Abstract
The plastid genome is transcribed by two distinct RNA polymerases, the PEP encoded by the plastid genome and the NEP encoded in the nucleus. Initial models of plastid transcription held that the NEP is responsible for the transcription of housekeeping genes needed early in development, and that the PEP transcribes genes required for photosynthesis. Recently, this model was challenged by the discovery that all plastid genes are transcribed by NEP in PEP-deficient tobacco plastids, suggesting that mRNA turnover may have a strong role in previously observed transcription patterns. In this study, we provide evidence that the NEP enzyme level decreases as plastids mature. In contrast, production of mRNAs by NEP increases as plastids mature, yet their accumulations remain constant. These results suggest that as plastids mature NEP may become more active, and that mRNA turnover varies between transcripts synthesized by NEP and PEP.
Keywords: maize, RpoTp, NEP, plastid, mRNA stability
Introduction
The plastid genome is transcribed by two distinct RNA polymerases (RNAPs). These are the plastid-encoded RNAP (PEP), which resembles eubacterial RNAPs in terms of its multisubunit architecture and requirement for sigma factors, and the nucleus-encoded polymerase (NEP), the core catalytic subunit of which resembles those of T3/T7 phage RNAPs (Liere & Maliga, 2001). So far, studies have shown that plastid T7-like RNAPs are nearly ubiquitous, with direct evidence in Physcomitrella patens (Richter et al, 2002), tobacco (Kobayashi et al, 2001; Hedtke et al, 2002), wheat (Ikeda & Gray, 1999) and maize (Chang et al, 1999), but apparently lacking in Chlamydomonas (Eberhard et al, 2002). Each RNAP recognizes its own family of promoters (Liere & Maliga, 2001). The PEP promoters generally have the bacterial σ70 recognition sequences composed of canonical −35 and −10 elements. NEP promoters generally feature a YRTA consensus; however, a second class of NEP promoters has been termed exceptional, due to their lack of a recognizable consensus sequence (Weihe & Börner, 1999), and may be transcribed by another form of NEP.
Accepted models of chloroplast transcription posit a developmentally regulated division of labour between the two enzymes. In this scenario, NEP promoters are most active in tissues with undifferentiated plastids, their usage declines as chloroplasts mature, and ultimately some NEP promoters may be silent in mature chloroplasts. In turn, PEP promoters become increasingly active, corresponding to the elaboration of the photosynthetic machinery. This model is supported by RNA gel blot analysis of PEP-deficient plants and cell lines, where only a few plastid transcripts are abundant, suggesting that PEP is required for the majority of transcription. Furthermore, these NEP-transcribed mRNAs were often more abundant in PEP-deficient than in wild-type cells, suggesting that there might be a developmental regulation of NEP, and/or a dynamic relationship between the abundance and/or activities of PEP and NEP (Han et al, 1993; Hess et al, 1994; Allison et al, 1996; Silhavy & Maliga, 1998a, 1998b; Kapoor & Sugiura, 1999).
Recent re-examination of transcription in PEP-deficient tobacco using a genomic microarray approach (Krause et al, 2000; Legen et al, 2002), however, demonstrated that all plastid genes could still be transcribed by NEP to some degree. This is inconsistent with a simple division of labour. Instead, it implies that the abundance of NEP-derived transcripts seen in PEP-deficient cells using RNA gel blot analysis might be due to a combination of relaxed promoter specificity, increased transcriptional readthrough and subsequent processing, and/or increased RNA stability. Without direct knowledge of NEP activity, and by using PEP-deficient mutants, it is difficult to reconcile models for NEP/PEP dynamics in one of which NEP is a general enzyme and in the other a specialized enzyme. Instability of some NEP-derived transcripts in immature plastids would be consistent with the fact that differential RNA stability related to plastid development is well known, both in monocots and dicots (Klaff & Gruissem, 1991; Kim et al, 1993).
Here, we have used maize to focus on how NEP is regulated during plastid development, in a wild-type rather than mutant context. Unlike dicot systems, maize has a single plastid NEP, termed RpoTp (Chang et al, 1999), and maize leaves have a developmental gradient of proplastids to chloroplasts from base to tip (Leech et al, 1973), allowing comparisons within a single organ. Our results show that the RpoTp protein level decreases with plastid maturity, yet transcription of RNAs with NEP promoters increases. These transcripts also accumulate relatively equally regardless of the state of plastid development. This suggests that these transcripts become increasingly less stable, the opposite of that seen for RNAs transcribed from genes with PEP promoters. In the light of these and other findings, we present a revised model for the developmental regulation of plastid transcription and RNA stability.
