Abstract
The objective of this study was to develop a molecular detection method that better estimates the potential risk associated with the presence of Vibrio vulnificus. For that purpose, we applied seminested reverse transcription-PCR (RT-PCR) to viable but nonculturable (VBNC) populations of V. vulnificus and targeted the cytotoxin-hemolysin virulence gene vvhA. Three strains, two environmental, IF Vv10 and IF Vv18, and one clinical, C7184, were used in this study. Artificial seawater, inoculated with mid-log-phase cells, was maintained at 4°C. VBNC cells resulted after 3, 6, and 14 days for C7184, IF Vv18, and IF Vv10, respectively. Our data indicate that seminested RT-PCR is sensitive for the detection of vvhA mRNA in artificial seawater when exclusively nonculturable bacteria are present. This is the first report of the expression of a toxin gene in VBNC V. vulnificus. Moreover, vvhA transcripts were shown to persist in nonculturable populations over a 4.5-month period, with a progressive decline of the signal over time. This result indicates that special attention should be given to the presence of potentially pathogenic VBNC cells in environmental samples when assessing public health risk.
Vibrio vulnificus is a gram-negative estuarine bacterium that has been shown to be responsible for human gastroenteritis, wound and/or soft tissue infections, and fatal primary septicemia, with mortality exceeding 50%. Infections are acquired after consumption of raw or undercooked contaminated seafood or after wound exposure to seawater, and they preferentially affect persons having underlying hepatic diseases or immunocompromised individuals (21).
It has been shown that V. vulnificus enters into a viable but nonculturable (VBNC) state in response to low-temperature incubation (33). When in this state, bacteria can no longer be cultured on the media normally used for their isolation but maintain respiratory and metabolic activities and may resuscitate (22, 32, 33). This physiological state is considered to be a survival strategy in response to adverse environmental conditions and may account for the difficulty in isolating V. vulnificus from water and shellfish during winter months (19).
This phenomenon was demonstrated for many bacterial species, including significant human pathogens such as Vibrio cholerae, Shigella dysenteriae, Escherichia coli O157:H7, and Campylobacter jejuni (18). Moreover, maintenance of the virulence potential of such pathogens in the VBNC state has been demonstrated in several studies (5, 7, 20, 24). Considering this potential, it is important that alternative methods for microbiological quality assessment of food and environmental samples include VBNC pathogens.
During the last few decades, rapid and specific PCR-based detection methods have been developed for several pathogens, including V. vulnificus (1, 12, 13). PCR amplification of the hemolysin gene (4) and random amplification of polymorphic DNA techniques (29) have been applied to VBNC cells of V. vulnificus, but for VBNC bacteria, both techniques suffered a loss of sensitivity. Moreover, even though they can detect VBNC cells, a major disadvantage of these DNA-based detection methods is that they also may amplify DNA from dead microorganisms (8). In many cases, a positive detection of DNA from pathogenic microorganisms may be considered a false-positive result if dead cells or free DNA that no longer present a risk for the consumers are present in the sample.
A molecular method that would better evaluate the potential health hazard associated with these pathogenic bacteria would detect only viable microorganisms (both culturable and nonculturable). Numerous studies have investigated the potential of mRNA detection using reverse transcription-PCR (RT-PCR) as a viability marker. Detection of mRNA is thought to be a good viability marker, due to its central role in cell metabolism and its very short half-life. Moreover, amplification of RNA by RT-PCR provides the advantages of PCR, namely, high specificity and sensitivity, if a suitable target gene is chosen. A good correlation between the detection of mRNA and the presence of culturable bacteria has been demonstrated for several pathogens, including V. cholerae (3), Listeria monocytogenes (9), and E. coli (27).
However, in previous studies, RT-PCR detection was applied exclusively to bacterial cultures or samples artificially contaminated with actively growing cells and thus did not reflect the physiological status of bacteria in the environment. Under natural conditions, and especially in seawater environments, bacteria face several stresses and develop survival strategies, including entry into the VBNC state. In this state, genetic expression may be modified, and it has been reported that DNA, RNA, and protein synthesis, as well as the concentration of ribosomal and nucleic acids, decreased drastically in VBNC cells of V. vulnificus (17). The novelty of our approach is the application of RT-PCR to nonculturable populations of V. vulnificus maintained in artificial seawater (ASW) under conditions resembling the natural environment.
Because V. vulnificus has been involved in severe septicemia and is responsible for most incidences of death associated with raw-oyster consumption, it is of major importance to assess the potential risk associated with the presence of these bacteria (even in the VBNC state) in the marine environment. The aim of this study was to develop a molecular detection method that would not only detect VBNC cells but also allow estimation of the potential virulence of the bacteria. For the latter purpose, we targeted mRNA of the vvhA virulence gene, specific for V. vulnificus (34). This gene encodes a hemolysin that is cytotoxic to Chinese hamster ovary cells and demonstrates cytolytic activity against mammalian erythrocytes (11). This hemolysin also induces vasodilatation and may be involved in the pathogenesis of hypotensive septic shock (10).
MATERIALS AND METHODS
Bacterial strains and growth media.
