Summary
A second member of the human APOBEC3 family is a Vif-blockable innate antiretroviral factor
Keywords: retroviruses, HIV, innate antivirals, APOBEC
The importance of innate intracellular immunity as a mechanism of protection against viruses is increasingly being recognized. For example, the Friend virus susceptibility 1 (Fv1) gene restricts the sensitivity of mice to the murine leukaemia virus (MLV; Best et al, 1996), and the cytoplasmic body-component tripartite-motif 5α (TRIM5-α) limits the susceptibility of non-human primates to infection by the human immunodeficiency virus (HIV; Stremlau et al, 2004). Inevitably, viruses have evolved strategies to overcome these obstacles. The virion-infectivity factor (Vif) protein of lentiviruses counters the antiviral action of the cellular enzyme apolipoprotein B mRNA-editing enzyme catalytic-polypeptide 3G (APOBEC3G; Sheehy et al, 2002). APOBEC3G belongs to the cytidine deaminase superfamily, the founding member of which is APOBEC1. Another well-characterized relative is activation-induced deaminase (AID), which is a B-cell protein that has an essential role in antibody diversification through governing somatic hypermutation, gene conversion and classswitch recombination at the immunoglobulin loci (Neuberger et al, 2003).
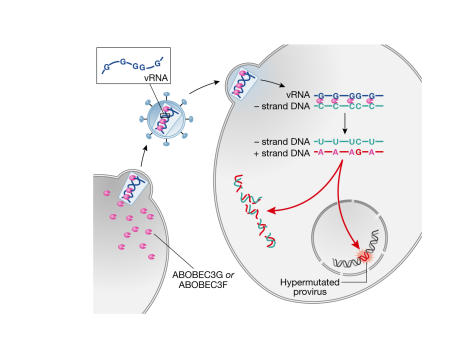
APOBEC3G is expressed notably in T lymphocytes and macrophages, which are the main targets of HIV. In the absence of Vif, it is packaged into virions during viral assembly and acts during reverse transcription to deaminate deoxycytidine residues to deoxyuridine (dU) in the growing minus-strand viral DNA (Sheehy et al, 2002; Harris et al, 2003; Lecossier et al, 2003; Mangeat et al, 2003; Zhang et al, 2003). These dU-rich transcripts are either degraded or yield G-to-A hypermutated proviruses that are largely non-functional (Fig 1). Vif fends off the attack from the host by binding to the cytidine deaminase, which prevents its virion incorporation and triggers its degradation in proteasomes (Conticello et al, 2003; Kao et al, 2003; Mariani et al, 2003; Marin et al, 2003; Sheehy et al, 2003; Stopak et al, 2003; Yu et al, 2003; Liu et al, 2004; Mehle et al, 2004). In the absence of Vif, APOBEC3G is active against a broad range of retroviruses and can also block the hepatitis B virus (HBV; Harris et al, 2003; Mangeat et al, 2003; Turelli et al, 2004).
Figure 1.
Innate immunity by deamination. Apolipoprotein B mRNA-editing enzyme catalytic-polypeptide 3G (APOBEC3G) and APOBEC3F are incorporated into nascent retroviral particles through as yet unknown mechanisms. During reverse transcription, they deaminate deoxycytidine residues to deoxyuridine (dU) in the minus-strand viral DNA. These dU-rich transcripts are either degraded by DNA-repair pathways or yield hypermutated proviruses. In the presence of Vif, the antiviral factors are sequestered and degraded in virus-producing cells.
