Summary
Symposium on Bacterial Chromosomes
Keywords: cell division, DNA replication, cytoskeleton, cell cycle
Introduction
From the study of bacteria we know a great deal about DNA replication, recombination and repair, transcription, translation and gene regulation, and cellular metabolism. However, despite powerful molecular genetics tools, our understanding of the organization and behaviour of bacterial chromosomes has lagged behind, simply because their dynamics have been difficult to observe directly. Bacteria lack a mitotic system, and an equivalent prokaryotic mechanism for the separation of duplicated chromosomes remains mysterious. Confounding the picture is the complexity of the bacterial cell cycle, which is unlike the orderly and interdependent progression of events in eukaryotes.

The Keystone Symposium on Bacterial Chromosomes was held between 7 and 12 February 2004 in Santa Fe, New Mexico, USA. The conference was organized by S. Gottesman, N. Kleckner and J. Roth.
The application of fluorescence microscopy techniques, flow cytometry and cell synchronization has revolutionized our understanding of chromosome dynamics and the cell cycle in bacteria. Much of this conference was devoted to how chromosomes are organized, faithfully replicated and segregated to daughter cells, a few highlights of which are presented here.
Dominating the discussion were studies of two model organisms—Bacillus subtilis and Escherichia coli—that diverged from a common ancestor over two billion years ago and are very different beasts. Despite this, these organisms share a number of common features. Both are rod-shaped cells, about the size of a yeast nucleus, that divide at midcell. Both initiate replication at a single origin (oriC) in a circular chromosome, and bidirectional forks converge near a terminus region (ter) that has special properties to ensure chromosome segregation to daughter cells. Although there is no true nucleus in bacterial cells, a region of condensed chromosomal DNA (called a 'nucleoid') divides and separates before cell fission. In slow-growing cells, replication is temporally separated from cell division, similar to the eukaryotic cell cycle with G1, S, G2 and M phases. In rapidly dividing cells, however, new rounds of replication ensue before completion of the previous round, so that daughter cells are born with forks already partially progressed.
Defining events in the bacterial cell cycle
D. Bates (in the laboratory of N. Kleckner, Cambridge, MA, USA) presented an elegant analysis of chromosome dynamics in E. coli. Synchronous cell populations were obtained by a new 'baby cell' method and were analysed for landmark events (DNA replication and septation) and for the number and locations of oriC, ter and interstitial regions by fluorescence in situ hybridization (FISH) and of the replisome component DnaX, labelled with green fluorescent protein (GFP). For simplicity, cells were grown at relatively long doubling times such that each cell contained only a single (or two duplicating or duplicated) chromosomes. These studies provide new information about the early marking of the midcell, replisome positioning, relationships between sister chromosomes and the mechanism of coordination between DNA replication and cell division.
Sister chromosome cohesion
The pioneering work of S. Hiraga (Kyoto, Japan) suggested that E. coli sister chromosomes remain associated for a considerable length of time after duplication (Sunako et al, 2001), which is reminiscent of sister chromatid cohesion in eukaryotic cells. In synchronized E. coli cultures, the duplication of oriC, ascertained by flow cytometry, clearly precedes the period when two distinct oriC FISH foci are apparent. However, this finding has been challenged by others (Roos et al, 2001). Using an approach similar to Hiraga, Bates and Kleckner also observed a period of cohesion of oriC and other chromosomal loci. The generality and consequence of this cohesion in prokaryotes is unclear. The molecular basis of E. coli chromosome cohesion is also yet to be determined, although the structural maintenance of chromosome (SMC)-like protein MukB is a candidate.
The replisome back on track?
