Abstract
DnaJ is a molecular chaperone and the prototypical member of the J-protein family. J proteins are defined by the presence of a J domain that can regulate the activity of 70-kDa heat-shock proteins. Sequence analysis on the genome of Saccharomyces cerevisiae has revealed 22 proteins that establish four distinguishing structural features of the J domain: predicted helicity in segments I–IV, precisely placed interhelical contact residues, a lysine-rich surface on helix II and placement of the diagnostic sequence HPD between the predicted helices II and III. We suggest that this definition of the J-protein family could be used for other genome-wide studies. In addition, three J-like proteins were identified in yeast that contain regions closely resembling a J domain, but in which the HPD motif is non-conservatively replaced. We suggest that J-like proteins might function to regulate the activity of bona fide J proteins during protein translocation, assembly and disassembly.
Keywords: Hsp40, J domain, molecular chaperones, protein folding, protein import, protein translocation
Introduction
Molecular chaperones enforce and maintain the native structures of other proteins. Two prototypical molecular chaperones are DnaK and DnaJ from the cytoplasm of Escherichia coli. DnaK is a member of the 70-kDa heat-shock protein (Hsp70) family. Other family members are found ubiquitously in cells from all kingdoms of life and are located within most subcellular compartments (Frydman, 2001). DnaJ is a member of the Hsp40 family of molecular chaperones, which is also called the J-protein family, the members of which regulate the activity of Hsp70s.
Hsp70s bind to short unfolded hydrophobic regions of substrate polypeptides; in the case of nascent polypeptide chains, this binding can then promote their assembly into higher order structures (Bukau & Horwich, 1998). Hsp70s are also involved in the disassembly of protein structures, such as the removal of clathrin coats from vesicles (Lemmon, 2001), and the translocation of proteins across membranes (Jensen & Johnson, 1999). Members of the J-protein family function as coupling factors to stimulate ATP hydrolysis by a partner Hsp70 when a polypeptide is present in its substrate-binding pocket.
A sequence-based classification system has been established whereby true homologues of DnaJ are referred to as type I J proteins (Cheetham & Caplan, 1998). Type I J proteins have all three of the structurally defined domains that are found in DnaJ: a compact helical J domain that is linked by a glycine-rich region to a zinc-finger domain followed by a carboxy-terminal domain. There is good evidence to indicate that the zinc-finger domain and the carboxy-terminal domain can interact with non-native substrates. This sequence-based classification system distinguishes type II J proteins as those that have a J domain linked by a glycine-rich region to a carboxy-terminal domain. Notably, type III J proteins have a J domain but lack the other sequence features that are found in type I and II members of the family.
A combination of biochemistry and sequence analysis has identified a growing number of proteins that might be related to DnaJ in the Archaea, and in other bacterial species and eukaryotic organisms. Unfortunately, many of the sequences that are annotated in genome databases as 'DnaJ homologue', 'Hsp40' or 'J protein' have been assigned this classification on the basis of limited sequence similarity with ill-defined stretches of DnaJ from E. coli, without consideration of the essential structural features of J proteins. For example, in the Plasmodium falciparum data set, 14 protein entries are annotated as DnaJ related, only 10 of which seem to carry J domains. Here, we suggest a stricter definition of the J domain, which could be used to better characterize J proteins from sequence data. Furthermore, the first comprehensive analysis of the yeast genome shows that the total number of J proteins encoded in a genome can be reliably defined, and that some subcellular compartments are rich in J proteins to diversify the functions of a given Hsp70.
The J domain as a signature motif for the family
The J domain of DnaJ from E. coli and the homologous human protein HDJ1 have a conserved structure (Szyperski et al, 1994; Huang et al, 1999). The domain consists of four helices, the second of which has a charged surface that includes at least one pair of basic residues that are essential for interaction with the ATPase domain of Hsp70 (Genevaux et al, 2002). Further residues on the same face of this helix show backbone amide-proton chemical shifts in the presence of Hsp70, which indicates that they form an entire surface that is occluded in the transient DnaJ–Hsp70 complex (Greene et al, 1998). Flexibility in the isolated J domain is constrained on docking with the surface of the Hsp70 partner, as the J domain assumes a fit with a bend induced in helix II (Landry, 2003). The turn between helix II and III has the sequence triptych HPD, which is essential for the interaction of the protein with Hsp70 (Greene et al, 1998). In both DnaJ and Hdj1, several hydrophobic residues on apposing faces of helix II and III allow for tight packing and are not exposed to solvents (Szyperski et al, 1994; Qian et al, 1996).
