Abstract
The histone H2ABbd is a novel histone variant of H2A with a totally unknown function. We have investigated the behaviour of the H2ABbd nucleosomes. Nucleosomes were reconstituted with recombinant histone H2ABbd and changes in their conformations at different salt concentrations were studied by analytical centrifugation. The data are in agreement with H2ABbd being less tightly bound compared with conventional H2A in the nucleosome. In addition, stable cell lines expressing either green fluorescent protein (GFP)–H2A or GFP–H2ABbd were established and the mobility of both fusions was measured by fluorescence recovery after photobleaching. We show that GFP–H2ABbd exchanges much more rapidly than GFP–H2A within the nucleosome. The reported data are compatible with a lower stability of the variant H2ABbd nucleosome compared with the conventional H2A particle.
Keywords: histone variant H2ABbd, nucleosomes, FRAP, analytical centrifugation, stability
Introduction
Within the eukaryotic nucleus, DNA is packaged into chromatin. The repetitive subunit of chromatin, the nucleosome, consists of an octamer of conventional core histones (two each of H2A, H2B, H3 and H4) around which two superhelical turns of DNA of ⩽80 base pairs (bp) each are wrapped (van Holde, 1988). In addition, eukaryotes express histone variants, which exhibit a distinct primary sequence and variable identity compared with their conventional counterparts (van Holde, 1988; Tsanev et al, 1993). As a whole, the functions of the histone variants such as H2A.Z, H2AX and macroH2A are unknown, but they have been implicated in several vital cellular processes. H2A.Z is essential for the survival of mouse and Drosophila (van Daal & Elgin, 1992; Clarkson et al, 1999; Faast et al, 2001). This protein is involved in both transcriptional activation (White & Gorovsky, 1988; Stargell et al, 1993; Santisteban et al, 2000; Suto et al, 2000; Larochelle & Gaudreau, 2003) and gene silencing (Dhillon & Kamakaka, 2000; Meneghini et al, 2003). H2AX is phosphorylated at its carboxy terminus immediately following doublestrand DNA breakage (Rogakou et al, 1998, 1999). This histone variant appears to prevent aberrant repair of DNA breakage and is implicated in the dosage-dependent suppression of genomic instability and tumours in mice (Bassing et al, 2003). Phosphorylation is essential for the function of H2AX, as these effects are abolished in mutants in which the conserved serine phosphorylation sites are substituted for alanine or glutamic acid residues (Celeste et al, 2003). MacroH2A, a histone variant with an unusual primary structure, is believed to have an important role in X-chromosome inactivation in mammals (Costanzi & Pehrson, 1998; Mermoud et al, 1999) and might be involved in the transcriptional repression of autosomes (Perche et al, 2000). Indeed, macroH2A interfered with both transcription factor binding and SWI/SNF nucleosome remodelling (Angelov et al, 2003).
Histone variants are used by the cell to build nucleosomes with specialized architectures with dedicated functions (Ausio & Abbott, 2002). The crystal structure of the H2A.Z nucleosome particle revealed localized changes that may destabilize (H2A.Z–H2B) dimer and (H3–H4)2 tetramer interactions (Suto et al, 2000). These subtle changes may explain the distinct physicochemical properties of H2A.Z arrays and their specific interactions with nuclear proteins (Suto et al, 2000; Abbott et al, 2001; Fan et al, 2002). In another study, the nucleosomes containing macroH2A exhibited an altered structure in the vicinity of the dyad axis, which was associated with the inability of SWI/SNF to remodel these particles (Angelov et al, 2003).
Recently, a novel histone variant H2ABbd (Barr body deficient), with an identity of 48% to H2A, was identified (Chadwick & Willard, 2001). Interestingly, H2ABbd is shorter than the conventional H2A, and it contains a row of six arginines at its amino terminus and lacks the nonstructured C-terminus typical of the other histones from the H2A family. The primary sequence of the histone fold domain of H2ABbd differed significantly from that of H2A (Chadwick & Willard, 2001). This variant is excluded from the inactive X chromosome, and its distribution overlaps with the regions of histone H4 acetylation in the nucleus (Chadwick & Willard, 2001). The function of this protein is totally unknown.
