Abstract
We developed a growth test to screen for yeast mutants defective in endoplasmic reticulum (ER) quality control and associated protein degradation (ERAD) using the membrane protein CTL*, a chimeric derivative of the classical ER degradation substrate CPY*. In a genomic screen of ∼5,000 viable yeast deletion mutants, we identified genes necessary for ER quality control and degradation. Among the new gene products, we identified Dsk2p and Rad23p. We show that these two proteins are probably delivery factors for ubiquitinated ER substrates to the proteasome, following their removal from the membrane via the Cdc48–Ufd1–Npl4p complex. In contrast to the ERAD substrate CTG*, proteasomal degradation of a cytosolic CPY*–GFP fusion is not dependent on Dsk2p and Rad23p, indicating pathway specificity for both proteins. We propose that, in certain degradation pathways, Dsk2p, Rad23p and the trimeric Cdc48 complex function together in the delivery of ubiquitinated proteins to the proteasome, avoiding malfolded protein aggregates in the cytoplasm.
Keywords: ER-associated degradation, proteasome, ubiquitin, Dsk2p, Rad23p
Introduction
The endoplasmic reticulum (ER) is the site of entry for the secretory pathway, which supplies the ER, Golgi apparatus, plasma membrane, lysosome (vacuole) and cellular exterior with the protein equipment necessary for proper cellular function. Once in the ER, proteins are folded and modified to acquire their native, functional conformations. Failure of this process results in inactive proteins forming insoluble aggregates, which ultimately lead to cellular malfunction and unhealthy cellular states (Kopito, 2000; Kostova & Wolf, 2003). The ER contains an efficient protein quality control system that recognizes malfolded and orphan proteins (Ellgaard et al, 1999) and targets them for elimination through a process known as ER-associated protein degradation (ERAD), to ensure delivery only of properly folded proteins to their site of action. Elimination is achieved via retrograde transport back to the cytoplasm through a channel possibly containing the Sec61p protein (Kostova & Wolf, 2003). While still associated with the ER membrane, proteins are polyubiquitinated. Thereafter, an AAA–ATPase complex composed of Cdc48p (p97 in mammalian cells), Ufd1p and Npl4p binds the polyubiquitinated proteins and mediates their release from the membrane. The 26S proteasome finally degrades the ubiquitinated substrates. The proteasome consists of the 20S cylindrical core, which is responsible for hydrolysis of ubiquitinated proteins, and the 19S regulatory particle that has a number of functions, including substrate recognition, binding and unfolding, opening of the 20S core and driving of the polypeptide chain through this opening (Kostova & Wolf, 2003). The identity of proteins that recognize polyubiquitinated substrates and deliver them to the proteasome for degradation is a subject of longstanding discussion. Earlier findings had suggested that Rpn10 (S5a in mammalian cells), a 19S cap subunit containing ubiquitin-interacting motif, might be the subunit binding polyubiquitinated substrates. However, deletion of this subunit is not lethal in several organisms (van Nocker et al, 1996; Fu et al, 1998). Crosslinking experiments have indicated that the Rpt5 (S6′) ATPase subunit of the 19S cap complex interacts with polyubiquitin chains (Lam et al, 2002). Recently, two other polyubiquitin-binding proteins, Dsk2p and Rad23p, have been identified (Wilkinson et al, 2001; Chen & Madura, 2002; Funakoshi et al, 2002; Hartmann-Petersen et al, 2003). Dsk2p and Rad23p are not 19S cap subunits, but participate in spindle-pole body duplication (Biggins et al, 1996) and nucleotide excision repair (Watkins et al, 1993), respectively. Rad23p has also been proposed to bind N-glycanase, an enzyme involved in deglycosylating carbohydrate-containing substrates of the proteasome (Suzuki et al, 2001). Dsk2p and Rad23p possess an N-terminal ubiquitin-like domain (UBL), which binds to a specific site on the 19S cap, and a C-terminal ubiquitin-associated domain(s) (UBA), capable of binding polyubiquitin chains (Wilkinson et al, 2001; Rao & Sastry, 2002; Hartmann-Petersen et al, 2003). These characteristics suggest that degradation substrates can bind to the Dsk2p or Rad23p UBA domain through their polyubiquitin chain and, consequently, can be delivered to the proteasome by means of the UBL–19S cap interaction. However, there is controversy about assigning this role to Dsk2p and Rad23p. Although degradation of ubiquitin-fusion degradation (UFD) substrates depends on these two proteins (Chen & Madura, 2002; Rao & Sastry, 2002), in some in vitro studies an inhibitory effect of Rad23p on proteasome activity has been noted (Raasi & Pickart, 2003).