Definitive features of the J domain
The defining structural features of the J domain outlined above can be interpreted reliably from sequence data alone. Table 1 shows 22 yeast proteins that carry a J domain. Each qualifies as a J protein for the following reasons: first, secondarystructure profiling predicts helical regions that align with the four segments of DnaJ and Hdj1 (Fig 1, grey boxes I–IV); second, the helix II region is rich in basic residues (Fig 1, pink letters) and helical wheel projections (not shown) indicate that these distribute as a charged surface; third, the predicted turn between helix II and III has the conserved triptych HPD; and fourth, in helix II and III, hydrophobic residues are conserved in positions where they stabilize packing of the helices in Hdj1 and DnaJ (Fig 1, asterisks).
Table 1.
Twenty-two open-reading frames in the genome of Saccharomyces cerevisiae with well-conserved J domains
| J protein | Gene | Length* | J domain† | Type | TM§ | Subcellular location | Function (known or predicted) |
|---|---|---|---|---|---|---|---|
| Ydj1 | Ynl064c | 409 | 4–72 | I | 0 | Cytoplasm | Protein folding |
| Xdj1 | Ylr090w | 459 | 7–79 | I | 0 | Cytoplasm | Protein folding |
| Apj1 | Ynl077w | 528 | 4–73 | I | 0 | Cytoplasm | Protein folding |
| Sis1 | Ynl007c | 352 | 4–70 | II | 0 | Cytoplasm | Translation initiation |
| Djp1 | Yir004w | 432 | 4–72 | II | 0 | Cytoplasm | Peroxisome biogenesis |
| Zuo1 | Ygr285c | 433 | 98–170 | III | 0 | Cytoplasm | Translation and folding |
| Swa2 | Ydr320c | 668 | 604–668 | III | 0 | Cytoplasm | Clathrin uncoating |
| Jjj1 | Ynl227c | 590 | 2–72 | III | 0 | Cytoplasm | Endocytosis (?) |
| Jjj2 | Yjl162c | 482 | 11–79 | III | 0 | Cytoplasm | Unknown |
| Jjj3 | Yjr097w | 172 | 6–78 | III | 0 | Cytoplasm | Unknown |
| Caj1 | Yer048c | 391 | 4–73 | II | 0 | Nucleoplasm | Ca2+-dependent function (?) |
| Cwc23 | Ygl128c | 347 | 84–153 | III | 0 | Nucleoplasm | mRNA splicing |
| Mdj1 | Yfl016c | 511 | 59–127 | I | 0 | Mitochondria | Protein folding |
| Mdj2 | Ynl328c | 146 | 90–146 | III | 1 | Mitochondria | Mitochondrial biogenesis |
| Pam18 | Ylr008c | 168 | 109–168 | III | 1 | Mitochondria | Protein translocation |
| Jac1 | Ygl018c | 184 | 18–84 | III | 0 | Mitochondria | Iron–sulphur cluster assembly |
| Jid1 | Ypr061c | 301 | 62–153 | III | 1 | Mitochondria | Unknown |
| Scj1 | Ymr214w | 404 | 48–117 | I | 0 | ER | Protein folding |
| Hlj1 | Ymr161w | 234 | 19–87 | II | 1 | ER | Protein degradation |
| Jem1 | Yjl073w | 645 | 536–610 | III | 0 | ER | Karyogamy, protein folding |
| Sec63 | Yor254c | 663 | 123–198 | III | 3 | ER | Protein translocation |
| Erj5 | Yfr041c | 295 | 42–110 | III | 2 | ER | Unknown |
Iterative BLAST searches, starting with the J domain of DnaJ.
*The number of amino-acid residues in the open-reading frame.
†The position of the J domain in terms of the first and the last residue.
§The number of transmembrane (TM) segments predicted for each protein using the DAS algorithm (see the DAS Transmembrane Prediction server online at http://www.sbc.su.se/%7Emiklos/DAS/maindas.html). The subcellular location is given, along with the function or predicted function (references compiled in the Saccharomyces Genome Database, available online at http://www.yeastgenome.org). ER, endoplasmic reticulum; mRNA, messenger RNA.