Three strains of V. vulnificus were used in this study. Two strains of environmental origin were isolated in our laboratory, strain IF Vv10 from a mussel sample in La Rochelle (Charente Maritime, France) and strain IF Vv18 from a water sample collected in the Le Blayais estuary (Gironde, France). Identification and classification of both isolates were performed by phenotypic analysis and confirmed by molecular methods (6). The clinical strain C7184 (avirulent, translucent morphotype) used in this study was kindly provided by J. D. Oliver, University of North Carolina, Charlotte, N.C. Cells were grown on Heart Infusion (HI) agar (Difco Laboratories, Detroit, Mich.) at 37°C or in HI broth, at 37°C with shaking.
Induction of VBNC populations. (i) Inoculation of ASW.
ASW flasks were inoculated with each of the three strains. ASW was prepared with synthetic marine salt (Instant Ocean, Aquarium Systems, Sarrebourg, France). The salinity was adjusted to 33‰, and the pH was adjusted to 8.0. The solution was filtered through a 0.22-μm-pore-size filter (Millipore Corp.) and sterilized at 120°C for 20 min. A 1-ml volume of an overnight culture was used to inoculate 100 ml of fresh HI broth. Cells were allowed to grow to mid-logarithmic phase (optical density at 610 nm, 0.15 to 0.20), centrifuged at 2,500 × g for 20 min at 4°C, and washed twice in sterile ASW at 4°C. Assays, which consisted of 1,500 ml of prechilled ASW in 5-liter flasks, were inoculated to a final 1% concentration with washed cells. This protocol yielded a population of approximately 106 cells/ml in the inoculated ASW.
(ii) Colony enumeration.
Cell culturability was determined using marine agar (MA) plates (Difco Laboratories). Preliminary experiments, using both HI and MA plates, showed that plate counts of V. vulnificus from ASW assays were higher and maintained for a longer time on MA media (data not shown). CFU were enumerated either by diluting samples in cold ASW and spreading 0.1 ml on MA, in triplicate, or by filtering duplicate 1- or 10-ml assay aliquots through 0.22-μm-pore-size filters (Millipore Corp.) and placing each filter on MA plates. The detection limit of the plate counts was 0.05 CFU/ml. Colonies were counted after incubation for 48 h at 37°C. Incubation of plates for an additional 5 days at 37°C or for 3 days at three different incubation temperatures (20, 30, or 37°C) did not increase or change the number of colonies.
(iii) Total cell count.
The total cell number was determined by direct counting after 4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, Saint Quentin Fallavier, France) staining (23). Briefly, 2% formalin-fixed samples were filtered using 0.2-μm-pore-size polycarbonate black filters (Dominique Dutscher S.A., Brumath, France) and stained for 20 min with a 2.5-μg/ml DAPI solution. The filters were rinsed with sterile ultrapure water, mounted on a glass slide, and viewed using low-fluorescence immersion oil under an Olympus epifluorescence microscope with UV excitation.
(iv) Viable-cell count.
Viable-cell counts were determined using the 5-cyano-2,3-ditolyl tetrazolium chloride (CTC; Polysciences Europe, Eppelheim, Germany) method (26); i.e., bacteria with a functioning electron transport chain reduce the CTC in CTC-formazan, forming a red fluorescent precipitate in the cell membrane. Samples were incubated with 3.0 mM CTC, in the presence of 0.025% yeast extract, overnight at room temperature and then fixed with formalin (2% final concentration). Samples were stained with DAPI, as described above. Cells exhibiting red precipitate under green excitation were counted.
(v) Monitoring entry to the VBNC state.
The flasks were incubated in the dark at 4°C with shaking. Entry to the VBNC state was monitored using CFU enumeration and total and viable counts, as described above. A VBNC time zero (Tvbnc 0) was defined for each assay as the first day for which no colonies were detected in 10-ml sample duplicates. For instance, for assays with cells which have been nonculturable for 10 days, the VBNC time is Tvbnc 10.
RT-PCR detection of VBNC cells. (i) Sampling.
Once populations had reached the VBNC state, samples were taken at intervals for total RNA extraction, in order to determine the presence of vvhA mRNA. To compare the results among the three strains, VBNC populations of the same age, i.e., at identical Tvbnc, were analyzed. Assays of strains C7184, IF Vv18, and IF Vv10 were maintained for 136, 139, and 146 days, respectively. At this stage, the bacteria had been VBNC for 133 days. During this long period, the assays were carefully monitored for possible contamination, and no bacteria were found when 10 ml of ASW was filtered and the filter was placed on Trypticase soy salt agar and MA and incubated at 20°C for 4 days.
(ii) RNA isolation.
Cells from the ASW assays were harvested by aseptic filtration (5- to 250-ml volumes) using a 47-mm-diameter autoclaved polycarbonate filter (0.2 μm pore size; Nuclepore) placed on a Millipore membrane. The polycarbonate filter was rolled up and transferred, using sterile forceps, into a 15-ml polypropylene tube containing 2 ml of chilled RNA-NOW solution (Biogentex, Seabrook, Tex.). The tubes were maintained on ice unless otherwise stated. The tube was vortexed vigorously for 1 min, and the filter was dried and removed using a sterile pipette. These RNA homogenates were stored at −80°C before being processed. Total RNA was purified as specified by the manufacturer (Biogentex). RNA precipitation was conducted overnight at −80°C. The RNA pellets were washed twice with 10 ml of 70% ethanol and dissolved in 25 or 50 μl of diethylpyrocarbonate-treated water (Sigma-Aldrich). To maximize solubilization of the RNA, the extracts were heated for 10 min at 60°C and stored at −80°C in the presence of 40 U of RNase Block Ribonuclease inhibitor (Stratagene Europe, Amsterdam, The Netherlands).
