Abstract
Actin proteins are present in pro- and eukaryotes, and have been shown to perform motor-like functions in eukaryotic cells in a variety of processes. Bacterial actin homologues are essential for cell viability and have been implicated in the formation of rod cell shape, as well as in segregation of plasmids and whole chromosomes. We have generated functional green fluorescent protein fusions of all three Bacillus subtilis actin-like proteins (MreB, Mbl and MreBH), and show that all three proteins form helical filaments underneath the cell membrane, the pattern of which is distinct for each protein. Time-lapse microscopy showed that the filaments are highly dynamic structures. A number of separate filaments of MreB and Mbl continuously move through the cell along helical tracks underneath the cell membrane. The speed of extension of the growing end of filaments is within the range of known actin polymerization (0.1 μm/s), generating a potential poleward or centreward pushing velocity at 0.24 μm/min for MreB or Mbl, respectively. During nutritional downshift and a block in topoisomerase IV activity, the filaments rapidly disintegrated, showing that movement occurs only in growing cells. Contrary to Mbl and MreBH filaments, MreB filaments were generally absent in cells lacking DNA, providing a further distinction between the three orthologues.
Keywords: actin, intracellular movement, Bacillus subtilis
Introduction
Actin proteins function as cytoskeletal components in eukaryotic cells, and as structural fibres in muscle contraction. Additionally, actin proteins have motor-like functions (Marx, 2003; Mogilner & Oster, 2003; Upadhyaya & van Oudenaarden, 2003), most notably in cell migration through pushing of membranes. Once stimulated, motility receptors turn on a WASP family protein (through Rho GTPases), the latter binding and activating the Arp2/3 complex, which in turn induces branching and growth of actin filaments (Marx, 2003). Even in vitro, actin filaments can deform vesicles and thus push membranes, providing the force to elongate cellular extensions such as pseudopods (Lauffenburger & Horwitz, 1996; Mitchinson & Cramer, 1996). Moreover, Listeria monocytogenes cells are rapidly propelled within macrophages by polymerization of actin at one end of the cell (Tilney & Portnoy, 1989). Actin polymerization in this case is catalysed by the bacterial ActA protein, present at only one cell pole, which through its interaction with the Arp2/3 complex promotes formation of actin bundles at one end of the cell (Loisel et al, 1999). Growth of the barbed ends of the filaments adjacent to the bacterial cells can even push Listeria cells through the membrane into a neighbouring cell.
Actin is a 43 kDa protein that polymerizes into a two-stranded right-handed helix through addition of ATP-bound actin monomers. Actin movement arises through growth at the barbed (plus) end of the filament, while actin is released from the pointed (minus) end following ATP hydrolysis (a process termed treadmilling). Active pushing occurs through binding of actin monomers to the tip of the filament when the object moves away, thus preventing backward movement, such that the object is driven by brownian diffusion, with the actin filament dictating a single direction (polymerization rachett; Mogilner & Oster, 2003). Actin homologues (ParM, MreB and Mbl) have also been identified in bacteria (Jones et al, 2001; van den Ent et al, 2001; Moller-Jensen et al, 2002), where they are implicated in the cell structure of rodshaped cells. Bacillus subtilis MreB and Mbl, and Escherichia coli MreB form helical filaments just underneath the cell membrane (Jones et al, 2001; Carballido-Lopez & Errington, 2003; Shih et al, 2003), and Mbl is thought to be involved in the insertion of new cell wall material into the growing peptidoglycan layer, which seems to also follow a helical pattern (Daniel & Errington, 2003). MreB has been localized through immunofluorescence in fixed cells (Jones et al, 2001), whereas a green fluorescent protein (GFP)–Mbl fusion was shown to form filaments that are continuously remodelled along their whole length using fluorescence recovery after photobleaching (Carballido-Lopez & Errington, 2003). Recently, the role of actin-like proteins in the segregation of a subclass of plasmids (Moller-Jensen et al, 2002) and whole chromosomes (Kruse et al, 2003; Soufo & Graumann, 2003) has been demonstrated. Plasmid-encoded ParR protein binds to a specific cis site on the duplicated plasmids, which are located close to the cell centre, and induces polymerization of the ParM actin homologue. ParM filaments contain plasmids at their poleward ends, so two oppositely orientated ParM filaments appear to push plasmids towards each cell pole (Moller-Jensen et al, 2003). During depletion of MreB or Mbl in B. subtilis, or during overproduction of a dominant-negative mreB allele in E. coli, origin regions on the chromosomes fail to separate properly. It is unclear whether actin-like proteins have a direct role, for example, as an active segregation motor as in the E. coli plasmid system, or an indirect influence on the segregation of chromosomes. Thus, in contrast to eukaryotic actin, the exact function(s) of bacterial actins is still mysterious.
