Abstract
We present data on the genetic diversity and phylogenetic affinities of N2-fixing unicellular cyanobacteria in the plankton of the tropical North Atlantic Ocean. Our dinitrogenase gene (nifH) sequences grouped together with a group of cyanobacteria from the subtropical North Pacific; another subtropical North Pacific group was only distantly related. Most of the 16S ribosomal DNA sequences from our tropical North Atlantic samples were closely allied with sequences from a symbiont of the diatom Climacodium frauenfeldianum. These findings suggest a complex pattern of evolutionary and ecological divergence among unicellular cyanobacteria within and between ocean basins.
Oligotrophic marine ecosystems are typically limited in their concentrations of reduced forms of nitrogen (10, 11, 19), and diazotrophic (N2-fixing) cyanobacteria appear to be at an advantage (4, 29). Among the marine phytoplankton, a large array of symbiotic, colonial, and unicellular diazotrophic cyanobacteria have been reported (4, 7, 27), and species in the colony-forming genus Trichodesmium appear to be the dominant diazotrophs in tropical and subtropical waters (3, 5, 6). However, present calculations of N2 fixation rates by known diazotrophic cyanobacteria (i.e., Trichodesmium sp.) cannot account for all of the reduced forms of N necessary to balance the global nitrogen budget (5, 10, 11). Among the unicellular forms, N2 fixation has been shown in strains of Synechococcus sp. (14, 15, 23), Cyanothece sp. (19, 21, 22), and Gloeocapsa sp. (17). These unicellular cyanobacteria lack heterocysts and are able to fix carbon and nitrogen by separating the two processes temporally: they photosynthesize during the day and fix nitrogen during the night in what seems to be a well-defined circadian rhythm (9, 12). Recent evidence suggests a significant role for unicellular cyanobacterial N2 fixers in the subtropical North Pacific (29).
During a research cruise in the tropical North Atlantic, we encountered a high abundance of unicellular cyanobacteria that were genetically distinct (nifH) from those previously reported for the subtropical North Pacific (28, 29). We also characterized cyanobacterial genetic diversity and measured the nitrogenase activities of enrichment cultures.
Samples were collected on a cruise aboard the research vessel Knorr between 25 July and 18 August 2001. Water samples from a range of depths were collected by using a conductivity, temperature, density-rosette system at stations 27 (11°46′16"N, 53°38′82"W), 30 (8°26′88"N, 49°50′55"W), 31 (6°17′12"N, 47°48′66"W), 33 (4°42′00"N, 42°50′00"W), 44 (7°11′12"N, 52°25′76"W), and 53 (11°40′50"N, 54°0.2′50"W). Water from 10-liter Niskin bottles was prefiltered through a 10-μm-pore-size PCTE membrane to remove larger plankton. Depth profiles for physical properties and nutrient concentrations are part of the biocomplexity project (http://biology.usc.edu/bc/) funded by the National Science Foundation.
Four liters of filtrate was concentrated, and the cells were resuspended in a nitrogen-free culture medium as described previously (24). Cultures were initially incubated in a flowing-seawater bath on deck under neutral-density screening at 0.1% of ambient irradiance and later transferred to a laboratory incubator at 26 to 29°C and 50 to 100 microeinsteins m−2 s−1, with a cycle of 14 h of light and 10 h of darkness. Culture aliquots were plated, and individual colonies were resuspended in SO medium. Cell abundance and appearance were monitored with a compound epifluorescence microscope, and 3 ml of culture was transferred into 50 ml of SO medium every 3 weeks. All of the cultures (CL1.2 from station 27, CL7.2 from station 33, and CL13.3 from station 44) are available from the corresponding author.
The procedure used for extraction of DNA is that of Madrid et al. (13) with modifications. Four liters of filtrate was concentrated onto 0.22-μm-pore-size Durapore membrane filters (47-mm diameter; Millipore Corporation, San Jose, Calif.) that were then shredded and placed in 15 ml of DNA extraction buffer (20 mM Tris-HCl [pH 7.5 to 8.2], 50 mM EDTA, 20 mM NaCl) and frozen at −20°C until extraction. Approximately 50 ml containing 107 cells from concentrated cultures (including WH8501 donated by J. Waterbury, Woods Hole Oceanographic Institution [WHOI]) were centrifuge pelleted at 4,000 × g for 10 min, resuspended in 10 times the pellet volume of DNA extraction buffer, and maintained overnight in a shaking incubator at 50°C.