Here, we report a genome-wide screen carried out to identify new yeast mutants with defects in ER protein quality control and degradation. We identify Dsk2p and Rad23p as new components with overlapping functions involved in the ERAD of two topologically different but otherwise related substrates. On the basis of our data and previous findings, we propose that Dsk2p and Rad23p function as delivery factors between the Cdc48 complex and the proteasome, and that they are specific to certain ubiquitin–protein degradation pathways.
Results And Discussion
To gain a deeper insight into the molecular mechanisms of protein quality control and ER-associated degradation, we undertook a genome-wide screen using the EUROSCARF yeast library, consisting of about 5,000 Saccharomyces cerevisiae strains, each deleted for a single non-essential gene. The use of this library for screening mutants of ERAD is possible because cells tolerate a defect in this process as long as the unfolded protein response is intact (Friedländer et al, 2000; Travers et al, 2000). Cell growth is one of the most sensitive indicators of alterations in cell physiology due to mutations. It is known that yeast cells expressing a mutated version of the Sec61p translocon, Sec61-2p, are not viable at elevated temperature (37°C) because Sec61-2p becomes an ERAD substrate (Biederer et al, 1996). Viability under these conditions is restored only in the absence of key ERAD components, like Der3/Hrd1p, Ubc6p and Ubc7p (Biederer et al, 1996; Bordallo et al, 1998). On the basis of this growth phenotype of yeast strains defective in ERAD, we devised a screen to identify new ERAD components. We have previously described the membrane-localized ERAD substrate CTG*, which consists of an ER-lumenal malfolded CPY* domain connected to green fluorescent protein (GFP) in the cytoplasm via a transmembrane domain (Taxis et al, 2003). To screen for new mutants, we replaced the cytoplasmic GFP moiety with the Leu2 protein (3-isopropylmalate-dehydrogenase) (Fig 1A). This new construct, called CTL*, was placed under the control of the weak GAL4 promoter (Griggs & Johnston, 1993). Low expression of CTL* was particularly required because yeast cells can sustain growth even in the presence of minimal amounts of leucine. Consequently, strains with leu2 auxotrophy but, otherwise, wild type for ERAD are unable to grow in media lacking leucine (Fig 1B). Only when ERAD is defective is CTL* stabilized and able to complement the leu2 deficiency (Fig 1B). The low expression then allows for sharp growth differences to be easily observed. We took advantage of this growth phenotype to screen the ∼5,000 individual deletion mutants of the EUROSCARF yeast library expressing CTL*. Strains defective in most of the known ERAD components resulted in growth in the absence of leucine (Table 1), highlighting the reliability of the selection procedure. The screen yielded a number of new mutants exhibiting a reproducible growth phenotype. Among these, we observed strong complementation in strains carrying the dsk2 and, to a lesser extent, rad23 deletions (Fig 1B). To test whether these two proteins are true components of the ER degradation machinery, we analysed the degradation of two well-known ERAD substrates, soluble CPY* (Hiller et al, 1996) and membrane-anchored CTG* (Taxis et al, 2003), in a set of isogenic strains deleted in either one or both genes. Pulse-chase analyses showed a strong stabilization of CPY* haemagglutinin (HA) and CTG* in the Δdsk2Δrad23 double mutant, indicating that these proteins participate directly in ERAD (Fig 2). CPY*HA and CTG* exhibited about 10% stabilization in Δdsk2 and Δrad23 single mutants, and 40% and 45% stabilization, respectively, in the Δdsk2Δrad23 double mutant with respect to wild-type cells (Fig 2A,B). This synergistic effect implies that Dsk2p and Rad23p function together in ERAD, consistent with reports implicating Dsk2p and Rad23p in the proteasomal turnover of the artificial substrates UbV76-V-β-gal and Ub-P-β-gal (Chen & Madura, 2002; Rao & Sastry, 2002) via the UFD pathway. Fractionation of microsomes derived from wild-type and ufd1-1 (a mutant allele of Ufd1p, a component of the AAA–ATPase Cdc48p–Ufd1p–Npl4p complex (Ye et al, 2001; Jarosch et al, 2002)) strains results in a considerable portion of ubiquitinated CPY* to be retained in the pellet, with only a minor amount present in the soluble fraction. In wild-type cells, the ubiquitinated material released by the trimeric Cdc48 complex into the supernatant is rapidly degraded by proteasomes. In ufd1-1 cells there is no cytoplasmic ubiquitinated protein product owing to a defective trimeric Cdc48 complex (Jarosch et al, 2002). To elucidate the step at which Dsk2p and Rad23p may be involved, we examined the localization of ubiquitinated material in wild-type and Δdsk2Δrad23 strains. As shown previously, we found little ubiquitinated CPY*HA in the soluble fraction of wild-type cells compared with the pellet (Fig 3, lanes 1 and 2). As expected, there was no CPY*HA visible in the supernatant owing to degradation via the proteasome. In comparison, a considerably larger amount of ubiquitinated protein was detected in the soluble fraction of the Δdsk2Δrad23 double mutant (Fig 3, lane 4; compare lanes 4 and 2). These findings indicate that ubiquitinated CPY*HA accumulates in the cytosol as a consequence of failed delivery to the proteasome, and are in agreement with a role for Dsk2p and Rad23p downstream of the Cdc48–Ufd1–Npl4 complex, targeting substrates for proteasomal degradation.