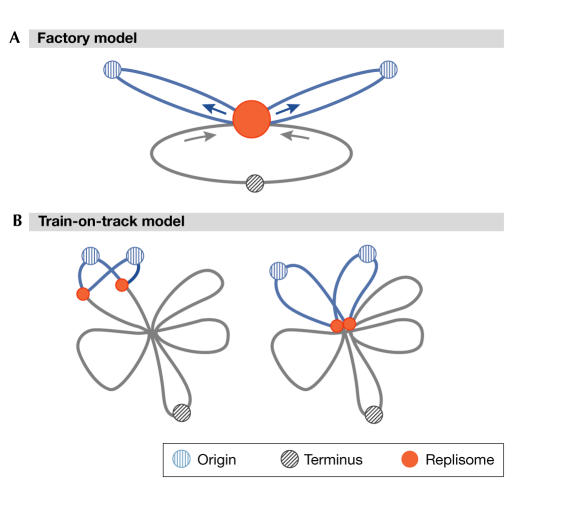
The early positioning of replication factors in Bacillus at the midcell led to the idea of a fixed replication 'factory' (Lemon & Grossman, 1998). This contrasts with the 'train-on-track' model, in which the replisome moves along a spatially fixed chromosome (Fig 1). One attractive feature of the factory model is that the movement of DNA from both replication forks through a fixed replisome could provide the required force and organization for the separation of newly replicated chromosomes (Lemon & Grossman, 2000). However, results presented at the conference call into question strict interpretations of the factory model. A. Wright (Boston, MA, USA) observed GFP fused either to DnaX of E. coli or to SSB—the singlestranded binding protein present at the fork—and concluded that the replisome is a more dynamic structure than previously thought. Wright observed that a single SSB focus could split into two foci and merge together over a time frame of several minutes. Results consistent with a dynamic character of the replisome were presented by several other speakers at the conference, including Bates for E. coli, and E. Harry (Sydney, Australia) for a B. subtilis replisome component. Wright urged reconsideration of the train-on-track model, although neither this nor the factory model may be strictly correct: the dynamic behaviour observed is also consistent with a mobile factory with weakly coupled replisomes.
Figure 1.
Models for replication. (A) Factory model. Replication occurs within a stationary complex (red circle). DNA is pulled into the complex and extruded, as shown by arrows. (B) Train-on-track model. Independent sister replication forks move around the cell in a dynamic manner, with variable overlap. (Figure provided by A. Wright.)
A bacterial centromere?
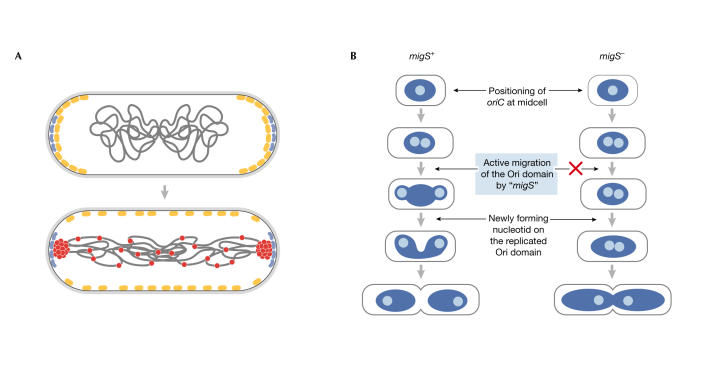
How do origin regions migrate to cell poles during the bacterial cell cycle? Their positioning does not depend on the presence of oriC per se (Gordon et al, 2002), which indicates that another site is necessary for the migration. H. Niki (Mishima, Japan) identified a potential 'centromere' in E. coli, which he termed migS. This 25-bp sequence comprises an imperfect stem–loop and is responsible for the bipolar positioning of the origin region (Yamaichi & Niki, 2004). Deletion of the migS sequence impairs the positioning of oriC (Fig 2B), whereas moving migS from its normal position—211 kb from oriC—to alternative positions in the chromosome, including near ter, results in the positioning of these regions near the cell poles. However, the loss of migS does not drastically affect either the viability or the number of anucleate E. coli cells. Studies are under way to identify the proteins that bind migS.
Figure 2.
Models of the B. subtilis and E. coli 'centromeres'. (A) In B. subtilis, RacA protein (red) interacts with DivIVA (blue) during sporulation to pull the origin regions to cell poles in the predivisional sporangium, as MinCD (yellow) moves away. (Figure provided by R. Losick; Ben-Yehuda et al, 2003.) (B) In E. coli, a migS+ and migS− E. coli cell model of the effect of migS on polarization of oriC. (Figure provided by H. Niki; Yamaichi & Niki, 2003.)