Samples were thawed and transferred to 50-ml polystyrene centrifuge tubes, 10% sodium dodecyl sulfate was added to a 1% final concentration, and proteinase K solution (10 mg ml−1) (Gibco, Carlsbad, Calif.) was added to a final concentration of 0.5 mg/ml−1. The mixture was incubated for 10 min at 50°C and frozen again for 15 min; this freeze-thaw cycle was repeated twice, and the sample was vortexed each time to facilitate cell lysis. DNA was extracted with phenol (twice) and chloroform (once) and precipitated with ethanol-sodium acetate under standard protocols (21).
PCR was used to amplify a conserved 16S ribosomal DNA (rDNA) cyanobacterial region (approximately 422 bp) with the CYA359F and CYA781bR cyanobacterial primers designed by Nübel et al. (16) and the nifH gene fragment (359 bp), with primers designed by Zehr and McReynolds (26). Reaction mixtures (50-μl final volume) contained the following: 33.5 μl of RNA- and DNA-free water, 5 μl of PCR buffer, 5 μl of MgCl2 (25 mM), 1 μl of a deoxynucleoside triphosphate mix, 2 μl of each primer (12.5 μM), 1 μl of the template, and 0.5 μl of Taq DNA polymerase (Promega, Madison, Wis.). The thermal cycle for amplification of 16S rDNA was as follows: 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min for 35 cycles. The thermal cycle for amplification of nifH was as follows: 93°C for 1.2 min, 50°C for 1 min, and 70°C for 1.5 min for 30 cycles. PCR products were separated according to size by using electrophoresis on a 1.8% agarose-Tris-borate-EDTA gel and purified with a QIAquick PCR purification kit (QIAGEN, Valencia, Calif.).
In order to detect nifH gene expression, RNA was extracted and cDNA was amplified according to the protocol of Zani et al. (25), with modifications. Two liters of filtrate was concentrated onto 0.22-μm-pore-size Durapore membrane filters (25-mm diameter; Millipore) at different depths during day and night casts. RNA was extracted with a ball-mill homogenizer and an RNeasy kit; residual amounts of DNA were removed with an RNase-free DNase set (QIAGEN). An Access reverse transcriptase PCR (RT-PCR) kit (Promega) was used to amplify cDNA for nifH genes according to the instructions of the manufacturer, and the primers of Zehr and McReynolds were used (26). In order to check for DNA contamination in RNA preparations, PCRs were performed without a prior RT step. No PCR product was detected, indicating that RT-PCR amplification products were derived from nifH transcripts. Amplified products were screened by electrophoresis on a 1.8% agarose-Tris-borate-EDTA minigel, and correct-size fragments were excised from the gel and purified from agarose with the QIAquick PCR purification kit. (QIAGEN).
Purified PCR products generated from environmental samples and cultures were ligated with the pGEM-T Easy vector system (Promega) according to the suggestions of the manufacturer and by following standard protocols (20). White colonies were restreaked onto new plates and PCR screened by using vector primers Sp6 and T7 (Promega).
Purified amplified products were sequenced with an ABI Prism BigDye terminator cycle sequencing ready-reaction kit (Perkin-Elmer, Foster City, Calif.) according to the manufacturer's protocols, with modifications. One microliter of each was loaded onto membrane combs (Gel Company, San Francisco, Calif.) and sequenced on an ABI 377 automated sequencer.
Forward and reverse sequences from each cloned insert were aligned and vector sequences were trimmed with Sequencher 3.1.1 (Gene Codes Corporation, Ann Arbor, Mich.). Resulting sequences were used to search the GenBank database, and the closest matches were downloaded and aligned with our clones to provide reference and outgroup sequences for phylogenetic reconstruction analyses. Published cyanobacterial 16S rDNA and nifH sequences from earlier studies were also downloaded and included in their respective data sets. The alignments were verified with MacClade 4.033 PCC software (Sinauer Associates Inc., Sunderland, Mass.), and PAUP∗ 4.0b8 (Sinauer Associates Inc.) was used for phylogenetic analysis. The resulting 31-taxon nifH data set included 392 characters, of which 147 were parsimony informative. The 16S rDNA data set of 30 reference taxons and 20 unknown taxons (see below) included 331 characters, of which 121 were parsimony-informative. Phylograms for the nifH gene were constructed by a maximum-parsimony bootstrap analysis with 250 replicates, random-addition initial trees, terminate branch and reconnect (TBR) branch rearrangement, and a grouping cutoff of 60%. For the 16S rDNA analysis we conducted a maximum-parsimony bootstrap analysis (250 replicates, random addition, TBR rearrangement) for individual clones and compared them to published sequences of known species in order to independently characterize the relationship of each of our unknown species to known species (1). We then constructed a summary tree indicating the range of bootstrap values for individual branches. For comparison with the parsimony-based results, we conducted a neighbor-joining distance (Kimura two-parameter) bootstrap analysis of both data sets in full and a maximum-likelihood bootstrap analysis of a reduced nifH data set (15 taxa, including half of the available sequences for each of the previously identified cyanobacterial clades, plus representative outgroups) using PAUP∗. A matrix of genetic distances (Kimura two parameters) for all pairwise comparisons was generated in PAUP∗; the mean distance and standard deviation were calculated from the matrix with Microsoft Excel.