Figure 1.
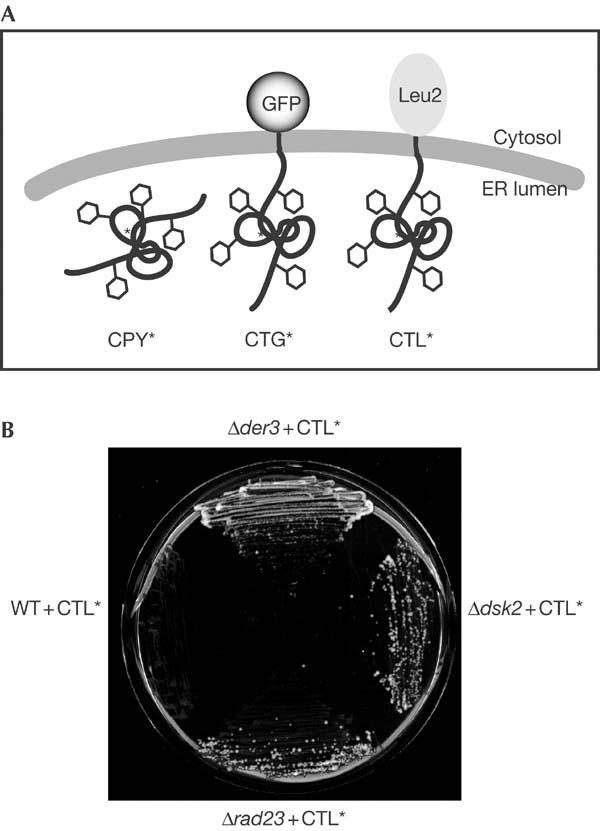
DSK2 and RAD23 are found in a screen for new ERAD components. (A) Schematic representation of the ERAD substrates CPY*, CTG* and CTL*. (B) The BY4743 wild-type (WT) strain expressing CTL* fails to grow on SC medium lacking leucine, whereas a strain deleted in the well-known ERAD component Der3p, used as a positive control, permits growth. The deletion strains Δdsk2 and Δrad23, expressing CTL*, complement the leucine auxotrophy of the strain.
Table 1.
Known ERAD components found in the screen
| ORF name | Gene name | Function |
|---|---|---|
| YOL013C |
HRD1/DER3 |
Ubiquitin-protein ligase |
| YLR207W |
HRD3 |
Der3p interacting protein |
| YHR204W |
HTM1/MNL1 |
ER-lumenal lectin |
| YNR030W |
ECM39/ALG12 |
N-glycan synthesis |
| YJR131W |
MNS1 |
ER α-mannosidase I |
| YMR022W |
UBC7 |
Ubiquitin-conjugating enzyme |
| YEL031W | SPF1/COD1 | ER calcium pump |
Figure 2.
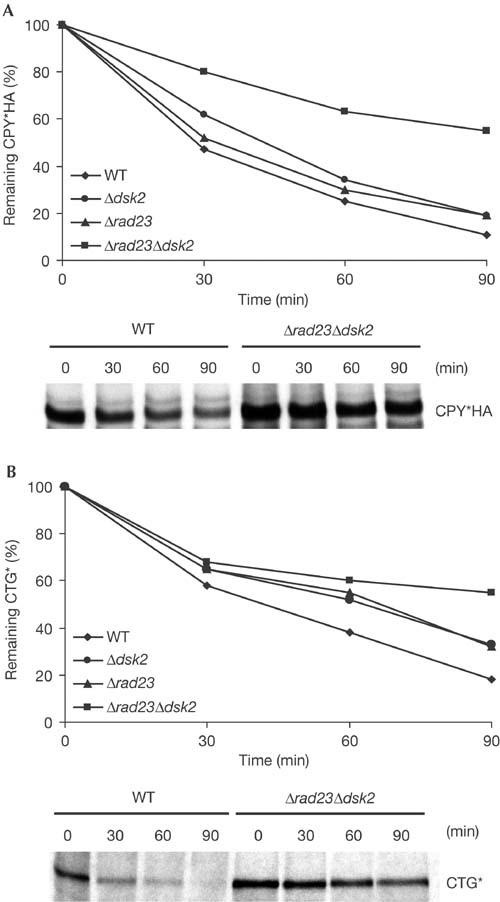
Mutants deleted in DSK2 and RAD23 are defective in the degradation of the ERAD substrates CPY*HA and CTG*. (A,B) Yeast strains expressing CPY*HA or CTG* were metabolically labelled, chased for the indicated times and immunoprecipitated using anti-HA or anti-CPY antibodies, respectively. Δdsk2 and Δrad23 single- and double-deletion mutants show synergistic effects in ERAD. Degradation of CPY*HA (A) and CTG* (B) is significantly delayed in a Δdsk2Δrad23 strain. Data represent the mean values of three (CPY*HA) and two (CTG*) independent experiments. WT, wild type.