Although G-to-A hypermutation, which is the consequence of C-to-U changes in the minus-strand DNA, has been noted in HIV-1 field isolates, not every occurrence can be attributed to APOBEC3G. Indeed, the first G of both GA and GG dinucleotides is commonly mutated, whereas APOBEC3G shows a marked preference for editing the second C of CC but not TC (Beale et al, 2004). In the 16 June issue of The EMBO Journal, Wiegand and colleagues have explained this conundrum by revealing that human T lymphocytes also produce APOBEC3F, which is a close relative of APOBEC3G (Wiegand et al, 2004). This enzyme has similar Vif-blockable anti-HIV properties, but favours TC as its target. In the same month, Zheng and colleagues also reported on the antiviral action of APOBEC3F in the Journal of Virology, and showed that APOBEC3F, like its sister protein, undergoes proteasomal degradation in the presence of HIV-1 Vif (Zheng et al, 2004). Interestingly, both APOBEC3F and APOBEC3G are expressed in the T-lymphoid cell lines that are most commonly used to study Vif function, which probably explains why RNA-interference-based evidence of the antiviral role of APOBEC3G was still missing from the otherwise prolific literature on this enzyme.
To act as antiretrovirals, APOBEC3F and APOBEC3G must be incorporated into particles. The determinants of this event are not yet known, but the fact that they apply to both of these proteins might facilitate their identification. In addition, the two antiviral APOBEC3 proteins will be able to provide information on the mechanism of Vif action. Aspartate at position 128 of APOBEC3G is crucial for Vif binding and constitutes an important barrier to the cross-species transmission of lentiviruses. Indeed, this is the binding site for Vif from HIV-1 and HIV-2, but not from simian immuno-deficiency virus from the African green monkey (SIVAGM), so this virus is restricted to human cells. Reciprocally, APOBEC3G from the African green monkey, which harbours a lysine at position 128, is recognized by Vif from SIVAGM but not HIV; it therefore inhibits the latter but not the former virus. Residue 128 in APOBEC3F is glutamate, and if this amino acid is introduced into APOBEC3G, it is compatible with HIV-1 but not SIVAGM Vif binding (Schrofelbauer et al, 2004). Like APOBEC3G, APOBEC3F must therefore contribute to the speciesspecific tropism of primate lentiviruses.
In the mouse, there is only one APOBEC3 orthologue and it is endowed with antiretroviral activity (Mariani et al, 2003). In humans, the APOBEC3 family has seven members (designated APOBEC3A through to APOBEC3G), which are clustered next to each other in the same orientation on chromosome 22 and probably evolved through consecutive duplications of a common ancestor (Jarmuz et al, 2002). APOBEC3E seems to be a pseudogene, whereas the other members of the family are expressed in various tissue-restricted ways. APOBEC3G and APOBEC3F are located some 25,000 base pairs apart and seem to be largely co-regulated, possibly through the sharing of common transcriptional control elements.
APOBEC1 edits the apolipoprotein B mRNA at a precise position, whereas the other APOBEC family members that have been characterized so far—including AID—target single-stranded DNA and act in a less sequence-specific manner. With these new studies, substrates have now been identified for two members of the human APOBEC3 group and in both cases, these substrates are of viral origin. The full extent of the APOBEC3F antiviral potential remains to be determined; however, an even more interesting question is whether either of these two cytidine deaminases can also edit cell-derived sequences. The fact that they are constitutively expressed rather than being virus induced suggests that this is the case. Considering their affinity for reverse transcripts, they might act on endogenous retroelements. These elements, whether endogenous retroviruses or non-long terminal repeat (LTR) retrotransposons, constitute up to 40% of the human genome. Although their contribution as facilitators of genome evolution is unquestionable, it is plausible that their retrotransposition activity is limited by innate mechanisms of the host. However, such a function would not fit with the narrow tissue distributions of APOBEC3F and APOBEC3G, which are expressed mainly in immune cells such as T and B lymphocytes and, at least for APOBEC3G, macrophages. This raises the intriguing possibility that there is an editing substrate that is essential for the function of all three of these cellular arms of the immune response. This warrants efforts aimed at identifying the cellular targets of APOBEC3G and APOBEC3F, and at determining whether these antiviral cytidine deaminases have a role beyond the blockade of exogenous retroelements.