(iii) Elimination of contaminating DNA.
To eliminate carryover DNA, 4 to 16 μl of RNA extract was incubated at 37°C for 1 h with 30 U of RNase-free DNase I (Roche Diagnostics, Meylan, France) and 40 U of RNase Block Ribonuclease inhibitor in a total volume of 20 μl. The DNase I was subsequently inactivated by heating the reaction mixture at 90°C for 5 min. To test for the absence of DNA in the DNase-treated RNA extracts, a conventional seminested PCR (without the RT step) was performed with the same primers and conditions used for seminested RT-PCR. RNA samples giving a positive amplification were not analyzed further. DNase digestion was performed again on a more diluted RNA extract, and the absence of DNA was retested.
(iv) Oligonucleotide primers.
The two primers VV1 (5′-GACTATCGCATCAACAACCG-3′, sense primer) and VV2R (5′-AGGTAGCGAGTATTACTGCC-3′, antisense primer) delineate a 704-bp region within the open reading frame of the cytotoxin-hemolysin gene vvhA that is unique to V. vulnificus (12). The internal primer VV3 (5′-GCTATTTCACCGCCGCTCAC-3′), used in the seminested PCR in conjunction with VV2R, generates a 604-bp fragment (13).
(v) Seminested PCR and seminested RT-PCR amplifications.
PCRs and RT-PCRs were conducted on a GeneAmp PCR system 2400 (Perkin Elmer) thermal cycler. To test for the absence of DNA, 4 μl of DNase-treated RNA extract was added to 46 μl of a PCR mixture containing 1× PCR buffer (10 mM Tris-HCl, 1.5 mM MgCl2, 50 mM KCl [pH 8.3]) (Roche Diagnostics), 200 μM each deoxynucleoside triphosphate (dNTP), 1.25 U of Taq DNA polymerase (Roche Diagnostics) and 0.5 μM each primer (VV1 and VV2-R). PCR amplification conditions consisted of a denaturation at 94°C for 5 min followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min. After a final extension at 72°C for 7 min, the tubes were cooled to 4°C. Then 2 μl of the first PCR amplification product was used as template for the seminested PCR, which was performed in a final volume of 40 μl with the same mixture conditions as the first PCR, except that VV3 primer replaced VV1. The PCR conditions of the seminested PCR were as follows: a first denaturation at 94°C for 5 min followed by 30 cycles of denaturation at 94°C for 30 s, annealing at 62°C for 30 s, and extension at 72°C for 45 s and a single final extension at 72°C for 7 min; the tubes were then cooled to 4°C.
The presence of mRNA encoded by the vvhA gene was analyzed by a two-step RT-PCR method in which the components of the GeneAmp RNA PCR kit (Perkin Elmer Applied Biosystems, Courtaboeuf, France) were used. DNase-treated RNA (4 μl) was reverse transcribed in a 20-μl volume with 5 mM MgCl2, 1× PCR buffer II (50 mM KCl, 10 mM Tris-HCl [pH 8.3]), 1 mM each dNTP, 1 U of RNase inhibitor per μl, 2.5 U of murine leukemia virus reverse transcriptase per μl, and 1 μM reverse primer VV2-R. The tubes were incubated at 42°C for 15 min, heated at 99°C for 5 min, and cooled to 5°C for at least 5 min. Half of the RT mixture was added to 40 μl of the PCR mixture. The final concentrations in the PCR mixture were as follows: 2 mM MgCl2, 1× PCR buffer II, 200 μM each dNTP, 1.25 U of AmpliTaq DNA polymerase per 50 μl, and 0.5 μM each primer (VV1 and VV2-R). The cycling parameters of this PCR and the seminested PCR were as described above.
Negative controls with no nucleic acid added were routinely incorporated. DNA extract from a culture of strain C7184 was used as a positive control for PCR and seminested PCR. Purified RNA from a stationary-phase culture of strain C7184 was used as a positive control for RT.
The amplification products (10 μl) were separated by horizontal gel electrophoresis in a 1.5% agarose gel and visualized by ethidium bromide staining and UV illumination.
Sequencing and DNA sequence analysis.
The partial nucleotide sequence of the vvhA gene of strain C7184 was determined after nucleic acid extraction from a culture (6) and seminested PCR amplification, under the conditions described above. Moreover, for each assay, the nucleotide sequences of the amplified products obtained from seminested RT-PCR were determined at the beginning (Tvbnc 3) and at the end (Tvbnc 91, 133, and 133 for strains IF Vv10, IF Vv18, and C7184, respectively) of the experiment. The 604-bp amplified fragments were purified from a 2.0% low-melting agarose gel (Nusieve GTG agarose; BMA, Rockland, Maine) by using columns from the QIAquick extraction kit (Qiagen, Courtaboeuf, France), as recommended by the suppliers. The purified products were sequenced by Genome Express (Meylan, France) on an ABI PRISM 373 automated DNA sequencer using the ABI PRISM Dye Terminator cycle-sequencing kit (PE Applied Biosystems) and primer VV3. Nucleotide sequences were aligned using the CLUSTALW (version 1.8) multiple-alignment program (28).