We have generated functional GFP fusions to all three B. subtilis actin-like proteins, MreB, Mbl and MreBH. We found that MreB and Mbl continuously move through the cells, that is, several distinct filaments move towards or away from the cell poles, respectively. Our findings show that intracellular actin movement exists in bacteria, which could provide directionality for cellular processes or drive transport of molecules and even of chromosomes.
Results And Discussion
Subcellular localization of B. subtilis actin proteins
We have created functional amino-terminal GFP fusions of all three B. subtilis actin-like proteins, MreB, Mbl and MreBH. Insertion of GFP–MreB at the original mreB locus (such that GFP–MreB driven by the xylose promoter is the only source of MreB in the cell) resulted in the formation of rod-shaped cells, the growth of which was indistinguishable from wild-type cells (Fig 1A). Likewise, a GFP–MreBH fusion fully complemented the wild-type gene, whereas a GFP–Mbl fusion was only partly functional, resulting in cells that were slightly wider than wild-type cells, infrequently showing abnormal morphology (in about 1% of the cells). Expression of GFP–Mbl from an ectopic site on the chromosome (creating a merodiploid cell) showed helical filaments indistinguishable from those in cells expressing solely GFP–Mbl that had normal cell morphology, suggesting that ectopic GFP–Mbl reflects the true localization of Mbl. Western blot analysis showed that all proteins were present as full-length GFP fusions (data not shown).
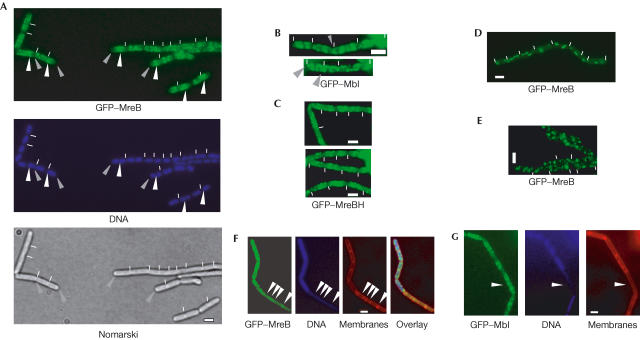
Figure 1.
Fluorescence microscopy of B. subtilis cells. Exponentially growing cells expressing (A) GFP–MreB, (B) GFP–Mbl or (C) GFP–MreBH. (D) Cells expressing GFP–MreB shifted from exponential growth in nutrient-rich medium to medium lacking nutrients for 5 min. (E) GFP–MreB-expressing cells from stationary phase. (F) Cells in which ParE (a subunit of topo IV) has been depleted for about 2–3 doubling times. GFP–MreB filaments are present only in cells containing DNA, and are absent in anucleate cells (indicated by arrowheads), which have normal rod shape. (G) Cells in which ParE has been depleted for 2–3 doubling times. GFP–Mbl is also present in anucleate cells (indicated by arrowhead). White lines indicate septa between cells (B. subtilis cells do not separate after division and form long chains of cells during exponential growth). Grey arrowheads in (A) point out cellular spaces close to the cell poles, and white arrowheads indicate the brightest signals of GFP–MreB filaments that are mostly close to or over the nucleoids. Grey arrowheads in (B) point out Mbl filaments that extend to the cell poles. Scale bars, 2 μm.