Characterizing the thermodynamic stability of variant nucleosomes is an important approach for understanding their inherent functions. Significantly, this analysis has only been performed on H2A.Z complexes in vitro (Abbott et al, 2001; Fan et al, 2002). Indeed, no other in vitro or in vivo studies have been documented for other variant nucleosome assemblies. In this work, we present both the in vitro and in vivo stability of H2ABbd nucleosomes as determined by analytical ultracentrifugation and fluorescence recovery after photobleaching (FRAP).
Results And Discussion
H2ABbd is able to replace H2A within the nucleosome
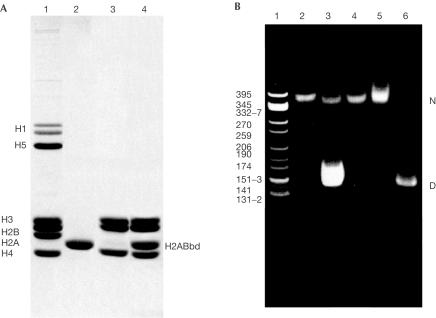
The low identity of H2ABbd primary structure compared with conventional H2A suggests that the incorporation of this histone variant into a histone octamer may probably result in a non-canonical nucleosome with a distinct structure and stability. To test this, we first cloned, expressed and purified H2ABbd to homogeneity (Fig 1A, lane 2). Next, H2ABbd, together with a native H2B, H3 and H4 complement (Fig 1A, lanes 3 and 4), was used to reconstitute nucleosomes onto a 146 bp random sequence DNA fragment isolated from chicken erythrocyte core particles (Fig 1B). The electrophoretic mobility shift assay (EMSA) demonstrates that H2ABbd is efficiently incorporated into the nucleosome core particles (Fig 1B, lanes 3–5), which is in agreement with the available data (Chadwick & Willard, 2001). Indeed, at an appropriate histone:DNA ratio, no free nucleosomal DNA was detected, demonstrating that complete reconstitution was achieved (Fig 1B, lanes 4 and 5).
Figure 1.
The histone variant H2ABbd is able to substitute conventional H2A in the nucleosome particles. (A) PAGE (18%) containing SDS of H2ABbd and the complement conventional histone proteins. Lane 1: chicken erythrocyte histone marker; lane 2: recombinant H2ABbd histone; lane 3: equimolar mixture of H2B, H3 and H4; lane 4: H2ABbd, H2B, H3 and H4 mixture used to reconstitute mononucleosomes. (B) Native gel electrophoresis of H2ABbd-containing mononucleosomes. Lane 1: CfoI digest of pBR 322 used as a DNA molecular marker; lane 2: purified chicken erythrocyte mononucleosomes; lanes 3–5: reconstituted mononucleosomes with H2ABbd. Lane 3 contains an excess of free DNA, lanes 4 and 5 are different loadings of the same sample, and lane 6 represents 146 bp random sequence nucleosomal DNA used as a reconstitution substrate for core particles. N, nucleosome; D, 146 bp DNA used in reconstitution experiments.
Sedimentation analysis of H2ABbd particles
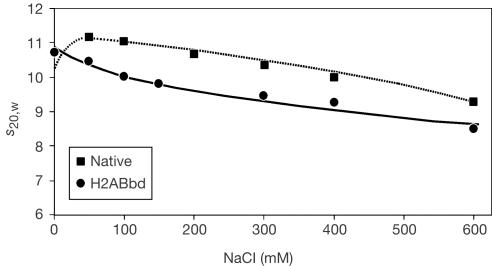
Changes in the conformation of nucleosomes as a result of variations in the ionic strength are informative for determining nucleosome stability in solution (Ausio et al, 1984; Greulich et al, 1985; Angelov et al, 2001). Within the studied range of salt concentrations (⩽0.6 M NaCl), the histone N-termini remain bound to DNA (Ausio et al, 1984; Greulich et al, 1985; Angelov et al, 2001). Such nucleosome conformational transitions may be physiologically relevant because they mimic nucleosome alterations that result from the interactions with transcription factors, chromatin remodelling complexes or RNA polymerases (Aalfs & Kingston, 2000; Abbott et al, 2001). With this in mind, we have characterized the sedimentation behaviour of H2ABbd nucleosome particles at NaCl concentrations between 0 and 600 mM and compared it with purified core particles from chicken erythrocytes (Fig 2). At low salt concentrations (<50 mM NaCl), both types of particles exhibit similar sedimentation coefficients. On raising the ionic strength, this parameter decreases for both particles, but the decrease of s20,W of the H2ABbd nucleosomes is clearly more pronounced (Fig 2). For example, at 200 mM NaCl, s20,W of the H2ABbd nucleosomes is 9.7 S compared with 11 S of conventional nucleosomes. This distinct drop in the sedimentation coefficient for H2ABbd particles is strongly suggestive of a more open structure, which exhibits a higher frictional coefficient compared with the conventional H2A particle. Thus, the increasing ionic strength of the solvent has a differential effect on the conformation of the two particles and suggests a lower inherent stability of the H2ABbd particle.