Lethal treatments.
At the end of the experiments (Tvbnc 133), cells of strain IF Vv18 were killed by boiling at 100°C for 10 min. Samples (50 ml) were filtered immediately after treatment and the RNA was extracted, following the protocol described above. As a positive control, the same volume of ASW was filtered, without treatment, immediately after sampling.
Nucleotide sequence accession numbers.
The partial nucleotide sequence of the vvhA gene from strain C7184 was deposited in GenBank under accession number AY046900. The accession numbers of the partial DNA sequences of the vvhA gene from strains IF Vv10 and IF Vv18 are AF376032 and AF376030, respectively.
RESULTS
VBNC populations.
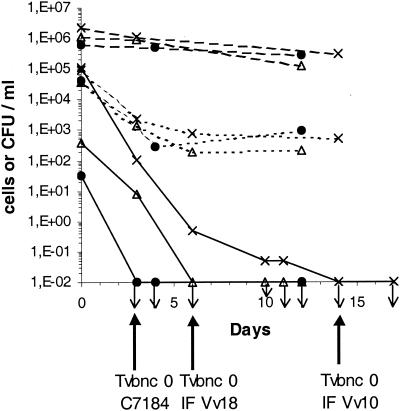
Figure 1 shows the responses of three strains of V. vulnificus to incubation at 4°C in ASW, i.e., the two environmental strains (IF Vv10 and IF Vv18) and the clinical strain (C7184). Three different methods were used to monitor the cells in the ASW: plate count, DAPI staining, and CTC viable count. A rapid loss in culturability was observed. Based on a detection limit of 0.05 CFU/ml (see Materials and Methods), cells of strains C7184, IF Vv18, and IF Vv10 became nonculturable within 3, 6, and 14 days, respectively. These results were verified in four separate experiments for strain C7184 and in two separate experiments for the environmental strains. Using an ASW assay containing VBNC cells of IF Vv18, it was verified that preincubation of the samples at 10% (vol/vol) in marine broth (Difco Laboratories) or in alkaline salt peptone water (30 g of NaCl per liter, 20 g of peptone per liter [pH 8.6]) for 6 h at 20 or 30°C prior to plating did not affect the culturability. The absence of any remaining culturable cells attached to the walls of the flasks was also checked. After the swabbing of an 18.75-cm2 area, preincubation of the swab under the same conditions as those described above, filtration, and plating on MA, no colony was detected. Whereas the cells had lost their culturability, total cell counts remained high and typical curved rods disappeared, with all cells becoming homogenously round, i.e., coccoid. In the three assays, CTC viable counts were nearly identical and declined similarly: counts declined by 2 log units within 4 to 6 days and remained fairly constant near the detection limit, i.e., approximately 500 to 1,000 CTC-respiring bacteria/ml. From these observations, we concluded that bacterial populations in the VBNC state were present in the three assays. Tvbnc 0 was defined for each strain (Fig. 1) and was subsequently used as a reference.
FIG. 1.
Entrance of logarithmic-phase cells of clinical and environmental strains of V. vulnificus into the VBNC state in ASW incubated at 4°C. Shown are the results of total direct count (- - -), CTC viable count (- - - - -), and plate count on MA (—). •, clinical strain C7184; ▵, environmental strain IF Vv18, ×, environmental strain IF Vv10. Downward-pointing arrows indicate a plate count value under the detection limit. For each strain, the first day of nonculturability was designated as time zero VBNC (Tvbnc 0) and was used as the time reference for subsequent analysis.
Detection of cytotoxin-hemolysin mRNA in VBNC populations of V. vulnificus.
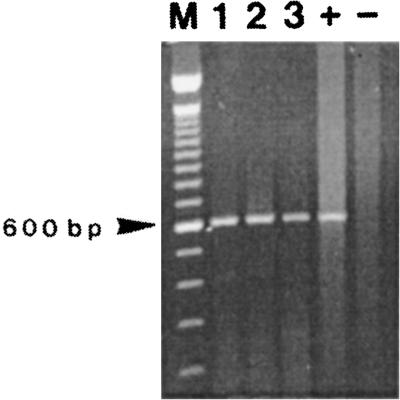
When no culturable bacteria could be detected in the ASW assays, samples were collected at Tvbnc 3, 11, 18, 25, 38, 67, 91, and 133 to study the expression of the cytotoxin-hemolysin gene (vvhA) in aging nonculturable populations. Total RNA was extracted from different filtered volumes, and the vvhA mRNA was amplified by seminested RT-PCR. vvhA mRNA was observed in nonculturable populations of the three strains of V. vulnificus studied for up to 4.5 months. To illustrate this result, Fig. 2 shows amplification products obtained from seminested RT-PCR performed with total RNA extracted from 25-day-old VBNC cells (Tvbnc 25) for the three strains C7184, IF Vv10, and IF Vv18.
FIG. 2.