Results
RpoTp becomes less abundant as plastids mature
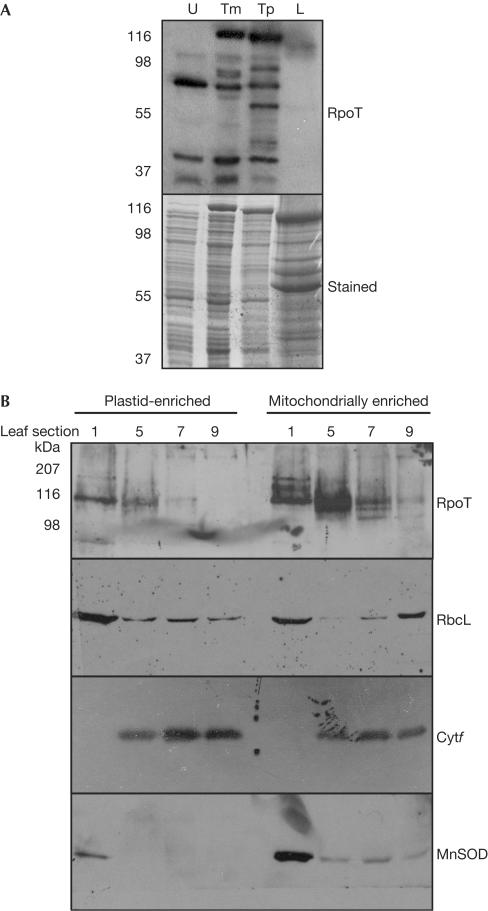
Current models suggest that NEP is more active in proplastids, implying that the catalytic subunit would be more abundant in immature or nonphotosynthetic plant tissues. As this has not been addressed experimentally, we examined the distribution of RpoTp in developing plastids using immunoblot analysis. To do so, we used an antibody raised against recombinant maize RpoTp (Chang et al, 1999). Because RpoTp is homologous to the mitochondrial enzyme RpoTm, we examined the specificity of the antibody using recombinant RpoTm and RpoTp, as shown in Fig 1A. The antibody readily recognized both proteins; however, no clear signal was detectable in leaf total protein extracts, as previously reported for RpoTm (Chang et al, 1999).
Figure 1.
RpoT protein levels decrease during plastid maturation. (A) Total protein from uninduced (U) Escherichia coli and cells induced by arabinose expressing recombinant RpoTm or RpoTp, or total leaf protein (L), was separated for immunoblot analysis and probed with a maize anti-RpoT antibody at 1:2,000 dilution. (B) Leaves from 6-week-old maize plants were divided into nine equal sections, and plastid and mitochondrially enriched fractions were extracted from the indicated sections. A 20 μg portion of protein from each fraction was used for immunoblot analysis using the antibodies shown on the right. Antibodies are described in Methods and were used at dilutions of 1:2,000 (RpoT and MnSOD) or 1:5,000 (RbcL and Cytf). Molecular mass standards are shown on the left.
To examine the accumulation of the RpoT proteins, partially emerged leaves were dissected from 6- to 8-week-old maize plants and divided into nine equal parts. Section 1 represents the base of the leaf (immature yellow plastids) and section 9 the tip of the leaf (mature chloroplasts). Tissues from sections 1, 5, 7 and 9 were macerated, and fractions enriched in either plastids or mitochondria were collected by differential centrifugation. Equal amounts of protein were analysed from each sample, as shown in Fig 1B.
The antibody detected immunoreactive species in both plastid and mitochondrial fractions, as expected, with migration in the 100–110 kDa range, again as previously reported for RpoTm and as predicted by the RpoTp amino-acid sequence (Chang et al, 1999). The results showed that RpoTp and RpoTm both decreased as leaf cells mature, becoming barely detectable in section 9. This observation agrees with the previous observation of RpoTp and RpoTm transcript accumulation across a similar developmental gradient (Chang et al, 1999). RpoTm apparently accumulates in several forms; the significance of this observation is unknown.