Figure 3.
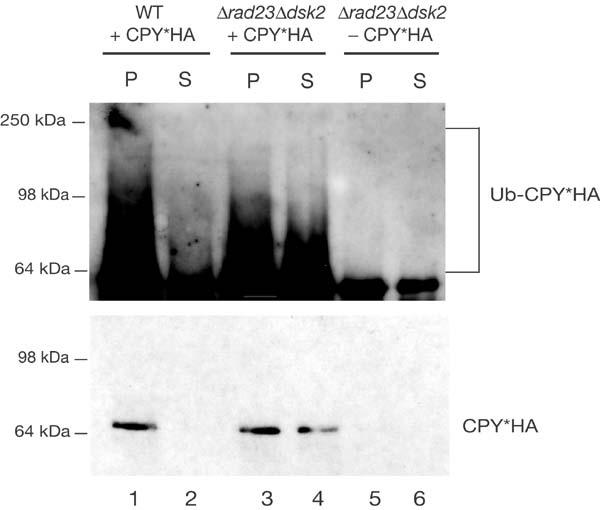
Polyubiquitinated CPY*HA accumulates in the cytosol of the Δdsk2Δrad23 mutant. Membrane association of ubiquitinated CPY*HA was investigated in respectively transformed wild type (WT) and Δdsk2Δrad23 strains. Microsomal extracts were separated into membrane-associated (P) and cytosolic (S) fractions. CPY*HA was precipitated with anti-CPY antibodies and analysed by immunoblotting with anti-ubiquitin antibodies. The pellet and supernatant fractions from non-transformed strain Δdsk2Δrad23 are included to reveal the background activity of the ubiquitin antibody. Immunoblotting with HA-antibody shows localization of CPY*HA in each fraction.
We also investigated whether Dsk2p and Rad23p are general components of the targeting/degradation machinery of ubiquitinated proteins. We constructed ΔssCPY*-GFP, which lacks the CPY signal sequence, as a representative soluble proteasomal substrate. Cell fractionation, protease protection and deglycosylation experiments verified that ΔssCPY*-GFP, which carries the same mutated motif as the ER-localized CPY* and CTG*, resides in the cytoplasm (Fig 4). Next, we determined whether cytoplasmic ΔssCPY*-GFP is a substrate of the 26S proteasome. We found that ΔssCPY*-GFP is rapidly degraded in wild-type cells, whereas degradation is abolished in a mutant (cim3-1) defective in one of the 19S cap AAA–ATPase subunits (Rpt6p) (Hiller et al, 1996) (Fig 5A). Cycloheximide chase analyses revealed that degradation of cytoplasmic ΔssCPY*-GFP is not affected by the absence of Dsk2p and Rad23p: the protein is degraded as rapidly in the Δdsk2Δrad23 double mutant as it is in wild-type cells (Fig 5B), underscoring the pathwayspecific requirement for Dsk2p and Rad23p. Interestingly, both the ERAD and UFD pathways, which require Dsk2p and Rad23p function, are also dependent on the Cdc48–Ufd1–Npl4p complex (Ye et al, 2001; Jarosch et al, 2002). In fact, Ufd1p, the central subunit of the trimeric Cdc48 complex, was first identified as a component of the UFD pathway (Johnson et al, 1995). Therefore, we tested whether Ufd1p is necessary for the degradation of cytoplasmic ΔssCPY*-GFP. Consistent with the fact that Dsk2p and Rad23p are not involved, we observed no difference in the degradation of ΔssCPY*-GFP in wild-type or ufd1-1 cells (Fig 5C). As a second, mainly cytoplasmic, substrate of the ubiquitin–proteasome system, we tested a fusion protein consisting of the Deg1 degradation domain of the MATα2 repressor protein and GFP, Deg1-GFP (Lenk & Sommer, 2000). The half-life of Deg1-GFP was determined by pulse-chase analysis in wild-type and ufd1-1 mutant cells as 22.5 and 24.5 min, respectively (data not shown), in agreement with published results (Lenk & Sommer, 2000). Cycloheximide chase experiments showed that Deg1-GFP is also not a target of Dsk2p and Rad23p (Fig 5D). On the basis of these findings, we propose that degradation pathways that require the AAA–ATPase complex Cdc48–Ufd1–Npl4p may also depend on Dsk2p and Rad23p, a concept that will require further exploration. In this scenario, substrates liberated from the trimeric Cdc48 complex are passed to Dsk2p and/or Rad23p, which, in turn, transfer them to the proteasome for degradation. We believe that, in this way, malfolded substrates of the ER are handed over to the proteasome following an uninterrupted path from the ER membrane, thus preventing the formation of insoluble protein aggregates in the cytoplasm.