GFP–MreB filaments were most prominent in the central part of the cell between or close to the nucleoids (white arrowheads) and were not detectable at the cell poles (Fig 1A, grey arrowheads). The average distance of the first visible GFP–MreB filament to the nearest cell pole was 0.44 μm (120 cells measured, shortest distance 0.24 μm in 12 cells (still a visible gap), longest distance 0.83 μm). Incubation of exponentially growing cells on an isotonic medium lacking nutrients abolished the helical pattern within a few minutes (Fig 1D), indicating that the filaments were highly dynamic. In stationary-phase cells, GFP–MreB localized as bright foci that were randomly positioned within the cells (Fig 1E), demonstrating that filaments are only present in growing cells. By contrast, GFP–Mbl filaments were frequently found throughout the cells, extending all the way to the cell poles (Fig 1B), as previously described (Carballido-Lopez & Errington, 2003). GFP–MreBH also formed helical filaments (Fig 1C), with a pattern somewhat between that of MreB and Mbl. MreBH was located close to the cell poles in about 25% of the cells (in 42 out of 160 cells analysed, there was no visible gap between MreBH filaments and the extreme poles, that is, the distance was less than 0.2 mm; Fig 1C), compared with 60–70% polewardly localized GFP–Mbl filaments (the average distance of GFP–MreBH filaments to the poles was 0.32 μm (105 cells measured), and that for GFP–Mbl 0.28 μm (90 cells measured)). Like MreB, Mbl and MreBH filaments rapidly disappeared on cessation of cell growth (data not shown). The number of helical turns per cell for the three actin homologues was highly variable between the cells, as was the intensity of individual helices and their position within a single cell (Fig 1A–C). Although large cells generally contained more helical turns (up to nine in cells of >4 μm) than small cells (a minimum of one helical turn was seen in the smallest cells of 2 μm, >250 cells analysed), cells with similar size frequently contained different numbers of helical turns of the filaments, which is most apparent for GFP–MreB (Fig 1A). Thus, helical filaments of the actin orthologues in live cells are more irregular than the previously described filaments in fixed cells.
Movement of MreB and Mbl along helical tracks
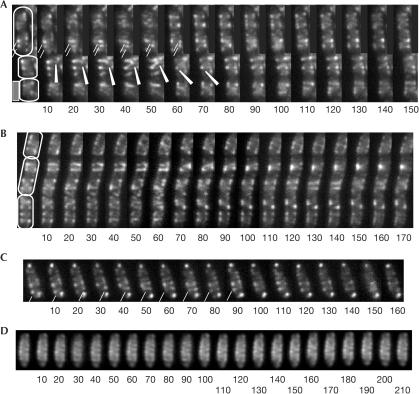
We turned to time-lapse microscopy to show whether there are any dynamic changes in the bacterial actin filaments. Different helical patterns are apparent between each panel in Fig 2A,B. GFP–MreB showed rapid movement of several distinct MreB filaments along a helical path in a scale of seconds (Fig 2A,B and movies 1–3 in supplementary material online, all frames taken in 10 s intervals). As the cells were immobilized on an agarose surface, it is clear that the rotational movement occurred within the cells, and was not due to whole-cell rotation. A potential problem with the dynamic movement within the cells is the fact that moving the focal plane through a helical object can create the optical illusion of movement. To rule out this possibility, we fixed growing cells with glutaraldehyde, and performed identical time-lapse experiments on the fixed cells. No movement was observed in these cells and the helical filaments remained static (Fig 2D), verifying the dynamics of MreB and Mbl (see below). In live cells, a single filament or more likely a bundle of filaments (as judged by the fluorescence intensity) was observed to perform a full turn around the diameter of the cell within 50–60 s (Fig 2A, between seconds 10 and 70, indicated by white lines and white arrowhead) (28 complete or half helical turns analysed). Given the distance of 3.46 μm for the circumference of B. subtilis cells (average diameter 1.099 μm), the leading edge of a bundle can travel at an average speed of 0.07 μm/s (not taking into account the helical curvature), which is in good agreement with previous measurements of isolated actin polymers (0.1–1 μm/s; Mogilner & Oster, 2003). We have measured the distance between individual helices, which averages at 0.50 μm (0.31–0.69 μm, 45 cells analysed). Thus, the net distance MreB filaments move relative to the cell length is 4.2 nm/s; so theoretically, MreB helices could push an object at a rate of 0.24 μm/min. However, movement of the filaments was not continuous over a prolonged period; frequently, full turns or 1.5 turns were observed, after which movement seemed to pause or cease (see supplementary movies 1,2,3 online). Thus, net movement through the cell is presumably lower than 0.24 μm/min, taking into account the putative pausing of filaments.