We tested the fidelity of our enriched fractions with antibodies raised against organelle-specific proteins (Rubisco LS and cytochrome f for plastids; MnSOD for mitochondria). RbcL accumulation is light independent (Nelson et al, 1984), and was thus detected in all of the plastid-enriched fractions. Because LS was also detected in the mitochondria-enriched fractions, these fractions were to some degree contaminated with plastid material. This was confirmed by analysis of Cytf, a developmentally regulated membrane subunit of the b6f complex, the accumulation of which increased across the developmental gradient and was also visible contaminating mitochondrial fractions. Mitochondrial contamination of plastids was assessed using anti-MnSOD, which recognizes the constitutive mitochondrial manganese superoxide dismutase encoded by Arabidopsis MSD1 (Kliebenstein et al, 1998). In our samples, MnSOD was detected in all of the mitochondrially enriched fractions but not in the plastid-enriched fractions from sections 5–9, although a slight contamination was evident in section 1, reflecting the difficulty of separating proplastids and mitochondria. This suggests that the amount of RpoTp in section 1 is slightly over-represented, but is still at a maximum in this section, decreasing thereafter.
Two mRNA patterns in developing chloroplasts
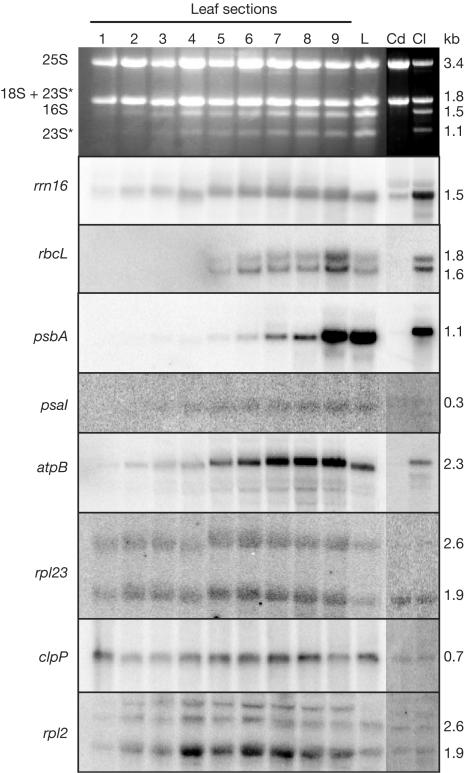
If the RpoTp levels detected in Fig 1B limit transcription activity, a substantial decrease in the accumulation of mRNAs driven by NEP promoters should occur during chloroplast maturation. Previously published results have suggested that in maize some plastid genes are preferentially transcribed by NEP (such as rpl2, rpl23, clpP and rps15), others by PEP (e.g. rrn16, rbcL, psbA, psaI, as well as the majority of remaining genes) and at least one by both (atpB) (Silhavy & Maliga, 1998a). To test transcript accumulation, RNA was analysed from sectioned leaves (as in Fig 1B), mature leaves and 7-day-old green or etiolated coleoptiles.
Two patterns of mRNA abundance are apparent from the filter hybridizations shown in Fig 2. Transcripts of rrn16, rbcL, psbA, psaI and atpB, transcribed exclusively or mainly by PEP, increase markedly as proplastids develop into chloroplasts, and in fact all except rrn16 and atpB are undetectable in the first leaf section. In contrast, accumulation of the NEP-transcribed RNAs rpl23, clpP and rpl2 changed little during plastid development, although rpl23 increased slightly in sections 4–8. Transcript levels in etiolated coleoptiles closely matched those seen in the immature leaf plastids. The stable NEP-derived transcript levels across the plastid developmental gradient suggest that the decrease in RpoTp seen in Fig 1B does not translate directly into reduced mRNA abundance.
Figure 2.
Two distinct patterns of plastid mRNA accumulation. RNA was extracted from leaf sections, leaves or coleoptiles as described in Methods. A 5 μg portion of each sample was analysed by filter hybridization using the gene-specific probes shown on the left. rRNAs (top panel) were visualized by ethidium bromide staining. Known or deduced transcript sizes, marked on the right, were derived by comparison to an RNA ladder. The asterisks mark in vivo breakdown products of 23S rRNA.
