 was introduced after PCR using the HindIII and EcoRI restriction sites. All deletion constructs and constructs with mutated upstream ATGs (ATG to ATA indicated by #) were generated by PCR. Furthermore, BACE1 with and without the 5′UTR or the 3′UTR were cloned into peak8 vector using appropriate restriction enzymes. Peak12/5′UTR-luciferase was obtained by cloning the 5′UTR of BACE1 into peak12/luciferase (Lichtenthaler et al, 2003). All constructs were verified by sequencing.
Cell culture and transfections
Human embryonic kidney 293 (HEK293) cells, African green monkey COS7 cells and human neuroglioma H4 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum, 1% penicillin/streptomycin and 2 mM glutamine. Transfections were carried out using LipofectAMINE 2000 (Invitrogen) according to the supplier's instructions.
Analysis of BACE1 expression
In a poly-L-lysine-coated 6-cm dish 2.5 × 106 HEK293-APP cells were plated and transiently transfected with 0.1 μg cDNA encoding BACE1 or any BACE1 variant and 0.1 μg pEGFP-N1 (Clontech) for a transfection control. At 24 h after transfection, cells were incubated for additional 16 h in 4 ml DMEM. Cell lysates were prepared and analysed for BACE1 by immunoblotting with anti-BACE-1 and for GFP with anti-GFP antibody. Detection was performed using enhanced chemiluminescence (ECL) or ECL Plus (Amersham Biosciences). Quantification was performed using the digital camera system FluorChem 8900 (Alpha Innotech) and AlphaEase FC software.
Alternatively, HEK293 cells were seeded in 24-well plates and transfected with 10 ng of peak8 vector encoding BACE1, 5′UTR-BACE1 or 3′UTR-BACE1 and secretory alkaline phosphatase (SEAP) as a transfection efficiency control. Cell lysates were prepared; protein levels were normalized for SEAP activity and analysed for BACE1 expression as described above.
Luciferase-reporter gene assay
For luciferase activity, HEK293 cells were seeded in 24-well plates and transfected as described above with peak12/5′UTR luciferase or peak12/luciferase and peak12/HA-SEAP. The medium was replaced after 24 h. Cell lysates were prepared with 25 mM glycyl-glycine, 25 mM MgSO4, 10 mM dithiothreitol (DTT) and 0.2% NP-40. A 10 μl portion of cell lysates was used for luciferase activity measurements in 100 μl 25 mM glycyl-glycine, 25 mM MgSO4, 10 mM DTT, 1 mM ATP and 35 μM D-luciferin. Quantification was performed using the luminometer 1420 Victor (Wallac). Luciferase activity was normalized for SEAP activity.
Northern blot analysis
At 48 h after transfection of HEK293-APP cells with 0.1 μg cDNA encoding BACE1, 5′UTR-BACE1 or empty vector total RNAs were extracted using the peqGOLD RNAPure kit (PeqLab, Erlangen, Germany). Total RNAs (10 μg) were separated on a 1.2% agarose gel in the presence of 6.5% formaldehyde, 20 mM MOPS (pH 7.0), 2 mM sodium acetate and 1 mM ethylenediaminetetraacetic acid (EDTA) and blotted onto a HyBond N membrane (Amersham Biosciences) by capillary transfer overnight. Subsequently, the blot was baked for 1 h at 80°C and UV crosslinked. After 2 h prehybridization at 65°C in 250 mM Na2HPO4, 250 mM NaH2PO4, 1 mM EDTA, 1% bovine serum albumin and 7% sodium dodecylsulphate (SDS), a 32P-dCTP (Amersham Biosciences)-labelled BACE1 or actin probe (Random Primers DNA Labeling System, Invitrogen) was added and hybridization was performed overnight at 65°C. Excess probe was removed by washing with 2 × SSC, 0.1% SDS at 25°C followed by washing with 0.1 × SSC, 0.1% SDS at 65°C. Signal was detected by exposure of the blot to Super RX film (Fuji) and quantified by using a PhosphorImager (Molecular Dynamics). Multiple tissue northern blot analysis was performed according to the supplier's protocol (Clontech).
In vitro translation
Capped mRNAs were generated using the mMessage mMachine kit (Ambion) and quantified by UV absorption at 260 nm. The size and integrity of transcripts were assessed by electrophoresis. In vitro translation reactions were performed with nuclease-treated rabbit reticulocyte lysate (Promega) according to the supplier's instructions.
In vitro assay for BACE1
Membranes from HEK293-APP cells transiently transfected with BACE1 were extracted according to a previously established protocol (Steiner et al, 2002) using 1% Triton X-100 and STE buffer instead of n-dodecyl-D-maltoside. The fluorimetric BACE1 activity assay was carried out as described previously (Capell et al, 2002).
Acknowledgments
We are grateful to the following institutions for financial support: the Fonds der Chemischen Industrie to S.F.L. and C.H., and the Deutsche Forschungsgemeinschaft through SFB 596 project A9 to C.H. and project B12 to S.F.L. and for a fellowship to S.S. through Graduiertenkolleg 688.