The rates of nitrogenase activity were determined for three concentrated cultures of unicellular cyanobacteria (CL1.2, CL7.2, and CL13.3) by using the acetylene (C2H2) reduction technique (2, 18). Four milliliters of culture was added to 7-ml serum vials in three replicates per culture. Next, 300 μl of C2H2 generated by the reaction of calcium carbide (CaC2) and water was injected in the vials. Small (100-μl) gas samples were withdrawn every 4 h and injected into a gas chromatograph for the measurement of ethylene (C2H4) production. These assays were done throughout the light-dark cycle of the incubators, and C2H4 production was registered by using an integrator.
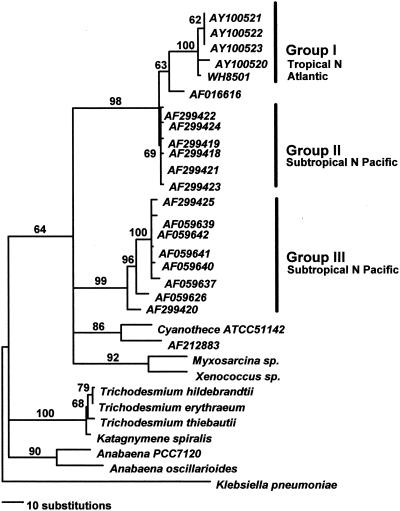
Most of our tropical North Atlantic nifH cDNA and DNA clones had sequences closely related to each other (>99% on a DNA level and 100% on an amino acid level), which grouped with a cultivated diazotrophic unicellular cyanobacterium isolated from the tropical North Atlantic, WH8501 (24) (group I) (Fig. 1). A sequence previously derived from the subtropical North Atlantic (GenBank accession number AF016616) (29) was basal to the clade of tropical North Atlantic sequences (Fig. 1). Group II is a set of unicellular cyanobacterial nifH sequences from the subtropical North Pacific (29) that group with group I sequences from the tropical North Atlantic at a high bootstrap support value (98%) (Fig. 1). Group III includes additional cyanobacterial sequences from the subtropical North Pacific (28, 29) that group together with high bootstrap support (99%) (Fig. 1) and are distinct from group I and II sequences from the tropical North Atlantic and subtropical North Pacific. The mean genetic distances (± standard deviations) for all nifH pairwise comparisons within groups were relatively small: 0.008 ± 0.003 (group I), 0.005 ± 0.0004 (group II), and 0.03 ± 0.004 (group III). The distances between groups I and III and groups II and III were much larger: 0.23 ± 0.002 (between groups I and III) and 0.22 ± 0.001 (between groups II and III). The genetic divergence between these sequences suggests that the tropical North Atlantic cyanobacteria that we sequenced (plus WH8501 and the cyanobacterium with accession number AF016616) and one set of cyanobacteria from the subtropical North Pacific are more closely related to each other than either is to the other set of subtropical North Pacific cyanobacteria (group III). Distance and maximum-likelihood-based analyses (data not shown) gave topologies nearly identical to results of the parsimony analyses, resulting in the same group I, II, and III relationships noted above.
FIG. 1.
Phylogram showing the relationships among different cyanobacterial nifH gene sequences constructed through maximum-parsimony bootstrap analysis (details in text). Fragments are approximately 359 bp in length.