Figure 1.
Multiple sequence alignment of the J domains of 22 yeast proteins. Colour coding indicates chemically similar residues as follows: orange, basic; blue, acidic; green, polar; red, non-polar. Grey boxes indicate the extent of helical segments predicted in each yeast protein (see the PredictProtein server online at http://www.cs.bgu.ac.il/~dfischer/predictprotein/#) that correspond to helices I–IV of DnaJ and Hdj1 (Szyperski et al, 1994; Qian et al, 1996). Asterisks represent hydrophobic residues that are important for helix packing and stability. The numbers indicate the length of the J domain from each protein.
Segments of various lengths are inserted in the helix I–II and II–III junctional turns. A particularly long insertion is found between helix I and II in the auxilin Swa2. In crystallized auxilin, this insertion forms a long loop that contains at least one basic residue that is essential for binding to Hsc70 (Jiang et al, 2003). Insertions of various lengths can be seen between helix II and III in several of the yeast proteins. The functional importance of this presumed interhelical loop is not clear, but it might influence interactions with an Hsp70 partner.
The next best thing: J-like proteins
Apart from these 22 proteins, three other open-reading frames in the yeast genome show marginal similarity to the J domain of one or more J proteins. However, none of these fulfils all of the sequence criteria for J domains outlined above. Jlp1, which is encoded by the gene Yll057c, is the best overall match, but it has a tyrosine in place of the histidine in the HPD consensus and has poor predicted helicity in the region of helix II. It is not clear in which subcellular compartment Jlp1 is located. Jlp2, which is encoded by the gene Ymr132c, is a cytosolic protein of unknown function with an alanine in place of the proline residue in the HPD consensus and few basic residues situated in the outer surface of helix II. Throughout its J-like domain it shows significant similarity to Sec63, which is a component of the yeast endoplasmic reticulum (ER) translocon. Jlp3, which is encoded by the gene Yjl104w, has 27% identity (46% similarity) with the helix II and III region of the mitochondrial J protein Pam18. Although it lacks the crucial HPD residues, the corresponding region of Jlp3 (sequence DKE) is predicted to form a turn between two helical segments, and the hydrophobic residues that would allow helix–helix packing are conserved.
Jlp3 was recently identified as Pam16/Tim16/Mia1, which is a subunit of the presequence-associated motor that includes the J protein Pam18 and the Hsp70 Ssc1. When bound to ATP, Ssc1 interacts only weakly with the translocase subunit Tim44 and with substrate polypeptides transiting through the translocase. The J domain of Pam18 can stimulate ATP hydrolysis by Ssc1 to drive protein translocation (Wiedemann et al, 2004). The J-like protein Pam16/Tim16/Mia1 is present in the same motor complex, in which it might promote Hsp70 recruitment without stimulating ATP hydrolysis or might compete with Pam18 to regulate Hsp70 activity.
The possibility that other J-like proteins might interact with an Hsp70 partner should be considered, although they lack the crucial HPD residues and would therefore be unlikely to stimulate ATP hydrolysis. The limited similarity shown by these 'next best' candidates indicates that the search of the genome is complete and that 22 is the total number of true J proteins in yeast.
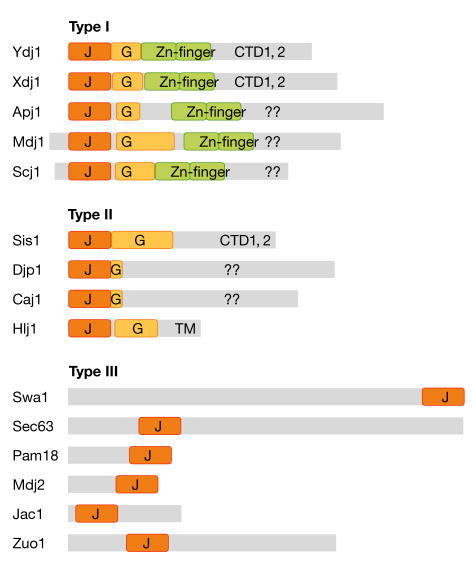
Type I family members and DnaJ-related functions
The additional domains of the yeast J proteins are suggestive of their in vivo functions. Five yeast J proteins are type I and have all three domains (and a glycine-rich region) in common with DnaJ. These are likely to represent true functional homologues of the bacterial chaperone (Fig 2). Two of these, Ydj1 and Xdj1, are closely related in sequence and might fulfil analogous cellular roles. The crucial functions of Ydj1 in the cytosol are associated with its ability to stimulate the ATPase activity of Ssa1/2 (the cytosolic Hsp70 of yeast) and, therefore, the re-folding of non-native protein substrates (Brodsky et al, 1998; Lu & Cyr, 1998). The precise function of Apj1 is not known but, similar to Ydj1, overexpression of Apj1 can cure growth defects in cells that express the yeast prion [PSI] (Kryndushkin et al, 2002). This indicates that Apj1 might also modulate folding reactions that are catalysed by Ssa1/2 in the cytosol.