A functionally related, but specialized, type of anchor is present in B. subtilis during sporulation. This involves an asymmetric division in which each cell must ensure that a copy of the chromosome is faithfully packaged into the small forespore compartment. R. Losick and his group (Boston, MA, USA) identified a protein, RacA, that binds preferentially near oriC during sporulation and 'tacks' the chromosome to the membrane by interacting with the DivIVA protein (Ben-Yehuda et al, 2003). Deletion of RacA both decreases the number of forespores in B. subtilis and impairs forespore maturation. Chromatin immunoprecipitation (ChIP) experiments identified several regions of more than 600 kb, predominantly to the left side of oriC, which interact with RacA. A subsequent bioinformatic analysis that compared 20 prominent RacA-binding regions identified a putative RacA-binding motif that consists of a 14-bp hairpin. Translocation of this motif to an origin-proximal region that is a cold spot for RacA interactions converted it into a hot spot in the ChIP experiment. RacA also nonspecifically binds to and compacts DNA. Losick presented a model in which RacA molecules that are bound to their preferred sites near the origin interact with each other and, directly or indirectly, with DivIVA (Fig 2A). This latter interaction promotes the positioning of oriC at the poles.
The dynamic bacterial cytoskeleton
Two homologous proteins, MreB and Mbl, constitute the bac-terial cytoskeleton and both their structures resemble actin. P. Graumann (Marburg, Germany) has studied their dynamics using GFP fusions to these proteins and time-lapse photography. He found that each is assembled into an apparently independent helical filament beneath the cellular membrane. These filaments move in opposite directions: MreB from the middle of the cell towards the poles, and Mbl from the pole to the centre. The MreB filaments disappear in the absence of DNA. Depletion of either MreB or Mbl causes aberrant partitioning of origins and a large fraction of anucleate cells (Soufo & Graumann, 2003). K. Marians (New York, NY, USA) has identified another E. coli protein, SetB, which assembles as a helical filament (Espeli et al, 2003). Although the MreB and SetB helical filaments coincide in small cells, they do not colocalize entirely in longer, filamentous cells. SetB is an integral membrane protein that interacts with MreB in yeast two-hybrid experiments. Although the mutation of SetB has only subtle effects, with mutants showing incomplete separation of nucleoids, its overexpression causes greater movement of origins and nucleoid phenotypes. The nature of the interactions between the cytoskeleton and oriC, however, remains to be determined. Possible intermediaries include the MukBEF complex (see below), SeqA or as-yet unidentified players.
Nucleoid organization
If the cytoskeleton is dynamic, how dynamic is the nucleoid? A. Segall (San Diego, CA, USA) and F. Boccard (Gif-sur-Yvette, France) independently presented their use of phage λ integrase-mediated rearrangements as a measure of ease of access between pairs of chromosomal attachment (att) sites. Results from both labs suggest that nucleoids are not infinitely fluid: the accessibility of one region from another depends on their location. Boccard and colleagues have found that interactions are restricted to each of four structured 'macrodomains'. Two of these macrodomains, one around oriC and the other around ter, coincide with the two chromosomal domains identified using FISH (Niki et al, 2000). As in E. coli, the Segall lab found large variations in accessibility in the Salmonella chromosome. In contrast to the Boccard lab findings, these variations were not strictly related to the size of the intervening chromosomal segments; intervals with one endpoint near the origin and the second near the terminus showed efficient interactions (Garcia-Russell et al, 2004). Although the overall message is the same—that the chromosome is ordered in some way—what accounts for the differences between the findings? The Boccard lab first screened for the occurrence of recombination by the appearance of a Lac+ phenotype, which required that inversions between the pairs of loci did not kill the cell. They subsequently investigated some of the identified intervals using quantitative PCR. By contrast, the Segall lab screened intervals using only a PCR assay, which did not require that cells bearing the inversions remained viable. The two emerging pictures are likely to give complementary views of the evolutionarily successful possibilities versus strictly physical limitations of nucleoid structure on interactions. In both views, such global limitations might be due to physical barriers imposed by interactions of the nucleoid with the cytoskeleton and/or the cell membrane, the segregation machinery, replication factories, or other factors.
N. Cozzarelli (Berkeley, CA, USA) presented a microarray-based investigation of nucleoid structure: domain boundaries were inferred by cutting the genome and monitoring the effect of the superhelical density change on neighbouring gene expression. The distance between the cut site and the affected genes indicated a domain size of roughly 10 kb, which coinicides with the size of topologically constrained loops that have been identified by electron microscopy. A search for 'domainins'—proteins involved in maintaining domains—has been initiated and two candidate proteins, the histone-like nucleoid structuring protein (H-NS) and DksA (a suppressor of dnaK and mukB mutations), have been identified. V. Rybenkov (Norman, OK, USA) presented data on the condensin activity of the MukB protein alone versus the MukBEF complex, and proposed that the former by itself acts as a condensin, whereas the entire complex might link the nucleoid to the cytoskeleton.