Figure 4.
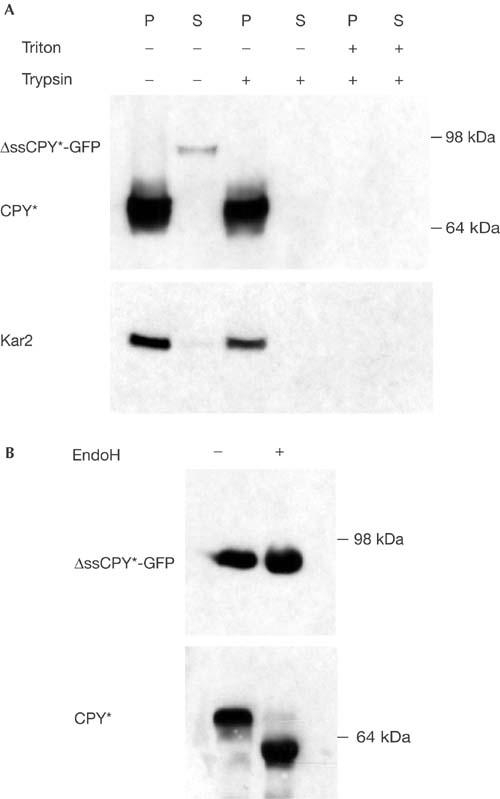
ΔssCPY*-GFP is located in the cytosol. (A) Spheroplasts from strain YCT397 transformed with ΔssCPY*-GFP and chromosomally expressing CPY* were isolated and fractionated into microsome pellet (P) and supernatant (S). Both fractions were treated with trypsin and triton as indicated. The ER lumenal Kar2p was used as a control for fractionation and protease protection accuracy. (B) ΔssCPY*-GFP is not glycosylated. ΔssCPY*-GFP and, as a control, glycosylated CPY* were immunoprecipitated using CPY antibodies from YCT397 expressing both proteins (see (A)), subjected to endoglycosidase H (EndoH) treatment and immunoblotted. ΔssCPY*-GFP was detected with anti-GFP; CPY* was detected with anti-CPY antibodies.
Figure 5.
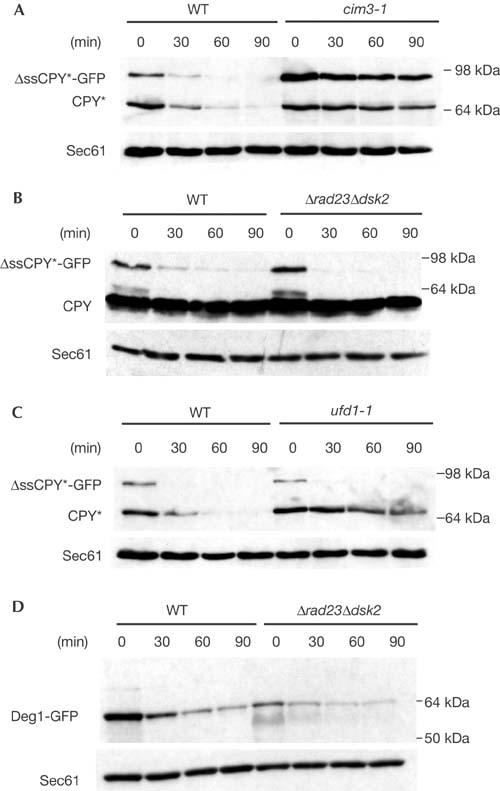
Degradation of cytosolic CPY*-GFP and Deg1-GFP is independent of Dsk2p and Rad23p. (A) Cycloheximide decay experiments performed in wild type (WT) and proteasomal mutant cim3-1 strains expressing both CPY* and ΔssCPY*-GFP show that ΔssCPY*-GFP is a substrate of the 26S proteasome. (B) Cycloheximide decay experiments performed in WT and Δdsk2Δrad23 strains expressing CPY and ΔssCPY*-GFP indicate that Dsk2p and Rad23p have no role in the degradation of cytosolic ΔssCPY*-GFP. (C) Degradation of ΔssCPY*-GFP is independent of Ufd1p. ER lumenal CPY*, conversely, is stabilized in the ufd1-1 mutant. (D) Degradation of Deg1-GFP is independent of Dsk2p and Rad23p. Sec61p was used as a control for protein loading.