Transcription activity increases as plastids mature
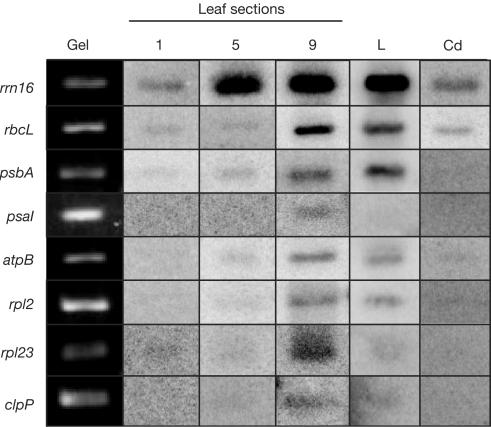
As RNA gel blots cannot distinguish between changes in transcription activity and RNA stability, an in vitro run-on assay was used to assess the production of transcripts in leaf sections, mature leaves and etiolated coleoptiles. As shown in Fig 3, as plastids mature, the transcription activity increases (columns 1, 5 and 9). On reaching maturity (column L), transcription stabilizes, with some genes remaining active (rrn16, rbcL and psbA) and others decreasing modestly (psaI, atpB, rpl2, rpl23 and clpP). Transcription in etiolated coleoptiles (column Cd) resembles that in the base of the immature leaf (column 1).
Figure 3.
Transcription increases as plastids mature. Fragments of maize plastid genes were either PCR-amplified or isolated from plasmids, separated in agarose gels and blotted onto nylon membranes. Blots were incubated with radiolabelled plastid run-on transcripts from the indicated tissues. The column labelled ‘gel' shows the stained gene fragments before blotting. Tissues tested include leaf sections (1, 5 and 9), and leaf (L) or etiolated coleoptile (Cd) material.
Increased transcription during plastid development is not surprising for the PEP-derived transcripts, as it closely parallels the accumulation seen in Fig 2 and is consistent with measures of total plastid transcription activity in greening spinach (Deng & Gruissem, 1987). By this same logic, however, NEP transcripts would be expected to have an even rate of production along the leaf gradient. As this is clearly not the case, it can be concluded that the two patterns of transcript accumulation seen in Fig 2 are due to two RNA turnover classes. In one case (genes with NEP promoters), transcription in immature plastids is relatively low, but stability of the transcripts is relatively high. As the plastids develop, transcription increases and stability decreases, keeping overall abundance constant. Conversely, genes with PEP promoters have relatively low transcription activities coupled with perhaps relatively high turnover in immature plastids, whereas transcription and stability increase as plastids mature.
Discussion
In this study, we have provided evidence that plastid transcripts fall into two categories of turnover during chloroplast biogenesis, through analysis of transcription activity and RNA accumulation. Earlier studies had focused on increased stability of photosynthetic transcripts as plastids matured and did not take into account the two-polymerase system that was discovered later.
The existing model of NEP–PEP dynamics has the logic of transcribing housekeeping genes in undifferentiated plastids, where metabolic activities do not include photosynthesis. Once PEP became active, NEP was assumed to be a relatively minor player required to maintain the levels of ribosomes, tRNAs and PEP (Liere & Maliga, 2001). Our results show that in maize, NEP abundance decreases; however, transcription of genes with NEP promoters increases as chloroplasts mature, suggesting that NEP is indeed under developmental regulation, perhaps mediated by accessory transcription factors.
The current model of chloroplast transcription relies heavily on analysis of PEP-deficient plants and cell lines and on developmentally regulated promoter recognition by the two different RNAPs, primarily using RNA gel blots. However, measurements of NEP itself and the use of a developmental series in wild-type plants are missing from these analyses. Initial examinations of PEP-deficient tobacco suggested a fairly strict division of labour between NEP and PEP (Allison et al, 1996; Hajdukiewicz et al, 1997), but a careful re-examination of such plants has revealed a more complex phenomenon (Krause et al, 2000; Legen et al, 2002). In particular, robust transcription but very low mRNA abundance for some PEP-driven genes was observed. This suggested that NEP was perfectly capable of transcribing genes believed to have only PEP promoters, but that the transcripts simply failed to accumulate, perhaps due to RNA processing defects. This phenomenon has been observed in other PEP-deficient mutants (Cahoon et al, 2003, and references therein) and shows that whereas depleting PEP can elegantly demonstrate the existence of several RNAPs, it may have limited application in the study of plastid gene regulation in wild-type plants.