Figure 2.
Sedimentation profile of nucleosome core particles reconstituted with H2ABbd or a full native histone complement under different ionic strengths (0–600 mM NaCl). The dotted line and squares are the predicted behaviour obtained from native nucleosome core particles (Ausio & van Holde, 1986).
GFP–H2ABbd has a higher mobility than GFP–H2A
The lower stability of H2ABbd nucleosomes reflects a potential perturbation of nucleosomal DNA and/or histone octamer interactions. Recent experiments described core histone exchanges in vivo, confirming a dynamic organization of the nucleosome (Kimura & Cook, 2001). If the structural lability of the reconstituted variant nucleosomes observed in vitro is maintained in vivo, one would expect a more dynamic exchange of H2ABbd compared with H2A.
The kinetics and interactions of a protein with its partners in living cells can be studied through expression of a green fluorescent protein (GFP) fusion construct. This chimaera is visualized by GFP autofluorescence. Irradiation with laser light results in a photobleached area with an observable loss of fluorescence. The subsequent fluorescence recovery kinetics of GFP fusion proteins can be measured by FRAP. Such kinetic experiments directly reflect the mobility of GFP fusion proteins, which in turn provide strong insights into the interaction of the protein with its partners in vivo (Lippincottschwartz et al, 2001).
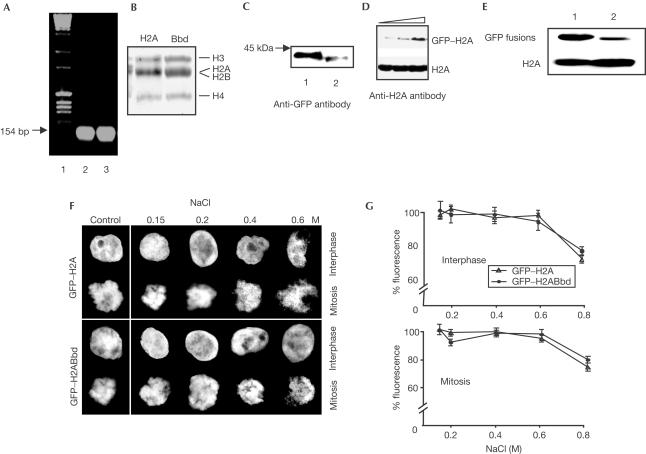
To measure the mobility of H2A and H2ABbd, we first established stable cell lines expressing GFP fused to either conventional H2A or to H2ABbd. In these cell lines, the GFP–histone fusions were assembled into nucleosomes (Fig 3). This was demonstrated as follows. Nuclei were prepared from the stable cell lines and were digested with micrococcal nuclease. Nucleosomes were isolated by centrifugation through a sucrose gradient containing 0.6 M NaCl (Mutskov et al, 1998). At this NaCl concentration, nucleosomes are depleted of linker histones and non-histone proteins; that is, only the histone octamers that were properly organized into nucleosomes were isolated (Fig 3A,B). The presence of GFP fusions within this preparation was demonstrated by using antibodies against GFP. Indeed, western blot analysis clearly shows that GFP–H2ABbd and GFP–H2A are both successfully incorporated in nucleosomes isolated from stable cell lines (Fig 3C). Importantly, the amount of expressed GFP–H2A was much lower (less than 10%) compared with that of the conventional H2A (Fig 3D), and that of GFP–H2ABbd was even lower (compare the signals for GFP–H2A and GFP–H2ABbd relative to those of H2A; Fig 3D,E). Thus, the expression of GFP fusions led to an insignificant increase in the total amount of H2A-type histones and no free GFP–histone fusions should be present. If this is correct, treatment of the cells with increasing NaCl concentrations up to 0.6 M should not release GFP–histone fusions from the cells and the intensity of the GFP signals should remain unchanged (Kimura & Cook, 2001). This was the case for both cell lines expressing the respective GFP–histone fusions (Fig 3F,G). We conclude that no detectable amount of free GFP–H2A or GFP–H2ABbd is present in the stable cell lines. This validates the use of the FRAP technique to measure the mobility of GFP–histone fusions.