Detection of vvhA mRNA in 25-day-old VBNC cells of V. vulnificus by seminested RT-PCR. Lanes: 1, clinical strain C7184; 2, environmental strain IF Vv18; 3, environmental strain IF Vv10; M, 100-bp DNA ladder size marker (Life Technologies); +, positive control with RNA extracted from a stationary-phase culture of strain C7184; −, negative control without RNA.
Several controls were tested to confirm this finding. First, all RNA samples were treated with RNase-free DNase I, and a seminested PCR amplification confirmed the absence of amplifiable DNA in the treated RNA extracts. In the absence of amplifiable DNA, the RT-PCR amplification products thus originated from retrotranscribed RNA.
Second, a multiple alignment of a 555-bp portion of the DNA sequence of the vvhA gene from the three strains was performed (data not shown). The percent identity between the clinical strain C7184 and strains IF Vv18 and IF Vv10 were 96.0 and 96.4%, respectively. The percent identity between the two environmental strains was 98.9%. The nucleotide differences allowed us to unambiguously differentiate the three strains. Thus, at the onset of the experiment (Tvbnc 3) and for the end positive results obtained (Tvbnc 133, 133, and 91 for strains C7184, IF Vv18, and IF Vv10, respectively), the RT-PCR-amplified products were sequenced. For each strain, a multiple sequence comparison was performed between the gene sequence and the sequences from the RT-amplified products, on gene portions from 312 to 561 bp, always including the most divergent region. We found that 100% identity was always obtained, demonstrating, for the three strains and over the period studied, the sequence identity of the RT-amplified fragments to the vvhA gene sequence obtained from the respective strain.
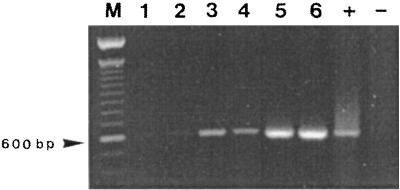
Finally, at each sampling date and for each assay, total RNA was extracted from different filtered volumes of ASW. On the one hand, we observed reproducibility of the results when seminested RT-PCR was performed on equivalent quantities of sample. On the other hand, we showed that the intensity of the RT amplification signal increased when higher concentrations of sample were used (Fig. 3).
FIG. 3.
Effect of filtered volume of ASW on the sensitivity of vvhA mRNA detection, using seminested RT-PCR. Shown are results from 11-day-old VBNC cells of clinical strain C7184. Lanes: 1 and 2, RNA extract from duplicate 5-ml samples; 3 and 4, RNA extract from 25-ml duplicate samples; 5 and 6, RNA extract from 50-ml duplicate samples; M, 100-bp DNA ladder size marker (Life Technologies); +, positive control with RNA extracted from a stationary-phase culture of strain C7184; −, negative control without RNA.
vvhA mRNA detection over time.
By using a standardized protocol as described in Table 1, a decline in signal intensity over a 4.5-month period was observed (Table 1). The ASW assay for strain IF Vv10, at Tvbnc 38, revealed a weak RT amplification signal, disappearing at and after Tvbnc 67. For the strain IF Vv18 assay, the signal intensity decreased at Tvbnc 67 and the signal disappeared at Tvbnc 133. For the C7184 assay, the signal disappeared at and after Tvbnc 67.
TABLE 1.
Cytotoxin-hemolysin mRNA detection by RT-PCR in VBNC populations of three strains of V. vulnificus maintained in ASW at 4°C over a 4.5-month period
| Strain |
vvhA mRNA detection in VBNC population of the following age (Tvbnc [days]):a
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 3 | 11 | 18 | 25 | 38 | 67 | 91 | 133b | |
| C7184 | + | + | + | + | + | − | ND | − |
| IF Vv10 | + | + | + | + | W | − | − | − |
| IF Vv18 | + | + | + | + | + | W | + | − |
Shown are results of seminested RT-PCR amplifications from a defined quantity of sample. A 25-ml volume of ASW was filtered, and the RNA pellet was dissolved in 25 μl of diethylpyrocarbonate-treated water and diluted twofold for DNase treatment. A 4-μl volume of DNase-treated RNA extract was used for RT. +, bright band; W, faint band; −, no band detected on the gel; ND, not determined.
At this stage of the experiment, a weak or a positive result was obtained by concentrating the sample 6- to 12-fold.
After 4.5 months of nonculturability, vvhA mRNA could no longer be detected in the three ASW flasks, using the protocol described in Table 1. However, when the sample was concentrated sixfold, vvhA mRNA was detected in the 133-day-old nonculturable populations of strains C7184 and IF Vv18 but not of strain IF Vv10 (data not shown). For strain IF Vv10, the sample had to be concentrated 12-fold to obtain even a faint band on the gel.
Detection of vvhA mRNA in heat-killed nonculturable cells.
At the end of the experiment (Tvbnc 133), using sixfold-concentrated samples, vvhA mRNA could clearly be detected in the strain IF Vv18 assay. At this stage of the experiment, CTC viable counts could no longer be used to detect viable cells, since the detection limit of the method was reached. To demonstrate that this positive amplification was associated with the presence of viable cells, samples were boiled for 10 min to kill the bacteria and the presence of vvhA mRNA was tested in the treated samples by using the same protocol as the one for the positive control. No signal was detected in the heat-killed samples. The same treatment (boiling for 10 min) applied to purified RNA did not affect RT-PCR amplification. Thus, loss of the signal must be attributed to the disappearance of mRNA due to its rapid degradation following cellular death.