Figure 2.
Time-lapse fluorescence microscopy. GFP–MreB filaments are highly dynamic and generally move away from mid-cell, whereas GFP–Mbl filaments move towards mid-cell. Frames are taken every 10 s, and all cells are equally scaled. (A,B,D) Cells expressing GFP–MreB, (A,B) exponentially growing cells, (C) single growing cell expressing GFP–Mbl and (D) fixed cell. White lines in (A) indicate two MreB bundles migrating in parallel towards the cell pole, and a white arrowhead indicates MreB filament visually making a full turn around the inner membrane periphery. White line in (C) indicates Mbl bundle migrating around the cell diameter, from cell pole towards mid-cell. Note that GFP–MreB filaments do not change in fixed cells in (D). Scale bar, 2 μm.
In small cells (Fig 2A, lower two cells), GFP–MreB filaments seemed to move through the cell from approximately one cell quarter to the other quarter (indicated by white arrowhead). In larger cells, we found that predominantly GFP–MreB filaments moved away from mid-cell towards opposite cell poles (Fig 2A and supplementary movie 1 online; >50 cells analysed). Frequently, several MreB filaments moved simultaneously towards the same pole, which can be seen in Fig 2A, indicated by two white lines. Thus, actin helices consist of several extending and retracting filaments, and not of static tubes. Like MreB, GFP–Mbl filaments were highly dynamic, performing a similar helical movement underneath the cell membrane (Fig 2C and supplementary movies 4 and 5 online). In contrast to GFP–MreB, GFP–Mbl foci were often present close to or at the cell poles, and bundles seemed to nucleate from this position and move towards the cell centre (Fig 2C, indicated by white line). Thus, the general direction of movement of MreB and Mbl filaments seemed to be opposite, whereas the speed of migration was similar.
MreB is influenced by the nucleoid
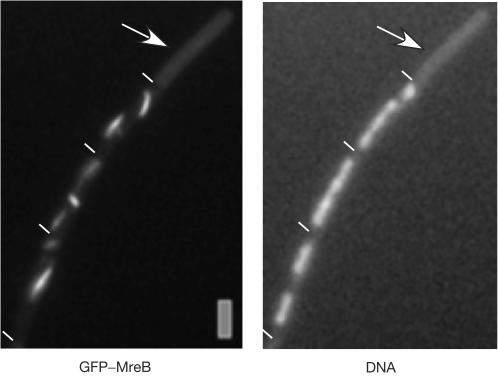
Depletion of MreB leads to a strong chromosome segregation defect, before a change in cell shape is visible, whereas depletion of Mbl leads to a milder segregation defect (formation of 25% anucleate cells compared with 5%, respectively; Soufo & Graumann, 2003). An even milder yet considerable segregation phenotype was observed during depletion of GFP–MreBH (1% anucleate cells, data not shown). Conversely, Mbl has been implicated in the insertion of cell wall precursors into the cell wall, which also seems to follow a helical path, whereas MreB does not seem to be associated with this process (Daniel & Errington, 2003). Thus, it is possible that the primary function of MreB is to separate replicated regions of the chromosome towards opposite cell poles, by means of its motor-like dynamics. To find out whether movement of MreB is affected by the state of the chromosomes, we moved the GFP–MreB fusion into a strain in which transcription of the parE gene (one subunit of topoisomerase IV (topo IV)) is driven by an inducible promoter. Depletion of ParE blocks decatenation of interlocked sister chromosomes, arresting chromosome segregation although cell growth continues, leading to the formation of anucleate cells (Kato et al, 1990; Figs 1F and 3, >15% anucleate cells observed 4–5 doubling times after depletion). During depletion of topo IV, GFP–MreB no longer formed dynamic helical filaments, but was present as slightly curved filaments (Fig 3; in 80% of 125 cells analysed, 20% contained only foci or no signal) that did not move when examined by time-lapse microscopy (data not shown). In contrast to the nutrient shift down experiment (Fig 1D), cell growth continued during depletion of topo IV, showing that a block in chromosome decatenation and/or supercoiling interferes with the movement of MreB. Moreover, GFP–MreB filaments were only seen in cells containing nucleoids, but not in those lacking any detectable DNA (Figs 1F and 3, indicated by arrows). Of 64 anucleate ParE-depleted cells analysed, none contained detectable MreB filaments, although background fluorescence was considerably higher than that in cells expressing MreB instead of GFP–MreB, showing that GFP–MreB is still present in anucleate cells (Fig 1F). Likewise, GFP–MreB filaments were absent in all of the 35 observed anucleate cells in a different (spo0J) mutant background (data not shown). Contrarily, GFP–Mbl or GFP–MreBH filaments were generally present in anucleate cells (in 66 out of 69 cells analysed, Fig 1G, and data not shown), providing a clear cytological distinction between the three actin orthologues and indicating that MreB is—directly or indirectly—associated with the chromosomes.