Figure 3.
GFP–H2A and GFP–H2ABbd histone fusions are assembled into nucleosomes. (A) Nuclei were isolated from stable A431 human cell lines expressing either GFP–H2A or GFP–H2ABbd, and nucleosomes were prepared as described in Methods. Agarose (1.5%) gel electrophoresis of DNA isolated from nucleosomes prepared from cell lines expressing GFP–H2A (lane 2) or GFP–H2ABbd (lane 3). Lane 1, molecular mass DNA markers. (B) SDS (18%) electrophoresis of histones isolated from nucleosomes prepared from GFP–H2A (lane 1) and GFP–H2ABbd (lane 2) stable cell lines. The positions of the core histones are designated on the right. Note that histones H2A and H2B co-migrate. (C) Western blot of proteins isolated from nucleosomes of cell lines expressing GFP–H2A (lane 1) and GFP–H2ABbd (lane 2). An anti-GFP antibody was used for the detection of GFP–histone fusions. The arrow shows the position of the protein marker with a molecular mass of 45 kDa. (D) GFP–H2A is expressed in trace amounts compared with H2A. Western blot of proteins isolated from cell lines expressing GFP–H2A. Increasing amounts of total cell extract were loaded on 15% SDS gel. An anti-H2A antibody was used for the detection of both H2A and GFP–H2A fusion proteins. (E) Western blot analysis of GFP–H2A and GFP–H2ABbd fusions. Total cell extracts from GFP–H2A (lane 1) or GFP–H2ABbd (lane 2) stable cell lines were loaded on a 15% SDS gel. The blot was cut into two parts. The lower part containing the blotted proteins with molecular masses below 30 kDa was revealed by using an anti-H2A antibody to detect histone H2A. The upper part of the blot was revealed by using an anti-GFP antibody. (F) The amount of GFP–H2A and GFP–H2ABbd fusions in the stable cell lines is not affected after treatment with elevated concentrations of NaCl. Interphase or mitotic cells expressing either GFP–H2A or GFP–H2ABbd were treated with buffered solutions containing NaCl at the concentrations indicated and the cells were imaged by using fluorescence microscopy. (G) Quantification of the data presented in (F). Values represent means and standard deviations for 45 cells. Note that the intensity of the GFP–H2A and GFP–H2ABbd signals did not change after treatment with NaCl concentrations up to 0.6 M.
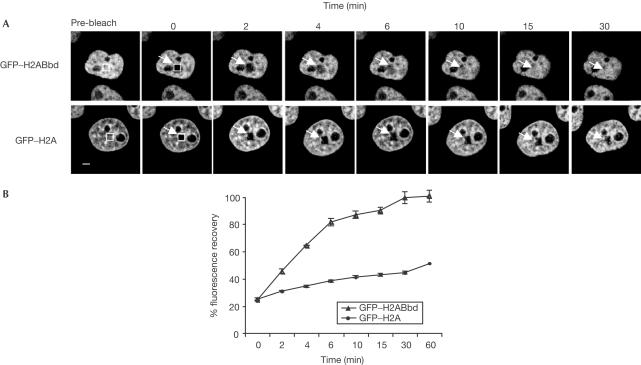
For the FRAP experiments, a small nuclear area was photobleached in single cells of both GFP–H2A and GFP–H2ABbd cell lines, and images were collected at concurrent intervals (Fig 4A). Clearly, the recovery kinetics differed in both samples (Fig 4A,B). Conventional GFP–H2A showed slower recovery, as even 30 min after photobleaching the fluorescence reaches only ⩽40% of its initial value (Fig 4A,B). This result agrees with the available data (Kimura & Cook, 2001). In contrast, the photobleaching recovery for the GFP–H2ABbd fusion was fast, as after 10–15 min an essentially complete recovery was observed (Fig 4A,B). Therefore, GFP–H2ABbd exchanges quicker than GFP–H2A within the nucleosome, an observation that is in agreement with the in vitro biophysical analysis that detected possible perturbations in the H2ABbd nucleosome structure.