In this study, we observed two classes of mRNA accumulation across the plastid developmental gradient. One class, corresponding to genes with NEP promoters, varied little with developmental stage, whereas the second class, corresponding to genes with PEP promoters, increased as chloroplasts matured. Run-on transcription, however, showed that the transcription of all the genes tested increased as the chloroplasts matured. This suggests that the accumulation of NEP promoter-derived mRNAs in immature plastids may be due to increased stability rather than altered NEP activity.
Speculation
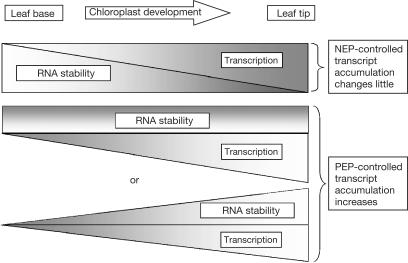
In the light of the data presented here, we propose a revised model for NEP and PEP in the context of maize chloroplast biogenesis (Fig 4). In this model, transcription activities of both NEP and PEP increase with chloroplast development, but stability of transcripts initiated from NEP promoters decreases to maintain constant accumulation. In the case of transcripts from PEP promoters, we cannot differentiate between unchanged stability, making their increased abundance a function solely of increased transcription rates, with concomitant increases in stability and transcription rate.
Figure 4.
Model for NEP–PEP dynamics in maize. The triangles represent increasing or decreasing amounts of polymerase activity and mRNA stability as it relates to chloroplast development along the leaf gradient. For transcripts with NEP promoters, transcription increases from the leaf base to the tip, whereas RNA stability decreases. For transcripts with PEP promoters, transcription rates increase as chloroplasts develop, whereas RNA stability remains constant or increases.
Changes in plastid mRNA stability during development have been previously reported in several species (Monde et al, 2000). However, the concept that two stability classes exist, corresponding to polymerase type, is new. A perspective on this finding can be gained if we make the following assumptions: (1) NEP and PEP initiate transcription from different promoter sets in wild-type plants, as previously postulated, and (2) accumulation of these two transcript categories varies according to plastid developmental stage. This implies that the polymerase type is not a fundamental regulator, but instead that functional classes of mRNAs are subjected to post-transcriptional regulation. Interestingly, genome-wide studies of mRNA stability in both yeast and Escherichia coli showed a wide range of mRNA lifetimes, but also a correlation between similar half-lives and functional relationship, such as genes encoding subunits of a given protein complex (Bernstein et al, 2002; Wang et al, 2002). Whereas the mechanism governing such a relationship is still obscure, although it is likely to involve the 5′ and/or 3′ untranslated regions, it is tempting to speculate that it has been conserved from prokaryotes to eukaryotes, including organelles.
Methods
Plant tissue and RNA analysis
Greenhouse-grown maize line B73 was used for all experiments. RNA was extracted from tissues frozen in liquid nitrogen and ground with a mortar and pestle, using the standard protocol for TriReagent (Molecular Research Center Inc., Cincinnati, OH, USA). Electrophoresis and filter hybridizations were as previously described (Cahoon et al, 2003).
Organelle enrichment
Organelle enrichment was performed as described by Gruissem et al (1986). Briefly, partially emerged maize leaves from 6- to 8-week-old plants were cut into nine equal sections. Sections were rinsed with water and macerated using scissors and a Waring blender in GM-mix, filtered through cheesecloth and Miracloth (Calbiochem, La Jolla, CA, USA), and centrifuged briefly in a Sorvall RB-5 GSA rotor at 4,500 r.p.m. followed by 5 min at 1,000 g to pellet plastids. The supernatant was collected for mitochondrial isolation. Plastids were resuspended in 500 μl GM-mix with a paintbrush and transferred to a microcentrifuge tube, pelleted and resuspended in an equal volume of buffer A (10 mM Tris pH 7.9, 1 mM EDTA and 5 mM dithiothreitol (DTT)), and stored at −80°C. Mitochondria were collected from the supernatant by centrifugation in a Sorvall GSA rotor at 8,000 r.p.m. for 30 min. The pellet was treated as for plastid fractions.
