Abstract
The crystal structure of the ligand-binding domain of RARβ, a suspect tumour suppressor, reveals important features that distinguish it from the two other RAR isotypes. The most striking difference is an extra cavity allowing RARβ to bind more bulky agonists. Accordingly, we identified a ligand that shows RARβ selectivity with a 100-fold higher affinity to RARβ than to α or γ isotypes. The structural differences between the three RAR ligand-binding pockets revealed a rationale explaining how a single retinoid can be at the same time an RARα, γ antagonist and an RARβ agonist. In addition, we demonstrate how to generate an RARβ antagonist by gradually modifying the bulkiness of a single substitution. Together, our results provide structural guidelines for the synthesis of RARβ-selective agonists and antagonists, allowing for the first time to address pharmacologically the tumour suppressor role of RARβ in vitro and in animal models.
Keywords: retinoic acid receptor, 3D structure, ligand-binding domain, ligand design, tumour suppressor
Introduction
The molecular mechanisms by which retinoic acid receptors (RARα, β, γ) regulate the transcription of target genes in a ligand-dependent manner are in principle understood (for review and references, see Laudet & Gronemeyer, 2002). In the absence of agonists, and in the presence of certain antagonists, RARs, which form heterodimers with RXRs, are believed to exist in association with target genes in a complex with co-repressors and associated factors, such as histone deacetylases (HDACs), the enzymatic activity of which results in local chromatin condensation and gene silencing. This co-repressor complex is tethered to the RAR–RXR heterodimer through a binding interface that is sensitive to allosteric changes induced by RAR agonists. Consequently, agonists dissociate the co-repressor complex and liberate an overlapping but distinct interaction surface for the binding of coactivator complexes. A major structural feature of the novel (holo-) surface is the contribution of the carboxy-terminally located helix H12, which is re-positioned on the ligand-binding domain (LBD). H12 can adopt several conformations depending on the type of ligand that is bound to the receptor and can be viewed as a positional interpreter of multiple types of agonists and antagonists. Several multiprotein machineries can bind to the holosurface of RAR–RXR heterodimers; their common feature is the presence of a so-called NR box in the tethering subunit. Coactivator complexes contain histone acetyltransferases (HATs), which allow relieving of the chromatin-mediated silencing induced by HDACs, and ATP-dependent chromatin remodelling machineries prepare the template for the action of the RNA polymerase holo-enzyme. The implication of the individual complexes in specific gene activation, the temporal order of their recruitment, and their promoter and cell specificity are still poorly understood, but the corresponding studies are rapidly advancing (Metivier et al, 2003).
The present challenge is to integrate the above structural and molecular biological information into studies on the (patho)physiological relevance of retinoid (and rexinoid) receptor signalling, to provide novel tools for research and therapy. One of the most interesting features of retinoid and rexinoid signalling is its well-documented cancer chemotherapeutic and preventive potential (for recent reviews, see Altucci & Gronemeyer, 2001; Sun & Lotan, 2002). A great number of gene ablation studies (reviewed in Laudet & Gronemeyer, 2002) have, at least to a certain degree, revealed the global impact of retinoid and rexinoid receptors in physiological processes, but can in no way replace receptor pharmacology. For example, in the case of the retinoid/rexinoid receptor heterodimer, multiple possibilities of modulating co-repressor and coactivator interaction have been reported (Germain et al, 2002); such modulation cannot be mimicked by genetic means. Indeed, the enormous potential of nuclear receptor-based drug design is only insufficiently explored (Gronemeyer et al, 2004).
A particularly interesting receptor for cancer research is RARβ. The RARβ gene is frequently deleted or its expression is epigenetically silenced during cancer progression and RARβ re-expression can restore retinoic acid-mediated growth control, suggesting that the anticancer action of retinoids is mediated by RARβ (Altucci & Gronemeyer, 2001; Widschwendter et al, 2001; Sirchia et al, 2002; Sun & Lotan, 2002). Consequently, RARβ has been viewed as a tumour suppressor. However, the mechanism underlying its antitumour action has remained elusive. One of the reasons for this is the lack of selective agonists and antagonists. The design of such compounds is extremely difficult, because only one residue in the ligand-binding pocket (LBP) of RARβ differs from that of its paralogue RARα, and two residues differ from RARγ. For this reason, we have set out to define the 3D structure of the RARβ LBD. Unexpectedly, the RARβ LBP presents features that allow one to develop specific ligands. We report here on a highly RARβselective agonist and postulate precise guidelines for the rational design of RARα, β, γ-selective agonists and antagonists.