Figure 4.
FRAP analysis of GFP–H2A and GFP–H2ABbd proteins. (A) Cells expressing either GFP–H2A or GFP–H2ABbd were imaged before bleaching (pre-bleach) or during recovery after bleaching. Images were recorded at the indicated times post-bleaching. The arrows designate the rectangular bleached area. Scale bar, 2.5 μm. (B) Quantification of the FRAP data. Values represent means and standard deviations for 11 nuclei from three independent experiments.
In conclusion, our data show that the variant H2ABbd nucleosome exhibits lower stability in vitro compared with the conventional nucleosome particle. Importantly, the FRAP experiments demonstrate that in vivo H2ABbd is more rapidly exchanged than conventional H2A histone. In addition, the available data indicate that this histone variant is tightly associated with acetylated euchromatic regions of the genome and possibly with active genes (Chadwick & Willard, 2001). This suggests that H2ABbd nucleosomes might be inherently specialized to enhance localized nucleosome disruption and facilitate transcription.
Methods
Nucleosome reconstitution By using appropriate primers, the H2ABbd histone variant cDNA (Chadwick & Willard, 2001) was PCR-amplified from the human testis Marathon-ready cDNA library, expressed in Escherichia coli and purified to homogeneity (Luger et al, 1999; Angelov et al, 2003). A titration was carried out using SDS–polyacrylamide gel electrophoresis (SDS–PAGE) to ensure that all histones in the final mixture were present in equimolar amounts. The histone mixture thus obtained was dialysed overnight against 2.0 M NaCl, 10 mM Tris (pH 7.5), 10 mM β-mercaptoethanol and 0.1 mM EDTA at 4°C, and was mixed with 146 bp random sequence DNA in the same buffer at a histone:DNA ratio of 1.13:1.0 (w/w). Nucleosome core particle reconstitution was carried out using salt gradient dialysis (Mutskov et al, 1998). The integrity of the core particles was analysed by 4% native PAGE and sedimentation velocity in the analytical ultracentrifuge (see below).
Analytical ultracentrifugation Reconstituted H2ABbd nucleosomes were dialysed against buffers of varying ionic strengths and were subjected to analytical ultracentrifuge analysis as described elsewhere (Ausio et al, 1989). Briefly, sedimentation velocity runs were performed in a Beckman XL-A ultracentrifuge using an An-55 aluminium rotor and doublesector cells with aluminium-filled Epon centerpieces. A value of 0.650 cm3/g was used for the partial specific volume of the nucleosome core particle.
Cell culture, transfection and immunoblotting Human A431 cells were grown on Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Biowhittaker, Europe). Cells were transfected with either GFP–H2A or GFP–H2ABbd constructs using FuGENE 6 reagent (Roche) according to the manufacturer's protocol. Stable transfected cells were selected by gentamicin (500 μg/ml). Cell clones were amplified and their positivity was checked by fluorescence.
Nuclei were isolated from stable A431 human cell lines expressing either GFP–H2A or GFP–H2ABbd. After appropriate digestion with micrococcal nuclease, linker histone-depleted nucleosomes were prepared by using sucrose gradients containing 0.6 M NaCl. Nucleosomal DNA was extracted and analysed by agarose gels and the protein composition of the particles was determined by SDS–PAGE. Alternatively, cells were harvested by trypsinization and lysed in Laemmli sample buffer containing 7 M urea. GFP fusion proteins were then detected by immunoblotting with a monoclonal anti-GFP antibody (BD Clontech). Blots were developed by using the ECL plus western blotting detection system (Amersham Bioscience).
Quantification of the GFP fluorescence of interphase and mitotic cells at different NaCl concentrations Cells were arrested at mitosis by treatment with nocodazole (40 ng/ml final concentration) for 16 h. Both mitotic and interphase cells were washed with PBS. After incubation at 23°C for 10 min with PBS-T-G buffer (PBS, 0.1% Triton X-100, 50% glycerol) containing NaCl at different concentrations, they were fixed with 4% paraformaldehyde. The cells used as a control were fixed immediately after washing with PBS. Fluorescence intensities were quantified by using home-made software. The intensity of fluorescence of the NaCl-treated cells was presented as a ratio (in percentages) to the fluorescence intensity of the control cells.