References
- Beale RC, Petersen-Mahrt SK, Watt IN, Harris RS, Rada C, Neuberger MS (2004) Comparison of the differential context-dependence of DNA deamination by APOBEC enzymes: correlation with mutation spectra in vivo. J Mol Biol 337: 585–596 [DOI] [PubMed] [Google Scholar]
- Best S, Le Tissier P, Towers G, Stoye JP (1996) Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382: 826–829 [DOI] [PubMed] [Google Scholar]
- Conticello SG, Harris RS, Neuberger MS (2003) The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr Biol 13: 2009–2013 [DOI] [PubMed] [Google Scholar]
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH (2003) DNA deamination mediates innate immunity to retroviral infection. Cell 113: 803–809 [DOI] [PubMed] [Google Scholar]
- Jarmuz A, Chester A, Bayliss J, Gisbourne J, Dunham I, Scott J, Navaratnam N (2002) An anthropoidspecific locus of orphan C to U RNA editing enzymes on chromosome 22. Genomics 79: 285–296 [DOI] [PubMed] [Google Scholar]
- Kao S, Khan MA, Miyagi E, Plishka R, Buckler-White A, Strebel K (2003) The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J Virol 77: 11398–11407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecossier D, Bouchonnet F, Clavel F, Hance AJ (2003) Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science 300: 1112. [DOI] [PubMed] [Google Scholar]
- Liu B, Yu X, Luo K, Yu Y, Yu XF (2004) Influence of primate lentiviral Vif and proteasome inhibitors on human immunodeficiency virus type 1 virion packaging of APOBEC3G. J Virol 78: 2072–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D (2003) Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424: 99–103 [DOI] [PubMed] [Google Scholar]
- Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR (2003) Speciesspecific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114: 21–31 [DOI] [PubMed] [Google Scholar]
- Marin M, Rose KM, Kozak SL, Kabat D (2003) HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat Med 9: 1398–1403 [DOI] [PubMed] [Google Scholar]
- Mehle A, Strack B, Ancuta P, Zhang C, McPike M, Gabuzda D (2004) Vif overcomes the innate antiviral activity of APOBEC3G by promoting its degradation in the ubiquitin-proteasome pathway. J Biol Chem 279: 7792–7798 [DOI] [PubMed] [Google Scholar]
- Neuberger MS, Harris RS, Di Noia J, Petersen-Mahrt SK (2003) Immunity through DNA deamination. Trends Biochem Sci 28: 305–312 [DOI] [PubMed] [Google Scholar]
- Schrofelbauer B, Chen D, Landau NR (2004) A single amino acid of APOBEC3G controls its speciesspecific interaction with virion infectivity factor (Vif). Proc Natl Acad Sci USA 101: 3927–3932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Choi JD, Malim MH (2002) Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418: 646–650 [DOI] [PubMed] [Google Scholar]
- Sheehy AM, Gaddis NC, Malim MH (2003) The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat Med 9: 1404–1407 [DOI] [PubMed] [Google Scholar]
- Stopak K, de Noronha C, Yonemoto W, Greene WC (2003) HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol Cell 12: 591–601 [DOI] [PubMed] [Google Scholar]
- Stremlau M, Owens CM, Perron MJ, Kiessling M, Autissier P, Sodroski J (2004) The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427: 848–853 [DOI] [PubMed] [Google Scholar]
- Turelli P, Mangeat B, Jost S, Vianin S, Trono D (2004) Inhibition of hepatitis B virus replication by APOBEC3G. Science 303: 1829. [DOI] [PubMed] [Google Scholar]
- Wiegand HL, Doehle BP, Bogerd HP, Cullen BR (2004) A second human antiretroviral factor, APOBEC3F, is suppressed by the HIV-1 and HIV-2 Vif proteins. EMBO J doi:10.1038/sj.emboj.7600246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF (2003) Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif–Cul5–SCF complex. Science 302: 1056–1060 [DOI] [PubMed] [Google Scholar]
- Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L (2003) The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424: 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng YH, Irwin D, Kurosu T, Tokunaga K, Sata T, Peterlin BM (2004) Human APOBEC3F is another host factor that blocks human immunodeficiency virus type 1 replication. J Virol 78: 6073–6076 [DOI] [PMC free article] [PubMed] [Google Scholar]