We also propose that, contrary to previous interpretations that Dsk2p and Rad23p are essential for proteasomal targeting of ubiquitinated substrates in general or that they act as inhibitors in these same processes, the action of Dsk2p and Rad23p is not all-or-none, but pathway specific and finely tuned.
Methods
Yeast strains and bacterial constructs. Molecular biological and genetic techniques were carried out using standard methods (Guthrie & Fink, 1991). Saccharomyces cerevisiae strains MY3589 (MATa, ura3-52, leu, ade2), MY3588 (MATa, ura3-52, trp1, ade2, his3, dsk2Δ:LEU2) and MY3592 (MATα, ura3-52, leu2, his3Δ200, ade2, dsk2Δ:LEU2, rad23Δ) were described in Biggins et al (1996). MY3587F (MATα, ura3-52, leu2, his3, ade2, rad23Δ) was described in Rao & Sastry (2002). Strains BY4743 (MATa/α, his3Δ1/his3Δ1, leu2Δ0/leu2Δ0, lys2Δ0/LYS2, MET15/met15Δ0, ura3Δ0/ura3Δ0), BY4743Δdsk2 (MATa/α, his3Δ1/his3Δ1, leu2Δ0/leu2Δ0, lys2Δ0/LYS2, MET15/met15Δ0, ura3Δ0/ura3Δ0, dsk2Δ:kanMX4/ dsk2Δ:kanMX4), BY4743Δrad23 (MATa/α, his3Δ1/his3Δ1, leu2Δ0/leu2Δ0, lys2Δ0/LYS2, MET15/met15Δ0, ura3Δ0/ura3Δ0, rad23Δ:kanMX4/rad23Δ:kanMX4) and BY4743Δder3/hrd1 (MATa/α, his3Δ1/his3Δ1, leu2Δ0/leu2Δ0, lys2Δ0/LYS2, MET15/met15Δ0, ura3Δ0/ura3Δ0, der3:kanMX4/ der3:kanMX4) are from the EUROSCARF Gene Deletion Bank (Frankfurt, Germany). YPH499Y (MATa, ura3-52, leu2Δ1, his3Δ200, trp1Δ63, lys2-801, ade2-101, prc1-1) and CMY762Y (MATa, ura3-52, leu2Δ1, his3Δ200, cim3-1, prc1-1) were previously described in Hiller et al (1996), whereas YCT397 (MATa, ura3-52, leu2-3,112, his4-519, ade1-100, prc1-1) and YCT415 (YCT397, ufd1-1) were described in Taxis et al (2003). Cells were grown at 25°C (ts strain cim3-1) or at 30°C in synthetic complete media.
3HA-tagged CPY* and CTG* were expressed from plasmids pCT42 and pMA1, respectively (Taxis et al, 2002, 2003). Deg1-GFP was expressed from plasmid pUL28 (Lenk & Sommer, 2000). pRS316 expressing Gal4-CTL* was cloned in a stepwise process: the LEU2 gene was PCR-amplified using the primers 5′lPac (GGATCCTTAATTAAGTCTGCCCCTAAGAAG) and 3′lSph (GTACAGGCATGCATAAATGTATGTAGATTG), and the 1.1 kb fragment was cloned into pNEB193 between PacI and SphI restriction sites. The 2.5 kb CT* fragment obtained by digesting pMA1 with KpnI and PacI was cloned into the respective sites of pNEB193-LEU2, yielding pNEB193-CTL*. CTL* was then transferred into pRS316 as a 3.7 kb HindIII fragment, giving rise to pRS316-tdh3-CTL*. The TDH3 promoter of CTL* was replaced by the GAL4 promoter as follows: the GAL4 promoter was PCR-amplified using the primers 5′GAL4CTLNotI (AAATATGCGGCCGAGGACCCTGACGGCGA) and 3′GAL4CTL (GTAAACTGGTGAATGCTTTCATCTTTCAGGCTTGCTTC), the latter being partially complementary both to GAL4 and the 5′ end of prc1-1. A 500 bp N-terminal fragment of CPY* was PCR-amplified with the primer set 5′CTLGAL4 (GAAGCAAGCCTCCTGAAAGATGAAAGCATTCACCAGTTTAC), having partial complementarity both to CPY* and the 3′ end of the GAL4 promoter, and 3′CTLBsu36I (GAAGTTATAGACATCCTTACC). These two PCR products were used as overlapping templates in a third PCR to generate a final promGAL4:prc1-1 fusion fragment, which was cloned into NotI–Bsu36I-digested pRS316-tdh3-CTL*, generating pRS316-gal4-CTL*. Cytosolic CPY*-GFP (ΔssCPY*-GFP) was cloned in two steps. First, we cloned pRS316CPY*-GFP via homologous recombination in yeast (details available upon request). Then, we removed the signal sequence from pRS316CPY*-GFP by a QuikChange (Stratagene) based PCR-mutagenesis approach, using the complementary primer set 5′PRC1(−)ss (GTATACATACGCTATGATCTCATTGCAAAGACCG) and 3′PRC1(−)ss (CGGTCTTTGCAATGAGATCATAGCGTATGTATAC).