Figure 3.
Fluorescence microscopy of B. subtilis cells expressing GFP–MreB, in which ParE (a subunit of topo IV) has been depleted for 5–6 doubling times. The arrow indicates anucleate cell. Scale bar, 2 μm.
Conclusions
Our data show for the first time, to our knowledge, that proteins move rapidly along defined tracks within a prokaryotic cell. Multiple filaments or more likely bundles of MreB and Mbl filaments move continuously through growing B. subtilis cells, possibly through a treadmilling mechanism. This process could constitute an ancestor of eukaryotic actin dynamics, such as pushing of membranes in directed cell movement. As actin bundles can push whole L. monocytogenes cells through the cytosol of macrophages (Marx, 2003) and beads through a viscous solution (Wiesner et al, 2003), it is possible that bacterial actin filaments push molecules through Bacillus cells. MreB has recently been shown to be essential for bipolar separation of origin regions on the chromosomes, and thus for chromosome segregation (Kruse et al, 2003; Soufo & Graumann, 2003). MreB could be an active segregation motor, similar to the actin homologue ParM that pushes plasmids away from mid-cell towards opposite cell poles in E. coli (Moller-Jensen et al, 2003). This is feasible because origin regions on the chromosome separate with an average velocity of 0.17 μm/min (Webb et al, 1998), whereas MreB can travel a distance of 0.24 μm/min towards the cell poles. Moreover, MreB filaments moved towards opposite cell poles, and movement of the filaments was lost after a block in chromosome segregation. Alternatively, MreB movement could position cellular components, such as the central replication machinery, or it could drive membrane proteins away from the middle of the cell to the cell poles, such as the polar localized chemotaxis receptors (Maddock & Shapiro, 1993). Conversely, Mbl could direct transport of peptidoglycansynthesizing enzymes through the membrane and thus mediate localization of such enzymes in a helical pattern (Scheffers et al, 2004). Indeed, MreB, but not Mbl or MreBH, was associated with the nucleoids, providing a clear distinction between the three actin orthologues, besides their distinct helical patterns. Our data also suggest that E. coli Min proteins involved in division site selection might perform dynamic movement analogous to that of B. subtilis actin-like proteins, as the former proteins also form helical filaments (Shih et al, 2003) and show oscillating movement (Raskin & de Boer, 1999). It is important to identify the direct targets that are possibly moved or positioned by bacterial actin proteins.
Methods
Growth conditions. E. coli XL1-Blue (Stratagene) or B. subtilis strains were grown in Luria–Bertani rich medium supplemented with 50 μg/ml ampicillin or other antibiotics, where appropriate.