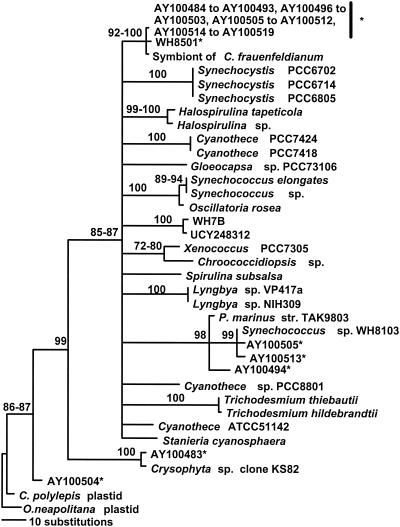
We created 16S rDNA libraries using cyanobacterium-specific primers for stations 30, 31, and 53. A summary phylogram for 16S rDNA sequences found in these libraries is shown (Fig. 2). Of a total of 190 clones in the three libraries, 99% contained 16S rDNA sequences of unknown free-living unicellular cyanobacteria, related to a symbiont of the planktonic diatom Climacodium frauenfeldianum. The symbiosis between C. frauenfeldianum and cyanobacteria has been previously described for both the tropical Atlantic and Pacific oceans (8). The close relationship of this group of free-living unicellular cyanobacteria with the symbiont sheds light on a novel and very poorly understood group of microorganisms that may be major contributors to N2 fixation both in symbiosis and in free- living Cyanothece spp. Other close relatives of these clones included WH8501 (24) and Cyanothece sp. strain ATCC 51142 (19), two strains of known diazotrophic unicellular cyanobacteria, the first of which was isolated from the same region of the tropical Atlantic where we carried out this research. The diazotrophic unicellular cyanobacteria in our enrichment cultures cluster with free-living C. frauenfeldianum symbiont-like cyanobacteria (Fig. 2). The 16S rDNA sequences of our isolates were nearly identical to each other, evidencing only a few independent 1-base differences, which may either reflect a minor divergence between isolates or result from the low fidelity of the Taq polymerase.
FIG. 2.
Summary phylogenetic reconstruction among cyanobacterial 16S rDNA sequences created through maximum-parsimony bootstrap analysis (details in text). Sequences marked with an asterisk were determined by this study, and fragments are approximately 422 bp in length.
Other cyanobacteria and chloroplast sequences were found in our libraries. 16S rDNA sequences, which clustered with Synechococcus sp. strain WH8103, were found in station 30 (15 m) and 31 (22 m) libraries (five and three clones, respectively). 16S rDNA sequences phylogenetically affiliated with Prochlorococcus marinus TAK983-2 were found in station 31 (22 m, three clones). The station 30 (15 m) library also contained 16 rDNA sequences affiliated with an uncultured stramenopile (clone KS82) from the tropical Pacific (one clone) and with a plastid of the prymnesiophyte Chrysochromulina polylepis (one clone). Nevertheless, most sequences derived from our 16S rDNA libraries were very similar: the mean genetic distance for all pairwise comparisons between our 16S rDNA clones was 0.008 substitution per nucleotide.
Given the nifH results reported above, it would be of great interest to analyze 16S rDNA sequences from the group II and group III subtropical North Pacific cyanobacteria characterized previously (29). However, given the relatively conservative nature of variation for 16S rDNA, further determination of the levels of divergence of cyanobacteria from within- and between-oceanic regions by use of 16S rDNA sequences may be difficult.
Since nitrogenase is extremely sensitive to oxygen (18), unicellular autotrophs like members of the group of cyanobacteria found in our samples are likely to fix carbon during the day and N2 at night, alternating both metabolic pathways under a circadian rhythm, as shown for other strains of unicellular cyanobacterial diazotrophs (9, 11, 14, 15, 17, 19, 21, 22, 23). In fact, we detected nifH gene transcripts (mRNA) of unicellular cyanobacteria only at night whereas nifH genes (DNA) were easily detectable at the same stations (stations 27, 30, and 31) both day and night (Table 1).
TABLE 1.
Stations and depths where nifH gene (cDNA and DNA) clones were detecteda
| Station | Depth (m) | No. of nifH cDNA clones obtained
|
No. of nifH DNA clones obtained
|
||
|---|---|---|---|---|---|
| Night | Day | Night | Day | ||
| 27 | 9 | 3 | 0 | 7 | 3 |
| 27 | 22 | 5 | 0 | 3 | 4 |
| 30 | 15 | 5 | 0 | 5 | 6 |
| 30 | 60 | 3 | 0 | 6 | 3 |
| 31 | 22 | 7 | 0 | 5 | 0 |
All of the cDNA clones were obtained during the night. For all clones, cDNAs and DNA for nifH were related to cyanobacterial nifH sequences.