Figure 2.
Structural classification of J proteins. Representation of type I, II and III J proteins from yeast aligned according to the amino-terminus of the mature protein. Mdj1 and Scj1 have targeting sequences processed in the mitochondria and endoplasmic reticulum, respectively. The grey boxes represent each polypeptide and show the scale of the J domain (J, orange), glycine-rich region (G, yellow) and zinc-finger domain (Zn-finger, green) found in some J proteins compared with the protein–protein-interaction domains that bind non-native substrate. CTD, carboxy-terminal domain; TM, transmembrane segment.
Mdj1 is a type I J protein that is involved in folding proteins that are imported into the mitochondrial matrix through binding substrate and stimulating the ATPase activity of the mitochondrial Hsp70 Ssc1 (Neupert, 1997). Scj1 is a type I J protein that regulates protein folding that is mediated by the Hsp70 Kar2 in the lumen of the ER (Silberstein et al, 1998). Scj1 remains an unusual example of a DnaJ homologue: although it carries a well-conserved zinc-finger domain for substrate binding, the conserved cysteines in this domain seem to be organized through disulphide bonds rather than the coordination of metal ions in the oxidizing environment of the ER.
Type I and II proteins are functionally similar
Sis1, Caj1, Djp1 and Hlj1 have obvious glycine-rich segments attached to their J domains, which designate them as type II proteins. As previously reviewed by Kelley (1998), the distinction between type I and II J proteins might not be meaningful when considering their function. For the few J proteins that have been studied in detail, it seems that both type I and II proteins bind to non-native substrates for presentation to their Hsp70 partners, and the carboxy-terminal domain of Sis1 has been shown to bind to substrate polypeptides as effectively as the combined zinc-finger/carboxy-terminal domain of Ydj1 (Fan et al, 2004). For prediction purposes, the presence of a glycine-rich region in a sequence might be a useful indicator of whether a new J protein carries the domains that allow non-native substrate binding, which are sometimes difficult to discern.
Type III J proteins are found at diverse intracellular sites
The type III J proteins represent a functionally distinct group. No type III J protein has been shown to bind to non-native polypeptides, and it is unlikely that these proteins function as molecular chaperones. Some type III J proteins assist the recruitment of a select isoform of Hsp70 to a discrete site. Two well-studied examples are Swa2 and Sec63. Swa2 is the yeast auxilin that is associated with clathrin-coated vesicles in the cytoplasm; its J domain allows the docking of Ssa1/2 for the disassembly of clathrin coats (Lemmon, 2001). Sec63 has three transmembrane domains that anchor it in the ER, and its lumenal J domain recruits the Hsp70 Kar2 to polypeptides that are in transit to the ER (Corsi & Schekman, 1997).
In other cases, the Hsp70 and type III J protein are recruited independently to the site of action, where the J protein productively stimulates ATP hydrolysis by the partner Hsp70. For example, the J protein Zuo1 and the Hsp70 Ssz1 are targeted independently to ribosomes, where they form the functionally active RAC complex (Craig et al, 2003). Pam18/Tim14 is little more than a J domain anchored to the inner face of the mitochondrial inner membrane. The Hsp70 Ssc1 also localizes there and is attached to the Tim23 translocase. Pam18/Tim14 stimulates the ATPase activity of Ssc1 to drive protein import into mitochondria (Neupert, 1997; Wiedemann et al, 2004). Another type III J protein is Cwc23, which co-purifies with the splicing machinery (Ohi et al, 2002). Although Hsp70 has yet to be ascribed a role in messenger RNA splicing, the assembly and disassembly of spliceosomes and accessory particles might benefit from the ATP-driven force that Hsp70 can generate, which is analogous to the disassembly of clathrin coats. Finally, Jem1 associates with integral proteins of the spindle-pole body, perhaps recruiting Hsp70 for its role in nuclear membrane fusion (Nishikawa et al, 2003).