Protein analysis
Organelle-enriched fractions were mixed with an equal volume of 2 × protein extraction buffer (100 mM Tris pH 8.0, 18% (w/v) sucrose, 40 mM β-mercaptoethanol, 1 mM phenylmethylsulphonyl fluoride and 4% sodium dodecyl sulphate) and heated to 95°C. Insoluble debris was pelleted and the supernatant was collected. Protein concentration was estimated using the Bio-Rad protein assay kit (Bio-Rad, Hercules, CA, USA). Proteins were separated in 10% polyacrylamide gels, blotted onto nitrocellulose, blocked with Tris-buffered saline containing 5% milk, incubated with primary and secondary antibodies, and detected using the ECL chemiluminescence kit (Amersham, Piscataway, NY, USA). Antibodies recognized RpoT (Chang et al, 1999), Rubisco LS (Agrisera; http://www.agrisera.se), cytochrome f (Chen et al, 1995) and MnSOD (Kliebenstein et al, 1998). Recombinant RpoTm and RpoTp were generated using the pBAD-Thio-Topo vector system (Invitrogen, Carlsbad, CA, USA) with induction by 0.02% arabinose.
Run-on transcription
Tissue was macerated in 1 × TAN buffer (Sakai et al, 1998) using a polytron grinder. Large debris was filtered out using Miracloth, and microscopic debris including intact cells and cellular organelles was pelleted. Run-on transcription assays were based on Klein & Mullet (1990) and performed within 10–15 min of macerating the tissue. Briefly, pellets were resuspended in 50 mM Hepes/KOH, pH 8.0, 10 mM MgCl2, 25 mM potassium acetate, 10 mM DTT, 2 mM spermidine, 125 μM each CTP, GTP and ATP, 10 μM unlabelled UTP plus 200 μCi [α32P]UTP, and incubated at 22°C for 30 min. RNA was extracted with TriReagent, pelleted with isopropanol, washed with 70% ethanol and resuspended in DEPC-treated water. Labelled RNAs were incubated with specific DNA probes (digested plasmids and PCR-amplified DNAs) blotted onto GeneScreen nylon filters. Blots were washed and visualized as described above for RNA analysis.
Acknowledgments
We thank I. Law for elaboration of the plastid transcription model and J. Brenchley for help with RNA isolation. A.B.C. was supported by USDA-NRI postdoctoral fellowship 2001-35301-10845. This work was also supported by NSF award 0090658 to D.B.S. Expression of recombinant RpoTm and RpoTp was carried out during a sabbatical visit by D.B.S. to the laboratory of Silva Lerbs-Mache at Univ. Josef-Fourier, Grenoble and was supported by a fellowship from the French Ministry of Education, Research and Technology.
References
- Allison LA, Simon LD, Maliga P (1996) Deletion of rpoB reveals a second distinct transcription system in plastids of higher plants. EMBO J 15: 2802–2809 [PMC free article] [PubMed] [Google Scholar]
- Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN (2002) Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci USA 99: 9697–9702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon AB, Cunningham KA, Bollenbach TJ, Stern DB (2003) Maize BMS cultured cell lines survive with massive plastid gene loss. Curr Genet 44: 104–113 [DOI] [PubMed] [Google Scholar]
- Chang C, Sheen J, Bligny M, Niwa Y, Lerbs-Mache S, Stern DB (1999) Functional analysis of two maize cDNAs encoding T7-like RNA polymerases. Plant Cell 11: 911–926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Kindle KL, Stern DB (1995) The initiation codon determines the efficiency but not the site of translation initiation in Chlamydomonas chloroplasts. Plant Cell 7: 1295–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng XW, Gruissem W (1987) Control of plastid gene expression during development: the limited role of transcriptional regulation. Cell 49: 379–387 [DOI] [PubMed] [Google Scholar]
- Eberhard S, Drapier D, Wollman FA (2002) Searching limiting steps in the expression of chloroplast-encoded proteins: relations between gene copy number, transcription, transcript abundance and translation rate in the chloroplast of Chlamydomonas reinhardtii. Plant J 31: 149–160 [DOI] [PubMed] [Google Scholar]
- Gruissem W, Greenberg BM, Zurawski G, Hallick RB (1986) Chloroplast gene expression and promoter identification in chloroplast extracts. Methods Enzymol 118: 253–270 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz PTJ, Allison LA, Maliga P (1997) The two RNA polymerases encoded by the nuclear and plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C-d, Patrie W, Polacco M, Coe EH (1993) Aberrations in plastid transcripts and deficiency of plastid DNA in striped and albino mutants of maize. Planta 191: 552–563 [Google Scholar]