Cultures of diazotrophic unicellular cyanobacteria were obtained for three stations from the tropical Atlantic Ocean: 27 (22 m), 33 (15 m), and 44 (13 m). The cultures were named CL1.2, CL7.2, and CL13.3, respectively. All cells in the cultures were small, coccoid, unicellular forms and measured 2.5 μm in diameter. Mean cell abundance in cultures ranged from 18 × 104 to 22 × 104 cells ml−1 for concentrated cultures, which were used for the nitrogenase activity assay. The highest rates of N2 fixation typically occurred around 02:00, 6 h after the beginning of the dark period, and ranged from 145 ± 69 to 115 ± 30 (means ± standard deviations) pmol of C2H4 ml−1 h−1. Rates for each culture were converted to N2 fixation rates by using a 3:1 ratio of C2H2 reduced to fixed N2 (19); rates ranged from 2.35 ± 0.83 to 2.64 ± 0.62 fmol of fixed N per cell h−1, comparable to rates published elsewhere (29).
The determination of the relationships within planktonic unicellular cyanobacteria suggested by these patterns of nifH and 16S rDNA genetic diversity is a preliminary step towards understanding this group of potentially important N2 fixers. We need a better grasp of evolutionary relationships and ecological roles within the unicellular cyanobacteria, as well as a finer-scale understanding of their spatial and temporal diversity within the major ocean basins. Studies of in situ rates of N2 fixation related to cyanobacterial sequence identity will further contribute to our understanding of the major players influencing the biogeochemistry of major oceanic regions.
The sequences that were generated from this study and derived from environmental samples have been deposited in the GenBank database under accession numbers AY100483 to AY100516, AY100520, AY100522, and AY100523; those from cultures have accession numbers AY100517 to AY100519 and AY100521.
Acknowledgments
This research was funded by the NSF. L.I.F. receives support from CONACyT, Mexico, the MSRC, SBU, and SFSU.
We are thankful to Vanessa Madrid (MSRC), Craig Reading (SFSU), Douglas Capone (USC), and the crew of the research vessel Knorr (WHOI) for their assistance in this project. We thank J. Waterbury (WHOI) for providing the WH8501 culture and two anonymous reviewers for their valuable comments on an early version of the manuscript.
REFERENCES
- 1.Baker, C. S., F. Cipriano, and S. R. Palumbi. 1996. Molecular genetic identification of whale and dolphin products from commercial markets in Korea and Japan. Mol. Ecol. 5:671-685. [Google Scholar]
- 2.Capone, D. G. 1993. Determination of nitrogenase activity in aquatic samples using the acetylene reduction procedure, p. 621-631. In P. F. Kemp, B. F. Sherr, E. B. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Press, Boca Raton, Fla.
- 3.Capone, D. G., J. Zehr, H. Paerl, B. Bergman, and E. J. Carpenter. 1997. Trichodesmium: a globally significant marine cyanobacterium. Science 276:1221-1229. [Google Scholar]
- 4.Capone, D. G., and E. J. Carpenter. 1999. Nitrogen fixation by marine cyanobacteria: historical and global perspectives. Bull. Inst. Océanogr. Monaco 19:235-256. [Google Scholar]
- 5.Carpenter, E. J., and C. C. Price. 1976. Marine Oscillatoria (Trichodesmium): explanation for aerobic nitrogen fixation without heterocysts. Science 191:1278-1280. [DOI] [PubMed] [Google Scholar]
- 6.Carpenter, E. J., and D. G. Capone. 1992. Nitrogen fixation in Trichodesmium blooms, p. 211-218. In E. J. Carpenter, D. G. Capone, and J. G. Reuter (ed.), Marine pelagic cyanobacteria: Trichodesmium and other diazotrophs. Kluwer Academic Publishers, London, England.
- 7.Carpenter, E. J., J. P. Montoya, J. Burns, M. R. Mulholland, A. Subramaniam, and D. G. Capone. 1999. Extensive bloom of a N2-fixing diatom/cyanobacterial bloom association in the tropical Atlantic Ocean. Mar. Ecol. Prog. Ser. 185:273-283. [Google Scholar]
- 8.Carpenter, E. J., and S. Janson. 2000. Intracellular cyanobacterial symbionts in the marine diatom Climacodium frauenfeldianum (Bacillariophyceae). J. Phycol. 36:540-544. [DOI] [PubMed] [Google Scholar]
- 9.Golden, S. S., M. Ishiura, C. H. Johnson, and T. Kondo. 1997. Cyanobacterial circadian rhythms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48:327-354. [DOI] [PubMed] [Google Scholar]
- 10.Gruber, N., and J. L. Sarmiento. 1997. Global patterns of marine nitrogen fixation and denitrification. Glob. Biogeochem. Cycles 11:235-266. [Google Scholar]
- 11.Hobbie, J. E., and M. M. Fletcher. 1988. The aquatic environment, p. 132-262. In J. M. Lunch and J. E. Hobbie (ed.), Micro-organisms in action: concepts and applications in microbial ecology. Blackwell Scientific Publications Ltd., Oxford, England.