Separation anxieties
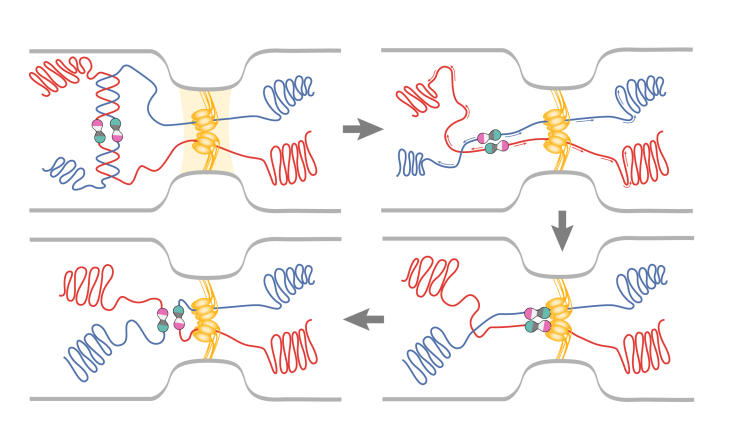
After replication, sister chromosomes are segregated to daughter cells usually, but not always, uneventfully. Recombinationdependent repair of collapsed replication forks sometimes generates chromosome dimers that are resolved into monomers by the action of XerC and XerD near the terminus of chromosome replication at a site known as dif. A nudge from FtsK, a DNA translocase, to XerD is necessary for this event (Massey et al, 2004). In addition, the two dif sites in the dimer chromosome must be juxtaposed near the septum. F. Cornet, J. Louarn and colleagues (Toulouse, France) have proposed that, to achieve this localization, FtsK interacts with short polarized recombination activating gene (RAG) sequences that are over-represented throughout the chromosome (Capiaux et al, 2002; Corre & Louarn, 2002). A chimeric FtsK102 protein that cannot activate E. coli Xer recombination, but can properly position the dif site, has been used to genetically separate these two functions (Yates et al, 2003). FtsK, through its interactions with FtsZ and DNA, is the physical link between the terminus of sister chromosomes and the cell-division machinery (Fig 3).
Figure 3.
Events at the terminus before division. During decatenation, dimer chromosomes are generated. FtsK (yellow ovals) interacts with polarized RAG sequences (arrows) to translocate the chromosomes in such a way that the dif sites (pink/green circles) are juxtaposed near FtsK. XerC/D recombination monomerizes the chromosomes for partition to daughter cells. (Figure provided by C. Lesterlin and F. Cornet.)
Determining the division site: nucleoid occlusion
In both B. subtilis and E. coli, the formation of a ring of the tubulin homologue FtsZ, is one of the early events that defines the future cell division plane (reviewed in Errington et al, 2003). Previous work suggests that two mechanisms regulate FtsZ ring formation. First, the presence of bulk DNA has an inhibitory effect on FtsZ ring formation, a phenomenon termed 'nucleoid occlusion' (Woldringh et al, 1991). Second, the MinCD proteins inhibit Z-ring formation in the nucleoid-free regions of the cell poles. In B. subtilis, DivIVA localized to the poles recruits MinCD (Marston et al, 1998); in E. coli, MinCD oscillates temporally between the two cell poles, aided by a third protein, MinE (Raskin & de Boer, 1999; Hu & Lutenhaus, 2001). J. Errington (Oxford, UK) presented the first candidate for a factor that mediates nucleoid occlusion, the nucleoid-localized Noc protein of B. subtilis. Disruption of noc caused replication-inhibited cells to divide through their nucleoid. Mutants in noc were synthetically lethal with minC or minD, but, surprisingly, these cells arrested without division. FtsZ appeared diffuse in these cells suggesting that, in the absence of these two systems, FtsZ apparently does not localize properly to initiate polymerization into a ring.
Is there a midcell 'mark'?
Harry has put forward the idea of a common midcell 'mark' for FtsZ-ring assembly and the replication factory: the replication factory masks the Z-ring assembly site at midcell during early stages of replication and later unmasks this site when the replisomes divide (Harry, 2001). At this conference, she reported on studies that followed the localization of PolC–GFP and FtsZ–YFP fusion proteins in B. subtilis. Although both at midcell, the PolC foci showed a much broader distribution than FtsZ. In addition, the resolution of the replication factory into two replisomes occurred throughout the round of replication. These findings are inconsistent with the midcell replication factory acting as a physical block to Z-ring assembly. However, the possibility remains that the replication factory and the Z ring respond differently to a common positioning mechanism.