In vivo experiments: FRAP Cells were grown on Lab-Tek chambered cover glass (Nalge Nunc International). For imaging, cells were maintained at 37°C on a temperature-controlled stage while medium was buffered with Hepes (10 mM, pH 7.5). Photobleaching and confocal microscopy were performed on a ZEISS LSM510 laser scanning confocal apparatus using a PlanApochromat × 40 water immersion objective. GFP was excited with a 488-nm Argon2 laser (power varying from 0.1 to 1%). FRAP experiments were carried out as follows: outlined regions were bleached by ten iterations of a full-power laser and subsequently recovery was monitored for about 1 h. Bleaching owing to the acquisition was corrected and was less than 10% in all experiments.
References
- Aalfs JD, Kingston RE (2000) What does ‘chromatin remodeling' mean? Trends Biochem Sci 25: 548–555 [DOI] [PubMed] [Google Scholar]
- Abbott DW, Ivanova VS, Wang X, Bonner WM, Ausio J (2001) Characterization of the stability and folding of H2A.Z chromatin particles: implications for transcriptional activation. J Biol Chem 276: 41945–41949 [DOI] [PubMed] [Google Scholar]
- Angelov D, Vitolo JM, Mutskov V, Dimitrov S, Hayes JJ (2001) Preferential interaction of the core histone tail domains with linker DNA. Proc Natl Acad Sci USA 98: 6599–6604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelov D, Molla A, Perche PY, Hans F, Cote J, Khochbin S, Bouvet P, Dimitrov S (2003) The histone variant macroH2A interferes with transcription factor binding and SWI/SNF nucleosome remodeling. Mol Cell 11: 1033–1041 [DOI] [PubMed] [Google Scholar]
- Ausio J, Abbott DW (2002) The many tales of a tail: carboxyl-terminal tail heterogeneity specializes histone H2A variants for defined chromatin function. Biochemistry 41: 5945–5949 [DOI] [PubMed] [Google Scholar]
- Ausio J, van Holde KE (1986) Histone hyperacetylation: its effects on nucleosome conformation and stability. Biochemistry 25: 1421–1428 [DOI] [PubMed] [Google Scholar]
- Ausio J, Seger D, Eisenberg H (1984) Nucleosome core particle stability and conformational change. Effect of temperature, particle and NaCl concentrations, and crosslinking of histone H3 sulfhydryl groups. J Mol Biol 176: 77–104 [DOI] [PubMed] [Google Scholar]
- Ausio J, Dong F, van Holde KE (1989) Use of selectively trypsinized nucleosome core particles to analyze the role of the histone ‘tails' in the stabilization of the nucleosome. J Mol Biol 206: 451–463 [DOI] [PubMed] [Google Scholar]
- Bassing CH, Suh H, Ferguson DO, Chua KF, Manis J, Eckersdorff M, Gleason M, Bronson R, Lee C, Alt FW (2003) Histone H2AX: a dosage-dependent suppressor of oncogenic translocations and tumors. Cell 114: 359–370 [DOI] [PubMed] [Google Scholar]
- Celeste A, Fernandez-Capetillo O, Kruhlak MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM, Nussenzweig A (2003) Histone H2AX phosphorylation is dispensable for the initial recognition of DNA breaks. Nat Cell Biol 5: 675–679 [DOI] [PubMed] [Google Scholar]
- Chadwick BP, Willard HF (2001) A novel chromatin protein, distantly related to histone H2A, is largely excluded from the inactive X chromosome. J Cell Biol 152: 375–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson MJ, Wells JR, Gibson F, Saint R, Tremethick DJ (1999) Regions of variant histone His2AvD required for Drosophila development. Nature 399: 694–697 [DOI] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR (1998) Histone macroH2A1 is concentrated in the inactive X chromosome of female mammals. Nature 393: 599–601 [DOI] [PubMed] [Google Scholar]
- Dhillon N, Kamakaka RT (2000) A histone variant, Htz1p, and a Sir1p-like protein, Esc2p, mediate silencing at HMR. Mol Cell 6: 769–780 [DOI] [PubMed] [Google Scholar]