- 12.León, C., S. Kumazama, and A. Mitsui. 1986. Cyclic appearance of aerobic nitrogenase activity during synchronous growth of unicellular cyanobacteria. Curr. Microbiol. 13:149-153. [Google Scholar]
- 13.Madrid, V. M., G. T. Taylor, M. I. Scranton, and A. Y. Chistoserdov. 2001. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 67:1663-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitsui, A., S. Kumazama, A. Takahashi, H. Ikemoto, S. Cao, and T. Arai. 1986. Strategy by which nitrogen-fixing unicellular cyanobacteria grow photoautotrophically. Nature 323:720-722. [Google Scholar]
- 15.Mitsui, A., S. Cao, A. Takahashi, and T. Arai. 1987. Growth of synchrony and cellular parameters of the unicellular nitrogen-fixing marine cyanobacterium Synechococcus sp. strain Miami BG 043511 under continuous illumination. Physiol. Plant. 69:1-8. [Google Scholar]
- 16.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paerl, H. W., B. M. Bebout, and L. E. Prufert. 1989. Naturally occurring patterns of oxygenic photosynthesis and N2 fixation in a marine microbial mat: physiological and ecological ramifications, p. 326-341. In Y. Cohen and E. Rosenberg (ed.), Microbial mats: physiological ecology of benthic microbial communities. American Society for Microbiology, Washington, D.C.
- 18.Postgate, J. 1978. Nitrogen fixation. Inst. Biol. Stud. Biol. Vol. 92.
- 19.Reddy, K. J., J. B. Haskell, D. M. Sherman, and L. A. Sherman. 1993. Unicellular, aerobic nitrogen-fixing cyanobacteria of the genus Cyanothece. J. Bacteriol. 175:1284-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Schneegurt, M. A., D. M. Sherman, S. Nayar, and L. A. Sherman. 1994. Oscillating behavior of carbohydrate granule formation and dinitrogen fixation in the cyanobacterium Cyanothece sp. strain ATCC 51142. J. Bacteriol. 176:1586-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schneegurt, M. A., D. M. Sherman, and L. A. Sherman. 1997. Composition of the carbohydrate granules of the cyanobacterium. Cyanothece sp. strain ATCC 51142. Arch. Microbiol. 167:89-98. [PubMed] [Google Scholar]
- 23.Spiller, H., and K. T. Shanmugam. 1987. Physiological conditions for nitrogen fixation in a unicellular marine cyanobacterium, Synechococcus sp. strain SF1. J. Bacteriol. 169:5379-5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Waterbury, J. B., and J. M. Willey. 1988. Isolation and growth of marine planktonic cyanobacteria. Methods Enzymol. 167:100-105. [Google Scholar]
- 25.Zani, S., M. T. Mellon, J. L. Collier, and J. P. Zehr. 2000. Expression of nifH genes in natural microbial assemblages in Lake George, New York, detected by reverse transcriptase PCR. Appl. Environ. Microbiol. 66:3119-3124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zehr, J. P., and L. A. McReynolds. 1989. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl. Environ. Microbiol. 55:2522-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zehr, J. P., and D. G. Capone. 1992. Problems and promises of assaying the genetic potential for nitrogen fixation in the marine environment. Microb. Ecol. 32: 263-281. [DOI] [PubMed] [Google Scholar]
- 28.Zehr, J. P., M. T. Mellon, and S. Zani. 1998. New nitrogen-fixing microorganisms detected in oligotrophic oceans by amplification of nitrogenase (nifH) genes. Appl. Environ. Microbiol. 64:3444-3450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zehr, J. P., J. B. Waterbury, P. J. Turner, J. P. Montoya, E. Omoregie, G. F. Steward, A. Hansen, and D. M. Karl. 2001. Unicellular cyanobacteria fix N2 in the subtropical North Pacific Ocean. Nature 412:635-638. [DOI] [PubMed] [Google Scholar]