D. Sherratt (Oxford, UK) has also championed the idea that the quarter-cell length position (the future midcell) acts as a mark at which oriC, the replisome, FtsZ, ter, FtsK and the septum sequentially reside (Sherratt, 2003). At the meeting, Sherratt reported that FtsZ localization can be seen in this region when the nucleoid is still present, which challenges the idea of nucleoid occlusion of FtsZ positioning.
Spontaneous replication fork reversal and restart
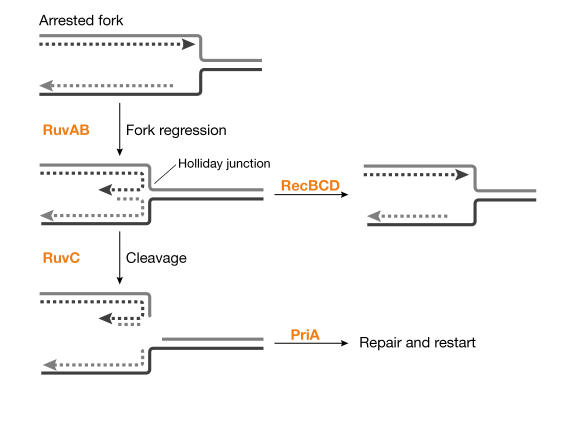
B. Michel (Jouy-en-Josas, France) presented several scenarios by which the fork can be arrested, and which can lead to different modes of processing. For example, arrest caused by defects in the replisome leads to replication fork reversal, such that nascent strands anneal and produce a Holliday junction structure in the retrograde fork (Fig 4). The nascent strand 'nub' of linear DNA is normally removed by RecBCD to allow restart; in the absence of the doublestranded break repair factor RecBCD, the fork can be cleaved by RuvABC, which produces linear chromosomes that are repaired by recombination and the PriA primosome to restore an intact fork (Seigneur et al, 1998). At the conference, Michel presented new data showing that most spontaneous replication arrests are processed similarly to that defined for replisome mutants, implying that replisome failure is a reasonably frequent spontaneous event. Because spontaneous events are difficult to observe, Michel used priA mutants, in which any arrested forks should be trapped. Linear DNA was detected in priA recB double mutants and was dependent on the RuvABC proteins, whereas RecBCD+ prevented such breakage in priA mutants.
Figure 4.
Spontaneous replication fork arrest and its processing. Spontaneous replication arrest events undergo fork reversal. The resulting Holliday junction structure is stabilized by RuvAB. The double-stranded nub can be digested by RecBCD nuclease, which backs up the fork. In the absence of RecBCD, the junction is cleaved by RuvC, producing a broken chromosome requiring repair. PriA is required to restart replication after completion of repair.
G-protein signalling of replication fork status
S.T.L. (Waltham, MA, USA) reported that certain mutants of an essential Ras-related G-protein, ObgE, are strongly sensitive to inhibitors of replication, and show abnormal septation and nucleoid separation. In the absence of RecA and RecBCD, obgE mutants were weakly viable, which indicates that ObgE acts to protect forks from breakage. Flow cytometry and SeqA–GFP localization suggested synchronous DNA replication or replication fork loss in obgE mutants. Lovett proposed that ObgE acts as a signalling factor for the deoxynucleotide pool status and progression of the replication fork and might regulate fork initiation, progression and stability. Furthermore, ObgE might regulate subsequent processes such as nucleoid segregation and cell division, which would be analogous to the S-phase checkpoint of eukaryotes.
Conclusion
This is an exciting time for studies of the bacterial chromosome, in which new visualization techniques can be combined with powerful molecular genetics. Details are beginning to emerge about how chromosomes are organized, replicated and segregated in these smallest of cells. There are still fundamentally important questions to address and we look forward to a mechanistic understanding of the dynamics of the prokaryotic chromosome.


Acknowledgments
The authors thank the organizers of this Keystone Symposium, which was sponsored in part by the Director's Sponsor Fund. We thank the speakers for allowing us to cite their results and regret that space limitations did not allow us to cover all the fine work discussed at the conference. S.T.L. and A.M.S. are supported by the National Institute of General Medical Sciences.