- Hedtke B, Legen J, Weihe A, Herrmann RG, Borner T (2002) Six active phage-type RNA polymerase genes in Nicotiana tabacum. Plant J 30: 625–637 [DOI] [PubMed] [Google Scholar]
- Hess WR, Hoch B, Zeltz P, Hubschmann T, Kossel H, Borner T (1994) Inefficient rpl2 splicing in barley mutants with ribosome-deficient plastids. Plant Cell 6: 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda TM, Gray MW (1999) Identification and characterization of T3/T7 bacteriophage-like RNA polymerase sequences in wheat. Plant Mol Biol 40: 567–578 [DOI] [PubMed] [Google Scholar]
- Kapoor S, Sugiura M (1999) Identification of two essential sequence elements in the nonconsensus type II PatpB-290 plastid promoter by using plastid transcription extracts from cultured tobacco BY-2 cells. Plant Cell 11: 1799–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M, Christopher DA, Mullet JE (1993) Direct evidence for selective modulation of psbA, rpoA, rbcL, and 16S RNA stability during barley chloroplast development. Plant Mol Biol 22: 447–463 [DOI] [PubMed] [Google Scholar]
- Klaff P, Gruissem W (1991) Changes in chloroplast mRNA stability during leaf development. Plant Cell 3: 517–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RR, Mullet JE (1990) Light-induced transcription of chloroplast genes. psbA transcription is differentially enhanced in illuminated barley. J Biol Chem 265: 1895–1902 [PubMed] [Google Scholar]
- Kliebenstein DJ, Monde RA, Last RL (1998) Superoxide dismutase in Arabidopsis: an eclectic enzyme family with disparate regulation and protein localization. Plant Physiol 118: 637–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y, Dokiya Y, Sugita M (2001) Dual targeting of phage-type RNA polymerase to both mitochondria and plastids is due to alternative translation initiation in single transcripts. Biochem Biophys Res Commun 289: 1106–1113 [DOI] [PubMed] [Google Scholar]
- Krause K, Maier RM, Kofer W, Krupinska K, Herrmann RG (2000) Disruption of plastid-encoded RNA polymerase genes in tobacco: expression of only a distinct set of genes is not based on selective transcription of the plastid chromosome. Mol Gen Genet 263: 1022–1030 [DOI] [PubMed] [Google Scholar]
- Leech RM, Rumsby MG, Thomson WW (1973) Plastid differentiation, acyl lipid, and fatty acid changes in developing green maize leaves. Plant Physiol 52: 240–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legen J, Kemp S, Krause K, Profanter B, Herrmann RG, Maier RM (2002) Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J 31: 171–188 [DOI] [PubMed] [Google Scholar]
- Liere K, Maliga P (2001) Plastid RNA polymerases in higher plants. In Regulation of Photosynthesis, Anderson B, Aro EM (eds) pp 29–39. Rotterdam: Kluwer [Google Scholar]
- Monde RA, Schuster G, Stern DB (2000) Processing and degradation of chloroplast mRNA. Biochimie 82: 573–582 [DOI] [PubMed] [Google Scholar]
- Nelson T, Harpster M, Mayfield S, Taylor W (1984) Light-regulated gene expression during maize leaf development. J Cell Biol 98: 558–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter U, Kiessling J, Hedtke B, Decker E, Reski R, Borner T, Weihe A (2002) Two RpoT genes of Physcomitrella patens encode phage-type RNA polymerases with dual targeting to mitochondria and plastids. Gene 290: 95–105 [DOI] [PubMed] [Google Scholar]
- Sakai A, Suzuki T, Miyazawa Y, Kuroiwa T (1998) Simultaneous isolation of cell-nuclei, plastid-nuclei and mitochondrial-nuclei from cultured tobacco cells; comparative analysis of their transcriptional activities in vitro. Plant Sci 133: 17–31 [Google Scholar]
- Silhavy D, Maliga P (1998a) Mapping of promoters for the nucleus-encoded plastid RNA polymerase (NEP) in the iojap maize mutant. Curr Genet 33: 340–344 [DOI] [PubMed] [Google Scholar]
- Silhavy D, Maliga P (1998b) Plastid promoter utilization in a rice embryogenic cell culture. Curr Genet 34: 67–70 [DOI] [PubMed] [Google Scholar]
- Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO (2002) Precision and functional specificity in mRNA decay. Proc Natl Acad Sci USA 99: 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weihe A, Börner T (1999) Transcription and the architecture of promoters in chloroplasts. Trends Plant Sci 4: 169–170 [DOI] [PubMed] [Google Scholar]