DISCUSSION
Because V. vulnificus is a natural inhabitant of marine environments, bacterial fecal indicators are useless to evaluate the risk associated with its presence and so specific detection methods must be developed. The classical method, which uses isolation on culture media, may underestimate the potential hazard of the samples, since it does not detect the VBNC forms of V. vulnificus. On the other hand, molecular detection methods based on PCR may overestimate the risk associated with the sample, since they may amplify free DNA or DNA from dead microorganisms (8).
The aim of this study was to develop a detection method to estimate the potential risk associated with samples, i.e., to detect VBNC bacteria and only the viable cells. Detection of mRNA using RT-PCR was previously shown to be a method of choice to detect specifically culturable microorganisms (3, 9, 27). We applied this technique to VBNC populations of three strains of V. vulnificus maintained in ASW. Moreover, to estimate the potential virulence of the populations tested, we targeted the mRNA of the vvhA toxin gene.
VBNC populations were obtained for strains C7184, IF Vv18, and IF Vv10 within 3, 6, and 14 days, respectively, by using mid-log-phase cells incubated in cold ASW. These rates of nonculturability are in agreement with previous reports of studies using a similar method to obtain VBNC V. vulnificus (22, 32).
Expression of the virulence gene vvhA, monitored for several months using seminested RT-PCR, was clearly demonstrated in VBNC populations of the three strains. Seminested RT-PCR is specific to RNA detection in the absence of DNA in the nucleic acid extracts. RNase-free DNase I treatment and control of elimination of DNA by using PCR were systematically accomplished in this study to ensure the specificity of the RNA detection. Moreover, sequencing of the amplified products at the onset and at the end of the experiment demonstrated identity of the amplicons to the vvhA gene and absence of external contamination or cross-contamination between assays.
mRNA detection is considered to be a good viability marker because of the very short half-life (a few minutes) of prokaryotic mRNA due to rapid degradation by the very organized RNA decay machinery (25). To confirm that vvhA mRNA detection in VBNC populations was correlated with the presence of viable cells with active gene expression, samples of the IF Vv18 assay mixture at Tvbnc 133 were boiled to kill the remaining viable cells and the total RNA was extracted and amplified. Loss of the RT-PCR signal was clearly shown in the heat-killed samples. Therefore, the validity of RT-PCR as a viability marker, demonstrated in previous studies using culturable bacteria killed by heat or ethanol treatments (9, 27), was confirmed for nonculturable populations of V. vulnificus. Recently, Lleo et al. (16) reported that nonculturable cells of Enterococcus faecalis expressed a gene involved in peptidoglycan synthesis over a 3-month period and that mRNA detection was correlated with different metabolic activities. Using a phylogenetically distant bacterium and a different target gene, we also demonstrate that VBNC bacteria maintain active gene expression for months, confirming the viable state of these nonculturable cells.
To investigate the expression of the vvhA gene, in a first step we amplified mRNA of the vvhA gene from stationary-phase cultures of strain C7184, which is in accordance with the maximum hemolytic activity observed during this growth phase (2, 11). Thus, RNA extracts from stationary-phase cultures were used as positive controls in our experiments. Transcripts of the vvhA gene were also amplified from the nonculturable populations of the three strains tested. To our knowledge, production of hemolysin or the presence of vvhA mRNA in VBNC cells of V. vulnificus has never been reported.
Production of the hemolysin is affected by environmental factors, such as temperature and salinity (14), and is repressed by glucose (2). Like other virulence factors of pathogenic bacteria, expression of the cytotoxin-hemolysin gene of V. vulnificus is undoubtedly highly regulated. At least two regulation systems have been identified, including cyclic AMP and the cyclic AMP receptor protein (2) and the transmembrane transcription activator ToxRS (14). Whether these regulation systems play a role in the expression of vvhA in VBNC populations has yet to be elucidated. Expression of the vvhA gene in the VBNC state, demonstrated in this study, is in accordance with glucose repression and maximum hemolysin production observed in the stationary phase, i.e., under nutrient limitation (2).
Retention of virulence properties by VBNC bacteria was studied previously. One study reported the production of Shiga toxin by 45-day-old VBNC cells of a virulent strain of Shigella dysenteriae type 1 and retention of adherence ability (24). In V. vulnificus, an initial study wherein mice were injected with 3-week-old VBNC bacteria suggested that VBNC cells had lost their virulence (15). However, in a more recent study, a small inoculum of younger VBNC cells was able to kill mice (20). Our results confirmed, at the molecular level, the pathogenic potential of VBNC V. vulnificus.
Although the method used was neither quantitative nor semiquantitative, by using a standardized protocol we were able to detect a decline in the RT-PCR amplification signal over time, with loss of the signal at Tvbnc 67, 67, and 133 for strains C7184, IF Vv10, and IF Vv18, respectively. Moreover, we observed that the intensity of the RT amplification signal increased when larger volumes of sample were used in the RNA extraction. Therefore, the intensity of the signal could be related to the number of cells and the decline of the signal over time could be interpreted as a decrease in the number of RNA-carrying cells in the sample.