The functions of the membrane-associated proteins Jid1 and Erj5, and three new type III proteins—Jjj1, Jjj2 and Jjj3—that are located within the cytoplasm, have yet to be determined. Careful localization of each protein is called for. Jid1 seems to be associated with mitochondria and Erj5 is associated with the ER (Huh et al, 2003). Jjj1 was identified in a screen for factors that are involved in fluid-phase endocytosis (Wiederkehr et al, 2001) and might therefore recruit Hsp70 to sites of membrane fission or fusion.
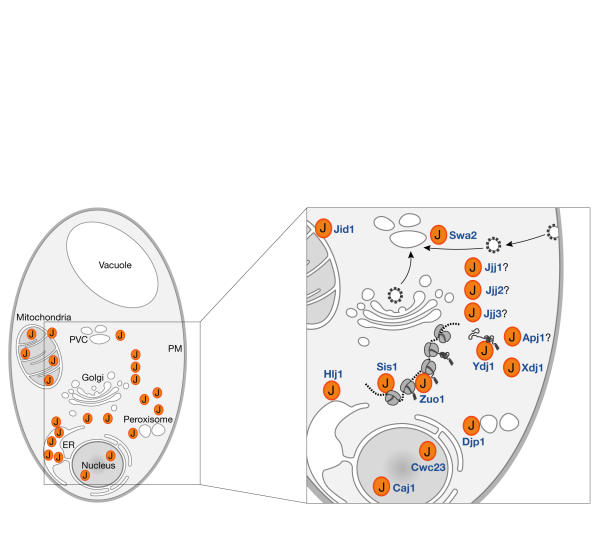
Partner selectivity: exclusive pairs or multiple partners?
There is selectivity in the interactions between J domains and isoforms of Hsp70. This is particularly apparent in the cytosol of yeast, in which three classes of Hsp70—Ssa, Ssb and Ssz—coexist with as many as 14 distinct J proteins (Fig 3). In some cases, a specific Hsp70 seems to interact only with one J protein; for example, the cytosolic Hsp70 Ssz1 associates with translating ribosomes and interacts exclusively with the J protein Zuo1. The Hsp70 Ssb1/2 is also associated with translating ribosomes and interacts with the ribosome-associated Zuo1–Ssz1 complex, but there is no evidence so far to show that it interacts with other J proteins either on the ribosome or free in the cytosol. Ssa1/2 is also present on translating ribosomes, but it does not interact with Zuo1 (Craig et al, 2003).
Figure 3.
Subcellular location and organization of J proteins in yeast. The location of the 22 proteins is shown on the left (see Table 1 for identities) and the inset section has been magnified to detail the J proteins in the cytosol. Most are free in the cytosol and dynamically associate with subcellular structures, although some, such as Hlj1 and perhaps Jid1 and Jjj1, are tethered to a specific membrane facing the cytosol.
How this selectivity is mediated is an intriguing question that will require structure-based studies. Our preliminary models of the surface-exposed residues on helix II of the J proteins that would dock with a Hsp70 partner do not indicate an obvious distinction between Zuo1 and the other cytoplasmic J proteins from yeast. However, many, perhaps all, of the other cytoplasmic J proteins in yeast (Fig 3) interact with the Ssa1/2 form of Hsp70.