Constructions of plasmids. Gfp mut1 including multiple cloning site was amplified from pSG1729 (Lewis & Marston, 1999) and was cloned in pSG1164 (Lewis & Marston, 1999) in which the gfp mut1 for carboxy-terminal fusion was excised using KpnI and SpeI. The resulting plasmid pHJDS1 was used for N-terminal GFP fusion at the original locus. To obtain inducible N-terminal GFP fusion alleles of mreB, mbl and mreBH at the original locus, the 5′ regions (350–500 base pairs) of the genes were PCR amplified and inserted in the EcoRI and ApaI sites of plasmid pHJDS1 to generate pJS12, pJS13 and pJS16, respectively. To create a fusion of GFP to the N terminus of mreB, mbl and mreBH at an ectopic site on the chromosome, the entire sequences of theses genes were PCR amplified and inserted in the EcoRI and ApaI sites of plasmid pSG1729 (Lewis & Marston, 1999) to generate pJS17, pJS18 and pJS21, respectively. To generate an isopropyl-β-D-thiogalactoside-inducible copy of gfp–mreB at the amylase locus, gfp–mreB was PCR amplified from pJS17 and was cloned as a HindIII–SphI fragment downstream of hyperspank promoter in plasmid pDR111 (a kind gift from D. Rudner, Harvard Medical School).
Bacterial strains. To express GFP–MreB, GFP–Mbl and GFP–MreBH at its original locus in Bacillus, pJS12, pJS13 and pSJ16 plasmids were transformed into wild-type B. subtilis (PY79) selecting for chloramphenicol resistance (Cm, 5 μg/ml) to generate strains JS12 (Pxyl–gfp–mreB), JS13 (Pxyl–gfp–mbl) and JS16 (Pxyl–gfp–mreBH), respectively. For GFP N-terminal fusions at the amy locus, plasmids pJS17, pJS18 and pJS21 for mreB, mbl and mreBH, respectively, were transformed into PY79 selecting for spectinomycin resistance (spec, 25 μg/ml) to generate strains JS17 (Pxyl–gfp–mreB:amy), JS18 (Pxyl–gfp–mbl:amy) and JS21 (Pxyl–gfp–mreBH:amy), respectively. To examine the subcellular localization of GFP–MreB or GFP–Mbl in the absence of ParE, chromosomal DNA from pJH1 (a kind gift from J. Mascarenhas, University of Marburg), in which the expression of ParE is under the control of hyperspank promoter, was used to transform JS17, JS18 or JS21 to produce strains JS22/25/26 (Pxyl–gfp–mreB/mbl/mreBH:amy, Phyperspank–parE).
Image acquisition. For microscopic analysis, Bacillus strains were grown in S750 defined medium (Jaacks et al, 1989) complemented with 1% casamino acids. Fluorescence microscopy was performed on an Olympus AX70 microscope. Cells were mounted on agarose gel pads containing S750 growth medium on object slides. For fixation of cells, a formaldehyde/glutaraldehyde mixture was added to exponentially growing cells, and cells were fixed for 30 min before time-lapse acquisition (Pogliano et al, 1995). Images were acquired with a digital CCD camera; signal intensities and cell length were measured using the Metamorph 4.6 program (Universal Imaging Corp.). DNA was stained with 4′,6-diamidino-2-phenylindole (final concentration 0.2 ng/ml) and membranes were stained with FM4-64 (final concentration 1 nM).
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/vaop/ncurrent/extref.7400209s1.pdf).
Supplementary Material
Supplementary material
movie 1
movie 2
movie 3
movie 4
movie 5
Acknowledgments
We thank J. Mascarenhas for providing valuable plasmids. This work was supported through an Emmy Noether fellowship of the Deutsche Forschungsgemeinschaft.