- Faast R, Thonglairoam V, Schulz TC, Beall J, Wells JR, Taylor H, Matthaei K, Rathjen PD, Tremethick DJ, Lyons I (2001) Histone variant H2A.Z is required for early mammalian development. Curr Biol 11: 1183–1187 [DOI] [PubMed] [Google Scholar]
- Fan JY, Gordon F, Luger K, Hansen JC, Tremethick DJ (2002) The essential histone variant H2A.Z regulates the equilibrium between different chromatin conformational states. Nat Struct Biol 9: 172–176 [DOI] [PubMed] [Google Scholar]
- Greulich KO, Ausio J, Eisenberg H (1985) Nucleosome core particle structure and structural changes in solution. J Mol Biol 186: 167–173 [DOI] [PubMed] [Google Scholar]
- Kimura H, Cook PR (2001) Kinetics of core histones in living human cells: little exchange of H3 and H4 and some rapid exchange of H2B. J Cell Biol 153: 1341–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larochelle M, Gaudreau L (2003) H2A.Z has a function reminiscent of an activator required for preferential binding to intergenic DNA. EMBO J 22: 4512–4522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincottschwartz J, Snapp E, Kenworthy A (2001) Studying protein dynamics in living cells. Nat Rev Mol Cell Biol 2: 444–456 [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Richmond TJ (1999) Expression and purification of recombinant histones and nucleosome reconstitution. Methods Mol Biol 119: 1–16 [DOI] [PubMed] [Google Scholar]
- Meneghini MD, Wu M, Madhani HD (2003) Conserved histone variant H2A.Z protects euchromatin from the ectopic spread of silent heterochromatin. Cell 112: 725–736 [DOI] [PubMed] [Google Scholar]
- Mermoud JE, Costanzi C, Pehrson JR, Brockdorff N (1999) Histone macroH2A1.2 relocates to the inactive X chromosome after initiation and propagation of X-inactivation. J Cell Biol 147: 1399–1408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutskov V, Gerber D, Angelov D, Ausio J, Workman J, Dimitrov S (1998) Persistent interactions of core histone tails with nucleosomal DNA following acetylation and transcription factor binding. Mol Cell Biol 18: 6293–6304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perche P, Vourch C, Souchier C, Robert-Nicoud M, Dimitrov S, Khochbin C (2000) Higher concentrations of histone macroH2A in the Barr body are correlated with higher nucleosome density. Curr Biol 10: 1531–1534 [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM (1998) DNA doublestranded breaks induce histone H2AX phosphorylation on serine 139. J Biol Chem 273: 5858–5868 [DOI] [PubMed] [Google Scholar]
- Rogakou EP, Boon C, Redon C, Bonner WM (1999) Megabase chromatin domains involved in DNA doublestrand breaks in vivo. J Cell Biol 146: 905–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santisteban MS, Kalashnikova T, Smith MM (2000) Histone H2A.Z regulates transcription and is partially redundant with nucleosome remodeling complexes. Cell 103: 411–422 [DOI] [PubMed] [Google Scholar]
- Stargell LA, Bowen J, Dadd CA, Dedon PC, Davis M, Cook RG, Allis CD, Gorovsky MA (1993) Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes Dev 7: 2641–2651 [DOI] [PubMed] [Google Scholar]
- Suto RK, Clarkson MJ, Tremethick DJ, Luger K (2000) Crystal structure of a nucleosome core particle containing the variant histone H2A.Z. Nat Struct Biol 7: 1121–1124 [DOI] [PubMed] [Google Scholar]
- Tsanev R, Russev G, Pashev I, Zlatanova J (1993) Replication and Transcription of Chromatin. Boca Raton, FI: CRC Press [Google Scholar]
- van Daal A, Elgin SC (1992) A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol Biol Cell 3: 593–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Holde K (1988) Chromatin. Berlin: Springer [Google Scholar]
- White EM, Gorovsky MA (1988) Localization and expression of mRNA for a macronuclearspecific histone H2A variant (hv1) during the cell cycle and conjugation of Tetrahymena thermophila. Mol Cell Biol 8: 4780–4786 [DOI] [PMC free article] [PubMed] [Google Scholar]