Antibodies. Polyclonal anti-CPY was used for immunoprecipitation of CTG*. Monoclonal anti-CPY and monoclonal anti-GFP (Molecular Probes) and polyclonal anti-sec61 (gift from T. Sommer) were diluted 1:10,000 for immunoblotting. Monoclonal anti-HA (Covance) was used at 1:1,000 dilution for immunoprecipitation and at 1:10,000 dilution for immunoblotting. Monoclonal anti-ubiquitin antibody (Covance) was used at 1:1,000 dilution for immunoblotting.
Pulse-chase analysis and immunoprecipitation. CPY* and CTG* pulse-chase experiments were performed as described previously (Taxis et al, 2002), with the exception that cells were grown in 2% glucose and labelled for 60 min.
Cycloheximide decay analysis. Cells were grown at 30°C to logarithmic phase in synthetic complete medium. The temperature-sensitive cim3-1 strain was grown at 25°C and then shifted to 30°C for 1 h. Deg1-GFP, expressed under the control of the CUP promoter, was induced for 1 h by the addition of 100 μM CuSO4. Cycloheximide (0.25 mg ml−1) was added and 2 OD600(6 × 107) of cells were taken at the indicated time points. Cell extracts were prepared by alkaline lysis and subjected to SDS–PAGE and western blotting as described in Taxis et al (2003).
Detection of ubiquitinated CPY*HA. Ubiquitinated CPY*HA was detected in pellet and supernatant fractions by following the procedure described in Hiller et al (1996).
Localization studies. For localization of ΔssCPY*-GFP protease protection (trypsin) and deglycosylation (endoglycosidase H), experiments were performed as described in Taxis et al (2002), (2003).
Acknowledgments
We thank H. Rao, M.D. Rose and T. Sommer for the generous gift of yeast strains and plasmids. We are grateful to members of the Wolf group for helpful discussions and E. Tosta for help with the preparation of the manuscript. This work was supported by a grant of the Deutsche Forschungsgemeinschaft (Bonn, Germany) and the Fonds der Chemischen Industrie (Frankfurt, Germany).