The decrease in the RT-PCR signal observed for V. vulnificus VBNC cells is in accordance with results of a study by Weichart et al. (30), who reported a progressive decline in the number of RNA- and DNA-carrying cells in VBNC populations of strain C7184 maintained in the cold for 5 months. Similarly, progressive loss of several viability markers over a 5-month period was described in VBNC populations of E. faecalis (16).
From these data, it appears that the VBNC state may be an intermediate physiological state between culturability and cell death, or a “sporelike” stage. Previous studies demonstrated that soon after entering nonculturability, VBNC cells maintain the ability to recover (32) and to infect mice (20), properties that may be lost after several weeks (15, 31). Alternatively, cells may not be responsive to the methods of resuscitation and of infection employed to date. Therefore, aged VBNC populations may not represent the same intensity of a health hazard as younger ones. The seminested RT protocol used in this study allowed the detection of very low levels of RNA, and it now appears critical that a quantitative or semiquantitative protocol be refined and the threshold beyond which positive amplification can be interpreted as representing a serious threat to public health be defined more precisely.
These experiments demonstrated that expression of a toxin gene is, indeed, detectable in VBNC cells and is maintained for at least 4.5 months under conditions that are not conducive to cell growth or reproduction. Therefore, nonculturable bacteria may not only be viable but may also express virulence factors. This result highlights the potential public health importance of VBNC pathogens in environmental and food samples, a phenomenon which has been recognized (7) and which may account for food-borne outbreaks of unknown origins. To reduce risk, methods other than traditional microbiological protocols must be developed.
Cytotoxin-hemolysin mRNA is an excellent candidate target for direct detection of viable V. vulnificus in environmental samples, since it allows both species-specific and sensitive detection and since it is present not only in culturable but also in VBNC bacteria.
RT-PCR is concluded to be a sensitive, rapid, and reliable method for monitoring viable pathogens in ASW. We currently are developing specific protocols for the molecular detection of viable natural populations of V. vulnificus in seawater and shellfish samples.
Acknowledgments
Michèle Gourmelon is gratefully acknowledged for her helpful discussions of stress and survival mechanisms in bacteria, and Maria del Mar Lleo from the Instituto di Microbiologia, Universita di Verona, Verona, Italy, is thanked for her excellent advice about assay preparation and RT-PCR development. The technical assistance of Annick Derrien is also acknowledged. We thank Jean Pierre Annezo for preparation of Fig. 2 and 3.
This work was supported by a postdoctoral fellowship from the Ministère de la Recherche, de la Technologie et de l'Enseignement Supérieur, Paris, France.
REFERENCES
- 1.Arias, C. R., E. Garay, and R. Aznar. 1995. Nested PCR method for rapid and sensitive detection of Vibrio vulnificus in fish, sediments, and water. Appl. Environ. Microbiol. 61:3476-3478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bang, Y. B., S. E. Lee, J. H. Rhee, and S. H. Choi. 1999. Evidence that expression of the Vibrio vulnificus hemolysin gene is dependent on cyclic AMP and cyclic AMP receptor protein. J. Bacteriol. 181:7639-7642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bej, A. K., W.-Y. Ng, S. Morgan, D. D. Jones, and M. H. Mahbubani. 1996. Detection of viable Vibrio cholerae by reverse-transcriptase polymerase chain reaction (RT-PCR). Mol. Biotechnol. 5:1-10. [DOI] [PubMed] [Google Scholar]
- 4.Brauns, L. A., M. C. Hudson, and J. D. Oliver. 1991. Use of the polymerase chain reaction in detection of culturable and nonculturable Vibrio vulnificus cells. Appl. Environ. Microbiol. 57:2651-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Colwell, R. R., P. R. Brayton, D. Herrington, S. A. Huq, and M. M. Levine. 1996. Viable but non-culturable Vibrio cholerae O1 revert to a cultivable state in the human intestine. World J. Microbiol. Biotechnol. 12:28-31. [DOI] [PubMed] [Google Scholar]
- 6.Hervio-Heath, D., R. R. Colwell, A. Derrien, A. Robert-Pillot, J. M. Fournier, and M. Pommepuy. 2002. Occurence of pathogenic vibrios in coastal areas of France. J. Appl. Microbiol. 92:1123-1135. [DOI] [PubMed] [Google Scholar]
- 7.Huq, A., I. N. G. Rivera, and R. R. Colwell. 2000. Epidemiological significance of viable but nonculturable microorganisms, p. 301-323. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, D.C.