The J proteins shown in the inset to Fig 3 might harness Ssa1/2 for as many as 12 cellular functions. Sis1 interacts with Ssa1/2 on ribosomes to assist aspects of translation (Horton et al, 2001), whereas Swa2 can recruit Ssa1/2 to disassemble clathrin coats from coated vesicles (Lemmon, 2001). Ydj1 could recruit Ssa1/2 for protein folding, as might Apj1 and Xdj1, although perhaps only under certain conditions and for select substrates (Kryndushkin et al, 2002). Hlj1 can recruit Ssa1/2 to the surface of the ER to facilitate the degradation of aberrant integral membrane proteins (Taxis et al, 2003; J. Brodsky, personal communication). Djp1 is another J protein that is found in the cytosol, which stimulates Ssa1/2 to assist in peroxisome biogenesis (Hettema et al, 1998). A population of Ssa2 has been found concentrated in the nucleoplasm (Huh et al, 2003). The function of Ssa1/2 in the nucleus is not known, but Caj1 and Cwc23 are available to assist it. Surprisingly, the nuclear J proteins Caj1 and Cwc23 are distinguished by a dyad HPD motif (HPDKHPD) in the loop that is predicted between helix II and III of their J domain. This motif is conserved in the mammalian Jdp1 protein; however, it is not clear where Jdp1 is located or what its function might be (Hahn et al, 1999).
Concluding remarks
The J proteins that are encoded in the yeast genome provide a general set of sequence-based principles with which to define the J domain and thereby identify the total J-protein family from genomic data. Whereas the structural distinctions between type I and II proteins are clear, the functional distinctions are less so. Type III J proteins fall into a functionally distinct category and stimulate Hsp70 activity at defined subcellular sites. Although they rely on other factors for substrate presentation, type III J proteins maximize the work done by the locally concentrated Hsp70. Structural studies will reveal how the various J proteins select their Hsp70 partners, and are starting to describe how J proteins and their Hsp70 partners cooperate to manipulate substrates. An exciting realization is that J-like proteins might structurally mimic J domains, which indicates further complexity in the regulation of Hsp70 function.
Acknowledgments
We thank J. Brodsky for critically reading the manuscript and gratefully acknowledge research support from the Australian Research Council.
References
- Brodsky JL, Lawrence JG, Caplan AJ (1998) Mutations in the cytosolic DnaJ homologue, YDJ1, delay and compromise the efficient translation of heterologous proteins in yeast. Biochemistry 37: 18045–18055 [DOI] [PubMed] [Google Scholar]
- Bukau B, Horwich AL (1998) The Hsp70 and Hsp60 chaperone machines. Cell 92: 351–366 [DOI] [PubMed] [Google Scholar]
- Cheetham ME, Caplan AJ (1998) Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress Chaperones 3: 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corsi AK, Schekman R (1997) The lumenal domain of Sec63p stimulates the ATPase activity of BiP and mediates BiP recruitment to the translocon in Saccharomyces cerevisiae. J Cell Biol 137: 1483–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig EA, Eisenman HC, Hundley HA (2003) Ribosome-tethered molecular chaperones: the first line of defense against protein misfolding? Curr Opin Microbiol 6: 157–162 [DOI] [PubMed] [Google Scholar]
- Fan CY, Lee S, Ren HY, Cyr DM (2004) Exchangeable chaperone modules contribute to specification of type I and type II Hsp40 cellular function. Mol Biol Cell 15: 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frydman J (2001) Folding of newly translated proteins in vivo: the role of molecular chaperones. Annu Rev Biochem 70: 603–647 [DOI] [PubMed] [Google Scholar]
- Genevaux P, Schwager F, Georgopoulos C, Kelley WL (2002) Scanning mutagenesis identifies amino acid residues essential for the in vivo activity of the Escherichia coli DnaJ (Hsp40) J-domain. Genetics 162: 1045–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greene MK, Maskos K, Landry SJ (1998) Role of the J-domain in the cooperation of Hsp40 with Hsp70. Proc Natl Acad Sci USA 95: 6108–6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn Y, Lee J, Seong C, Yoon J, Chung JH (1999) Structural analysis of phylogenetically conserved J domain protein gene. Biochim Biophys Acta 1447: 325–333 [DOI] [PubMed] [Google Scholar]