References
- Basi G et al. (2003) Antagonistic effects of βsite amyloid precursor protein-cleaving enzymes 1 and 2 on β-amyloid peptide production in cells. J Biol Chem 278: 31512–31520 [DOI] [PubMed] [Google Scholar]
- Capell A, Meyn L, Fluhrer R, Teplow DB, Walter J, Haass C (2002) Apical sorting of βsecretase limits amyloid β-peptide production. J Biol Chem 277: 5637–5643 [DOI] [PubMed] [Google Scholar]
- Clemens MJ, Bommer UA (1999) Translational control: the cancer connection. Int J Biochem Cell Biol 31: 1–23 [DOI] [PubMed] [Google Scholar]
- Fukumoto H, Cheung BS, Hyman BT, Irizarry MC (2002) βsecretase protein and activity are increased in the neocortex in Alzheimer disease. Arch Neurol 59: 1381–1389 [DOI] [PubMed] [Google Scholar]
- Gray NK, Hentze MW (1994) Regulation of protein synthesis by mRNA structure. Mol Biol Rep 19: 195–200 [DOI] [PubMed] [Google Scholar]
- Haass C (2004) Take five-BACE and the γsecretase quartet conduct Alzheimer's amyloid β-peptide generation. EMBO J 23: 483–488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C et al. (1992) Amyloid β-peptide is produced by cultured cells during normal metabolism. Nature 359: 322–325 [DOI] [PubMed] [Google Scholar]
- Hernandezsanchez C, Mansilla A, de la Rosa EJ, Pollerberg GE, Martinez-Salas E, de Pablo F (2003) Upstream AUGs in embryonic proinsulin mRNA control its low translation level. EMBO J 22: 5582–5592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holsinger RM, McLean CA, Beyreuther K, Masters CL, Evin G (2002) Increased expression of the amyloid precursor βsecretase in Alzheimer's disease. Ann Neurol 51: 783–786 [DOI] [PubMed] [Google Scholar]
- Horvath P, Suganuma A, Inaba M, Pan YB, Gupta KC (1995) Multiple elements in the 5′ untranslated region down-regulate csis messenger RNA translation. Cell Growth Differ 6: 1103–1110 [PubMed] [Google Scholar]
- Kitazume S, Tachida Y, Oka R, Shirotani K, Saido TC, Hashimoto Y (2001) Alzheimer's βsecretase, β-site amyloid precursor protein-cleaving enzyme, is responsible for cleavage secretion of a Golgi-resident sialyltransferase. Proc Natl Acad Sci USA 98: 13554–13559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazume S et al. (2003) Characterization of α 2,6sialyltransferase cleavage by Alzheimer's β-secretase (BACE1). J Biol Chem 278: 14865–14871 [DOI] [PubMed] [Google Scholar]
- Kozak M (1987) An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res 15: 8125–8148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Park EH, Couture G, Harvey I, Garneau P, Pelletier J (2002) An upstream open reading frame impedes translation of the huntingtin gene. Nucleic Acids Res 30: 5110–5119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler SF et al. (2003) The cell adhesion protein Pselectin glycoprotein ligand-1 is a substrate for the aspartyl protease BACE1. J Biol Chem 278: 48713–48719 [DOI] [PubMed] [Google Scholar]
- Morris DR, Geballe AP (2000) Upstream open reading frames as regulators of mRNA translation. Mol Cell Biol 20: 8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preece P et al. (2003) βsecretase (BACE) and GSK-3 mRNA levels in Alzheimer's disease. Brain Res Mol Brain Res 116: 155–158 [DOI] [PubMed] [Google Scholar]
- Selkoe DJ (2001) Alzheimer's disease: genes, proteins, and therapy. Physiol Rev 81: 741–766 [DOI] [PubMed] [Google Scholar]
- Steiner H et al. (2002) PEN-2 is an integral component of the γsecretase complex required for coordinated expression of presenilin and nicastrin. J Biol Chem 277: 39062–39065 [DOI] [PubMed] [Google Scholar]
- Vassar R (2002) βsecretase (BACE) as a drug target for Alzheimer's disease. Adv Drug Deliv Rev 54: 1589–1602 [DOI] [PubMed] [Google Scholar]
- Vassar R et al. (1999) βsecretase cleavage of Alzheimer's amyloid precursor protein by the transmembrane aspartic protease BACE. Science 286: 735–741 [DOI] [PubMed] [Google Scholar]
- Willis AE (1999) Translational control of growth factor and proto-oncogene expression. Int J Biochem Cell Biol 31: 73–86 [DOI] [PubMed] [Google Scholar]
- Wood MW, VanDongen HM, VanDongen AM (1996) The 5′-untranslated region of the N-methyl-D-aspartate receptor NR2A subunit controls efficiency of translation. J Biol Chem 271: 8115–8120 [DOI] [PubMed] [Google Scholar]
- Yan R et al. (1999) Membrane-anchored aspartyl protease with Alzheimer's disease βsecretase activity. Nature 402: 533–537 [DOI] [PubMed] [Google Scholar]
- Yang LB et al. (2003) Elevated βsecretase expression and enzymatic activity detected in sporadic Alzheimer disease. Nat Med 9: 3–4 [DOI] [PubMed] [Google Scholar]
- Yasojima K, McGeer EG, McGeer PL (2001) Relationship between β amyloid peptide generating molecules and neprilysin in Alzheimer disease and normal brain. Brain Res 919: 115–121 [DOI] [PubMed] [Google Scholar]