References
- Biederer T, Volkwein C, Sommer T (1996) Degradation of subunits of the Sec61p complex, an integral component of the ER membrane, by the ubiquitin–proteasome pathway. EMBO J 15: 2069–2076 [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Ivanovska I, Rose MD (1996) Yeast ubiquitin-like genes are involved in duplication of the microtubule organizing center. J Cell Biol 133: 1331–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordallo J, Plemper RK, Finger A, Wolf DH (1998) Der3p/Hrd1p is required for endoplasmic reticulum-associated degradation of misfolded lumenal and integral membrane proteins. Mol Biol Cell 9: 209–222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Madura K (2002) Rad23 promotes the targeting of proteolytic substrates to the proteasome. Mol Cell Biol 22: 4902–4913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellgaard L, Molinari M, Helenius A (1999) Setting the standards: quality control in the secretory pathway. Science 286: 1882–1888 [DOI] [PubMed] [Google Scholar]
- Friedländer R, Jarosch E, Urban J, Volkwein C, Sommer T (2000) A regulatory link between ER-associated protein degradation and the unfolded-protein response. Nat Cell Biol 2: 379–384 [DOI] [PubMed] [Google Scholar]
- Fu H et al. (1998) Multiubiquitin chain binding and protein degradation are mediated by distinct domains within the 26S proteasome subunit Mcb1. J Biol Chem 273: 1970–1981 [DOI] [PubMed] [Google Scholar]
- Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H (2002) Budding yeast Dsk2p is a polyubiquitin-binding protein that can interact with the proteasome. Proc Natl Acad Sci USA 99: 745–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griggs DW, Johnston M (1993) Promoter elements determining weak expression of the GAL4 regulatory gene of Saccharomyces cerevisiae. Mol Cell Biol 13: 4999–5009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR (1991) Guide to Yeast Genetics and Molecular Biology. Method in Enzymology, Vol 194. San Diego, CA, USA: Academic [PubMed] [Google Scholar]
- Hartmann-Petersen R, Seeger M, Gordon C (2003) Transferring substrates to the 26S proteasome. Trends Biochem Sci 28: 26–31 [DOI] [PubMed] [Google Scholar]
- Hiller MM, Finger A, Schweiger M, Wolf DH (1996) ER degradation of a misfolded luminal protein by the cytosolic ubiquitin-proteasome pathway. Science 273: 1725–1728 [DOI] [PubMed] [Google Scholar]
- Jarosch E et al. (2002) Protein dislocation from the ER requires polyubiquitination and the AAA–ATPase Cdc48. Nat Cell Biol 4: 134–139 [DOI] [PubMed] [Google Scholar]
- Johnson ES, Ma PC, Ota IM, Varshavsky A (1995) A proteolytic pathway that recognizes ubiquitin as a degradation signal. J Biol Chem 270: 17442–17456 [DOI] [PubMed] [Google Scholar]
- Kopito RR (2000) Aggresomes, inclusion bodies and protein aggregation. Trends Cell Biol 10: 524–530 [DOI] [PubMed] [Google Scholar]
- Kostova Z, Wolf DH (2003) For whom the bell tolls: protein quality control of the endoplasmic reticulum and the ubiquitin-proteasome connection. EMBO J 22: 2309–2317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam YA, Lawson TG, Velayutham M, Zweier JL, Pickart C (2002) A proteasomal ATPase subunit recognizes the polyubiquitin degradation signal. Nature 416: 763–767 [DOI] [PubMed] [Google Scholar]
- Lenk U, Sommer T (2000) Ubiquitin-mediated proteolysis of a short-lived regulatory protein depends on its cellular localization. J Biol Chem 275: 39403–39410 [DOI] [PubMed] [Google Scholar]
- Raasi S, Pickart CM (2003) Rad23 ubiquitin-associated domains (UBA) inhibit 26 S proteasome-catalyzed proteolysis by sequestering lysine 48-linked polyubiquitin chains. J Biol Chem 278: 8951–8959 [DOI] [PubMed] [Google Scholar]
- Rao H, Sastry A (2002) Recognition of specific ubiquitin conjugates is important for the proteolytic functions of the ubiquitin-associated domain proteins Dsk2 and Rad23. J Biol Chem 277: 11691–11695 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Park H, Kwofie MA, Lennarz W (2001) Rad23 provides a link between the Png1 deglycosylating enzyme and the 26S proteasome in yeast. J Biol Chem 276: 21601–21607 [DOI] [PubMed] [Google Scholar]
- Taxis C, Hitt R, Park SH, Deak PM, Kostova Z, Wolf DH (2003) Use of modular substrates demonstrates mechanistic diversity and reveals differences in chaperone requirement of ERAD. J Biol Chem 278: 35903–35913 [DOI] [PubMed] [Google Scholar]
- Taxis C, Vogel F, Wolf DH (2002) ER-Golgi traffic is a prerequisite for efficient ER degradation. Mol Biol Cell 13: 1806–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P (2000) Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258 [DOI] [PubMed] [Google Scholar]
- van Nocker S et al. (1996) The multiubiquitin-chain-binding protein Mcb1 is a component of the 26S proteasome in Saccharomyces cerevisiae and plays a nonessential, substratespecific role in protein turnover. Mol Cell Biol 16: 6020–6028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins JF, Sung P, Prakash L, Prakash S (1993) The Saccharomyces cerevisiae DNA repair gene RAD23 encodes a nuclear protein containing a ubiquitin-like domain required for biological function. Mol Cell Biol 13: 7757–7765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson CR et al. (2001) Proteins containing the UBA domain are able to bind to multi-ubiquitin chains. Nat Cell Biol 3: 939–943 [DOI] [PubMed] [Google Scholar]
- Ye Y, Meyer HH, Rapoport TA (2001) The AAA ATPase Cdc48/p97 and its partners transport proteins from the ER into the cytosol. Nature 414: 652–656 [DOI] [PubMed] [Google Scholar]