- Hettema EH, Ruigrok CC, Koerkamp MG, van den Berg M, Tabak HF, Distel B, Braakman I (1998) The cytosolic DnaJ-like protein djp1p is involved specifically in peroxisomal protein import. J Cell Biol 142: 421–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horton LE, James P, Craig EA, Hensold JO (2001) The yeast hsp70 homologue Ssa is required for translation and interacts with Sis1 and Pab1 on translating ribosomes. J Biol Chem 276: 14426–14433 [DOI] [PubMed] [Google Scholar]
- Huang K, Ghose R, Flanagan JM, Prestegard JH (1999) Backbone dynamics of the N-terminal domain in E. coli DnaJ determined by 15N- and 13CO-relaxation measurements. Biochemistry 38: 10567–10577 [DOI] [PubMed] [Google Scholar]
- Huh WK, Falvo JV, Gerke LC, Carroll AS, Howson RW, Weissman JS, O'Shea EK (2003) Global analysis of protein localization in budding yeast. Nature 425: 686–691 [DOI] [PubMed] [Google Scholar]
- Jensen RE, Johnson AE (1999) Protein translocation: is Hsp70 pulling my chain? Curr Biol 9: R779–R782 [DOI] [PubMed] [Google Scholar]
- Jiang J, Taylor AB, Prasad K, Ishikawa-Brush Y, Hart PJ, Lafer EM, Sousa R (2003) Structure–function analysis of the auxilin J-domain reveals an extended Hsc70 interaction interface. Biochemistry 42: 5748–5753 [DOI] [PubMed] [Google Scholar]
- Kelley WL (1998) The J-domain family and the recruitment of chaperone power. Trends Biochem Sci 23: 222–227 [DOI] [PubMed] [Google Scholar]
- Kryndushkin DS, Smirnov VN, Ter-Avanesyan MD, Kushnirov VV (2002) Increased expression of hsp40 chaperones, transcriptional factors, and ribosomal protein rpp0 can cure yeast prions. J Biol Chem 277: 23702–23708 [DOI] [PubMed] [Google Scholar]
- Landry SJ (2003) Swivels and stators in the Hsp40–Hsp70 chaperone machine. Structure (Camb) 11: 1465–1466 [DOI] [PubMed] [Google Scholar]
- Lemmon SK (2001) Clathrin uncoating: auxilin comes to life. Curr Biol 11: R49–R52 [DOI] [PubMed] [Google Scholar]
- Lu Z, Cyr DM (1998) Protein folding activity of Hsp70 is modified differentially by the hsp40 co-chaperones Sis1 and Ydj1. J Biol Chem 273: 27824–27830 [DOI] [PubMed] [Google Scholar]
- Neupert W (1997) Protein import into mitochondria. Annu Rev Biochem 66: 863–917 [DOI] [PubMed] [Google Scholar]
- Nishikawa S, Terazawa Y, Nakayama T, Hirata A, Makio T, Endo T (2003) Nep98p is a component of the yeast spindle pole body and essential for nuclear division and fusion. J Biol Chem 278: 9938–9943 [DOI] [PubMed] [Google Scholar]
- Ohi MD, Link AJ, Ren L, Jennings JL, McDonald WH, Gould KL (2002) Proteomics analysis reveals stable multiprotein complexes in both fission and budding yeasts containing Myb-related Cdc5p/Cef1p, novel pre-mRNA splicing factors, and snRNAs. Mol Cell Biol 22: 2011–2024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian YQ, Patel D, Hartl FU, McColl DJ (1996) Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain. J Mol Biol 260: 224–235 [DOI] [PubMed] [Google Scholar]
- Silberstein S, Schlenstedt G, Silver PA, Gilmore R (1998) A role for the DnaJ homologue Scj1p in protein folding in the yeast endoplasmic reticulum. J Cell Biol 143: 921–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szyperski T, Pellecchia M, Wall D, Georgopoulos C, Wuthrich K (1994) NMR structure determination of the Escherichia coli DnaJ molecular chaperone: secondary structure and backbone fold of the N-terminal region (residues 2-108) containing the highly conserved J domain. Proc Natl Acad Sci USA 91: 11343–11347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taxis C, Hitt R, Park SH, Deak PM, Kostova Z, Wolf DH (2003) Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J Biol Chem 278: 35903–35913 [DOI] [PubMed] [Google Scholar]
- Wiedemann N, Frazier AE, Pfanner N (2004) The protein import machinery of mitochondria. J Biol Chem 279: 14473–14476 [DOI] [PubMed] [Google Scholar]
- Wiederkehr A, Meier KD, Riezman H (2001) Identification and characterization of Saccharomyces cerevisiae mutants defective in fluid-phase endocytosis. Yeast 18: 759–773 [DOI] [PubMed] [Google Scholar]