References
- Carballido-Lopez R, Errington J (2003) The bacterial cytoskeleton: in vivo dynamics of the actin-like protein Mbl of Bacillus subtilis. Dev Cell 4: 19–28 [DOI] [PubMed] [Google Scholar]
- Daniel RA, Errington J (2003) Control of cell morphogenesis in bacteria: two distinct ways to make a rodshaped cell. Cell 113: 767–776 [DOI] [PubMed] [Google Scholar]
- Defeu Soufo HJ, Graumann PL (2003) Actin-like proteins MreB and Mbl from Bacillus subtilis are required for bipolar positioning of replication origins. Curr Biol 13: 1916–1920 [DOI] [PubMed] [Google Scholar]
- Jaacks KJ, Healy J, Losick R, Grossman AD (1989) Identification and characterization of genes controlled by the sporulation regulatory gene spo0H in Bacillus subtilis. J Bacteriol 171: 4121–4129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones LJ, Carballido-Lopez R, Errington J (2001) Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104: 913–922 [DOI] [PubMed] [Google Scholar]
- Kato J, Nishimura Y, Imamura R, Niki H, Hiraga S, Suzuki H (1990) New topoisomerase essential for chromosome segregation in E. coli. Cell 63: 393–404 [DOI] [PubMed] [Google Scholar]
- Kruse T, Moller-Jensen J, Lobner-Olesen A, Gerdes K (2003) Dysfunctional MreB inhibits chromosome segregation in Escherichia coli. EMBO J 22: 5283–5292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger DA, Horwitz AF (1996) Cell migration: a physically integrated molecular process. Cell 84: 359–369 [DOI] [PubMed] [Google Scholar]
- Lewis PJ, Marston AL (1999) GFP vectors for controlled expression and dual labelling of protein fusions in Bacillus subtilis. Gene 227: 101–110 [DOI] [PubMed] [Google Scholar]
- Loisel TP, Boujemaa R, Pantaloni D, Carlier MF (1999) Reconstitution of actin-based motility of Listeria and Shigella using pure proteins. Nature 401: 613–616 [DOI] [PubMed] [Google Scholar]
- Maddock JR, Shapiro L (1993) Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259: 1717–1723 [DOI] [PubMed] [Google Scholar]
- Marx J (2003) How cells step out. Science 302: 214–216 [DOI] [PubMed] [Google Scholar]
- Mitchinson TJ, Cramer LP (1996) Actin-based cell motility and cell locomotion. Cell 84: 371–379 [DOI] [PubMed] [Google Scholar]
- Mogilner A, Oster G (2003) Polymer motors: pushing out the front and pulling up the back. Curr Biol 13: R721–R733 [DOI] [PubMed] [Google Scholar]
- Moller-Jensen J, Jensen RB, Lowe J, Gerdes K (2002) Prokaryotic DNA segregation by an actin-like filament. EMBO J 21: 3119–3127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller-Jensen J, Borch J, Dam M, Jensen RB, Roepstorff P, Gerdes K (2003) Bacterial mitosis. ParM of plasmid R1 moves plasmid DNA by an actin-like insertional polymerization mechanism. Mol Cell 12: 1477–1487 [DOI] [PubMed] [Google Scholar]
- Pogliano K, Harry E, Losick R (1995) Visualization of the subcellular location of sporulation proteins in Bacillus subtilis using immunofluorescence microscopy. Mol Microbiol 18: 459–470 [DOI] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA (1999) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA 96: 4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffers DJ, Jones LJ, Errington J (2004) Several distinct localization patterns for penicillin-binding proteins in Bacillus subtilis. Mol Microbiol 51: 749–764 [DOI] [PubMed] [Google Scholar]
- Shih Y-L, Le T, Rothfield L (2003) Division site selection in Escherichia coli involves dynamic redistribution of Min proteins within coiled structures that extend between the two cell poles. Proc Natl Acad Sci USA 100: 7865–7870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Portnoy DA (1989) Actin filaments and the growth, movement, and spread of the intracellular bacterial parasite, Listeria monocytogenes. J Cell Biol 109: 1597–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya A, van Oudenaarden A (2003) Biomimetic systems for studying actin-based motility. Curr Biol 13: R734–R744 [DOI] [PubMed] [Google Scholar]
- van den Ent F, Amos LA, Lowe J (2001) Prokaryotic origin of the actin cytoskeleton. Nature 413: 39–44 [DOI] [PubMed] [Google Scholar]
- Webb CD, Graumann PL, Kahana J, Teleman AA, Silver P, Losick R (1998) Use of time-lapse microscopy to visualize rapid movement of the replication origin region of the chromosome during the cell cycle in Bacillus subtilis. Mol Microbiol 28: 883–892 [DOI] [PubMed] [Google Scholar]
- Wiesner S, Helfer E, Didry D, Ducouret G, Lafuma F, Carlier MF, Pantaloni D (2003) A biomimetic motility assay provides insight into the mechanism of actin-based motility. J Cell Biol 160: 387–398 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
movie 1
movie 2
movie 3
movie 4
movie 5