- 8.Josephson, K. L., C. P. Gerba, and I. L. Pepper. 1993. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl. Environ. Microbiol. 59:3513-3515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein, P. G., and V. K. Juneja. 1997. Sensitive detection of viable Listeria monocytogenes by reverse transcription-PCR. Appl. Environ. Microbiol. 63:4441-4448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kook, H., S. E. Lee, Y. H. Baik, S. S. Chung, and J. H. Rhee. 1996. Vibrio vulnificus hemolysin dilates rat thoracic aorta by activating guanylate cyclase. Life Sci. 59:41-47. [DOI] [PubMed] [Google Scholar]
- 11.Kreger, A. S., and D. Lockwood. 1981. Detection of extracellular toxin(s) produced by Vibrio vulnificus. Infect. Immun. 33:583-590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lee, J. Y., J. B. Eun, and S. H. Choi. 1997. Improving detection of Vibrio vulnificus in Octopus variabilis by PCR. J. Food Sci. 62:179-182. [Google Scholar]
- 13.Lee, S. E., S. O. Kim, S. J. Kim, H. S. Kim, J. H. Shin, S. H. Choi, S. S. Chung, and J. H. Rhee. 1998. Direct identification of Vibrio vulnificus in clinical specimens by nested PCR. J. Clin. Microbiol. 36:2887-2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee, S. E., S. H. Shin, S. Y. Kim, Y. R. Kim, D. H. Shin, S. S. Chung, Z. H. Lee, J. Y. Lee, K. C. Jeong, S. H. Choi, and J. H. Rhee. 2000. Vibrio vulnificus has the transmembrane transcription activator ToxRS stimulating the expression of the hemolysin gene vvhA. J. Bacteriol. 182:3405-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Linder, K., and J. D. Oliver. 1989. Membrane fatty acid and virulence changes in the viable but nonculturable state of Vibrio vulnificus. Appl. Environ. Microbiol. 55:2837-2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lleo, M. D., S. Pierobon, M. C. Tafi, C. Signoretto, and P. Canepari. 2000. mRNA detection by reverse transcription-PCR for monitoring viability over time in an Enterococcus faecalis viable but nonculturable population maintained in a laboratory microcosm. Appl. Environ. Microbiol. 66:4564-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oliver, J. D. 1993. Formation of viable but nonculturable cells, p. 239-272. In S. Kjelleberg (ed.), Starvation in bacteria. Plenum Press, New York, N.Y.
- 18.Oliver, J. D. 2000. The public health significance of viable but nonculturable bacteria, p. 277-300. In R. R. Colwell and D. J. Grimes (ed.), Nonculturable microorganisms in the environment. ASM Press, Washington, D.C.
- 19.Oliver, J. D. 1995. The viable but non-culturable state in the human pathogen Vibrio vulnificus. FEMS Microbiol. Lett. 133:203-208. [DOI] [PubMed] [Google Scholar]
- 20.Oliver, J. D., and R. Bockian. 1995. In vivo resuscitation, and virulence toward mice, of viable but nonculturable cells of Vibrio vulnificus. Appl. Environ. Microbiol. 61:2620-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oliver, J. D., and J. B. Kaper. 1997. Vibrio species, p. 228-264. In M. P. Doyle, L. R. Beuchat, and T. J. Montville (ed.), Food microbiology: fundamentals and frontiers. ASM Press, Washington, D.C.
- 22.Oliver, J. D., L. Nilsson, and S. Kjelleberg. 1991. Formation of nonculturable Vibrio vulnificus cells and its relationship to the starvation state. Appl. Environ. Microbiol. 57:2640-2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Porter, K. G., and Y. S. Feig. 1980. The use of DAPI for identifying and counting aquatic microflora. Limnol. Oceanogr. 25:943-948. [Google Scholar]
- 24.Rahman, I., M. Shahamat, M. A. R. Chowdhury, and R. R. Colwell. 1996. Potential virulence of viable but nonculturable Shigella dysenteriae type 1. Appl. Environ. Microbiol. 62:115-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rauhut, R., and G. Klug. 1999. mRNA degradation in bacteria. FEMS Microbiol. Rev. 23:353-370. [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez, G. G., D. Phipps, K. Ishiguro, and H. F. Ridgway. 1992. Use of a fluorescent redox probe for direct visualization of actively respiring bacteria. Appl. Environ. Microbiol. 58:1801-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sheridan, G. E. C., C. I. Masters, J. A. Shallcross, and B. M. Mackey. 1998. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl. Environ. Microbiol. 64:1313-1318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Warner, J. M., and J. D. Oliver. 1998. Randomly amplified polymorphic DNA analysis of starved and viable but nonculturable Vibrio vulnificus cells. Appl. Environ. Microbiol. 64:3025-3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weichart, D., D. McDougald, D. Jacobs, and S. Kjelleberg. 1997. In situ analysis of nucleic acids in cold-induced nonculturable Vibrio vulnificus. Appl. Environ. Microbiol. 63:2754-2758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weichart, D., J. D. Oliver, and S. Kjelleberg. 1992. Low temperature induced nonculturability and killing of Vibrio vulnificus. FEMS Microbiol. Lett. 100:205-210. [DOI] [PubMed] [Google Scholar]
- 32.Whitesides, M. D., and J. D. Oliver. 1997. Resuscitation of Vibrio vulnificus from the viable but nonculturable state. Appl. Environ. Microbiol. 63:1002-1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf, P. W., and J. D. Oliver. 1992. Temperature effects on the viable but nonculturable state of vibrio vulnificus. FEMS Microbiol. Ecol. 101:33-39. [Google Scholar]
- 34.Yamamoto, K., A. C. Wright, J. B. Kaper, and J. G. Morris. 1990. The cytolysin gene of Vibrio vulnificus: sequence and relationship to V. cholerae El Tor hemolysin gene. Infect. Immun. 58:2706-2709. [DOI] [PMC free article] [PubMed] [Google Scholar]