References
- Ben-Yehuda S, Rudner DZ, Losick R (2003) RacA, a bacterial protein that anchors chromosomes to the cell poles. Science 299: 532–536 [DOI] [PubMed] [Google Scholar]
- Capiaux H, Lesterlin C, Perals K, Louarn JM, Cornet F (2002) A dual role for the FtsK protein in Escherichia coli chromosome segregation. EMBO Rep 3: 532–536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corre J, Louarn JM (2002) Evidence from terminal recombination gradients that FtsK uses replichore polarity to control terminus positioning at division in Escherichia coli. J Bacteriol 184: 3801–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J, Daniel RA, Scheffers DJ (2003) Cytokinesis in bacteria. Microbiol Mol Biol Rev 67: 52–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli O, Nurse P, Levine C, Lee C, Marians K (2003) SetB: an integral membrane protein that affects chromosome segregation in E. coli. Mol Microbiol 50: 495–509 [DOI] [PubMed] [Google Scholar]
- Garcia-Russell N, Harmon TG, Le TQ, Amaladas NH, Mathewson RD, Segall AM (2004) Unequal access of chromosomal regions to each other in Salmonella: probing chromosome structure with phage λ integrase-mediated long-range rearrangements. Mol Microbiol 52: 329–344 [DOI] [PubMed] [Google Scholar]
- Gordon GS, Shivers RP, Wright A (2002) Polar localization of the Escherichia coli oriC region is independent of the site of replication initiation. Mol Microbiol 44: 501–507 [DOI] [PubMed] [Google Scholar]
- Harry EJ (2001) Bacterial cell division: regulating Z-ring formation. Mol Microbiol 40: 795–803 [DOI] [PubMed] [Google Scholar]
- Hu Z, Lutkenhaus J (2001) Topological regulation of cell division in E. coli spatiotemporal oscillation of MinD requires stimulation of its ATPase by MinE and phospholipid. Mol Cell 7: 1337–1343 [DOI] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD (1998) Localization of bacterial DNA polymerase: evidence for a factory model of replication. Science 282: 1516–1519 [DOI] [PubMed] [Google Scholar]
- Lemon KP, Grossman AD (2000) Movement of replicating DNA through a stationary replisome. Mol Cell 6: 1321–1330 [DOI] [PubMed] [Google Scholar]
- Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J (1998) Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev 12: 3419–3430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey TH, Aussel L, Barre F-X, Sherratt DJ (2004) Asymmetric activation of Xer sitespecific recombination by FtsK. EMBO Rep 5: 399–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niki H, Yamaichi Y, Hiraga S (2000) Dynamic organization of chromosomal DNA in Escherichia coli. Genes Dev 14: 212–223 [PMC free article] [PubMed] [Google Scholar]
- Raskin DM, de Boer PA (1999) Rapid pole-to-pole oscillation of a protein required for directing division to the middle of Escherichia coli. Proc Natl Acad Sci USA 96: 4971–4976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos M, van Geel AB, Aarsman ME, Veuskens JT, Woldringh CL, Nanninga N (2001) The replicated ftsQAZ and minB chromosomal regions of Escherichia coli segregate on average in line with nucleoid movement. Mol Microbiol 39: 633–640 [DOI] [PubMed] [Google Scholar]
- Seigneur M, Bidnenko V, Ehrlich SD, Michel B (1998) RuvAB acts at arrested replication forks. Cell 95: 419–430 [DOI] [PubMed] [Google Scholar]
- Sherratt DJ (2003) Bacterial chromosome dynamics. Science 301: 780–785 [DOI] [PubMed] [Google Scholar]
- Soufo HJ, Graumann PL (2003) Actin-like proteins MreB and Mbl from Bacillus subtilis are required for bipolar positioning of replication origins. Curr Biol 13: 1916–1920 [DOI] [PubMed] [Google Scholar]
- Sunako Y, Onogi T, Hiraga S (2001) Sister chromosome cohesion of Escherichia coli. Mol Microbiol 42: 1233–1241 [DOI] [PubMed] [Google Scholar]
- Woldringh CL, Mulder E, Huls PG, Vischer N (1991) Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol 142: 309–320 [DOI] [PubMed] [Google Scholar]
- Yamaichi Y, Niki H (2004) migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome. EMBO J 23: 221–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates J, Arroyo M, Sherratt DJ, Barre FX (2003) Species specificity in the activation of Xer recombination at dif by FtsK. Mol Microbiol 49: 241–249 [DOI] [PubMed] [Google Scholar]