Results And Discussion
Structure determination and refinement
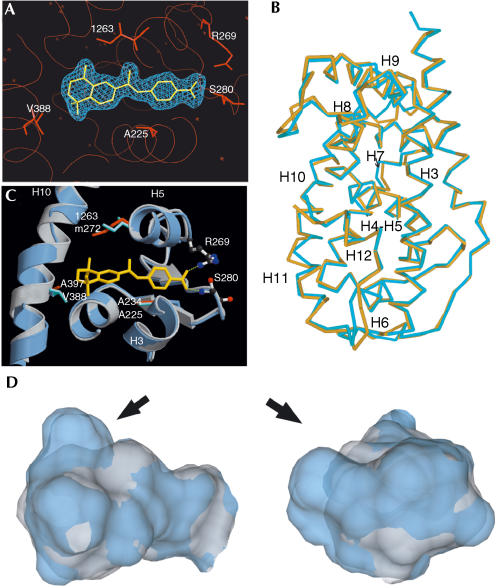
The 3D structure of the RARβ LBD–TTNPB complex was solved from a crystal (space group P 212121) that diffracted to 2.1 Å resolution. The RARγ LBD–9-cis retinoic acid (9-cis RA) complex (Klaholz et al, 1998) (PDB, 3lbd) was used as a starting model for molecular replacement. The quality of the electron density map allowed to accurately define the position of the ligand in its binding pocket (Fig 1A). Supplementary Table 1 online summarizes the corresponding crystallographic data. The final model encompasses residues A177–N409; no clear densities were observed for the His tag, the amino-terminal residues 173–176 and for residues 353–355 (loop H9–H10).
Figure 1.
Crystal structure of the RARβ LBD–TTNPB complex reveals an additional cavity in the RARβ LBP. (A) Electron density map of TTNPB in its pocket. 2Fo–Fc map at 2.1 Å resolution contoured at 1σ. The three isotypespecific residues (A225, I263 and V388) and residues anchoring the carboxylate (R269 and S280) are indicated. Illustration by PYMOL. (B) Superposition of the holo-RARβ–TTNPB (yellow) and RARγ–9-cis RA (blue) LBDs. Illustration by SETOR. (C) Superposition of TTNPB–RARβ (blue) and 9-cis RA–RARγ (grey) LBDs. The isotype-specific residues are shown in cyan (RARβ) and orange (RARγ) and TTNPB in yellow. The carboxylate anchoring residues are illustrated as ball-and-sticks. H bonds are represented as dashed lines. The figure was prepared by MOLSCRIPT and RASTER3D. (D) Superimposition of RARβ (blue) and RARγ (grey) LBPs. The arrow points to the additional cavity in RARβ.
Overall description
The RARβ holo-LBD exhibits the canonical antiparallel α-helical ‘sandwich' fold of nuclear receptors comprising 12 α-helices and one antiparallel β-sheet; its conformation corresponds to that of agonist complexes (Bourguet et al, 2000a). Superposition of TTNPB–RARβ and 9-cis RA–RARγ LBD (Renaud et al, 1995; PDB, 3lbd) complexes using the LSQ options from O revealed a generally high degree of structural overlap (r.m.s. deviation, 0.75 Å; Fig 1B). Although the observed shift of RARγ helices H10 and H11 towards the surface may be because of the bulkier volume of TTNPB compared with 9-cis RA, possibly stabilized by crystal packing contacts not observed in the 9-cis RA–RARγ LBD crystal, we do not exclude a RARβspecific conformation of this region.
Ligand-binding pocket
RARβ accommodates TTNPB in the canonical cavity between the β-turn and H11 in one, and H5 and H3/H12 in the other direction (Fig 1C). The hydrogen bond network anchoring the ligand carboxylate is identical to that formed in RARγ (Renaud et al, 1995; Klaholz et al, 2000b). Briefly, one of the carboxylate oxygen atoms interacts with Nɛ of RARβ R269 (3.08 Å) of the loop connecting H5 to the N-terminal βstrand, whereas the other oxygen is involved in a hydrogen bond network with Nα of S280 (2.69 Å) of the β-turn and one invariant water molecule (2.71 Å; Fig 1C; supplementary Fig S1 online). All TTNPB carbon atoms establish van der Waals contacts with residues lining the LBP (supplementary Fig S1 online). Compared with 9-cis RA-bound RARγ, no unexpected interactions were noted for TTNPB binding to RARβ, in keeping with the fact that only two residues differ between the two LBPs (Renaud et al, 1995; Gehin et al, 1999; Bourguet et al, 2000b).
Superposition with RARγ reveals that the LBP of RARβ is significantly larger (Fig 1D). Using VOIDOO, a volume of 503.5 Å3 was determined, whereas that in the 9-cis RA–RARγ complex is 429.4 Å3. This difference is not a mere conformational adaptation to the ligand but reflects a different architecture of the two LBPs, because (i) the maximal observed volume for the RARγ LBP occupied by various ligands was 457.2 Å3 (Klaholz et al, 2000b), thus >9% smaller than the RARβ cavity, and (ii) the greater volume of the RARβ pocket cannot be explained by steric repulsions or crystal packing effects. Instead, different conformations of one of the two divergent LBP residues account for the different RARβ and RARγ LBPs. In RARβ, the I263 side chain is positioned such that an additional cavity is generated between H5 and H10 (Fig 1C). This cavity does not exist in RARγ as M272 points inside the LBP (Klaholz et al, 1998). These structural features suggest that ligands occupying the additional space (arrow in Fig 1D) may acquire RARβ selectivity.
Structural basis of an RARβ agonist–RARγ antagonist
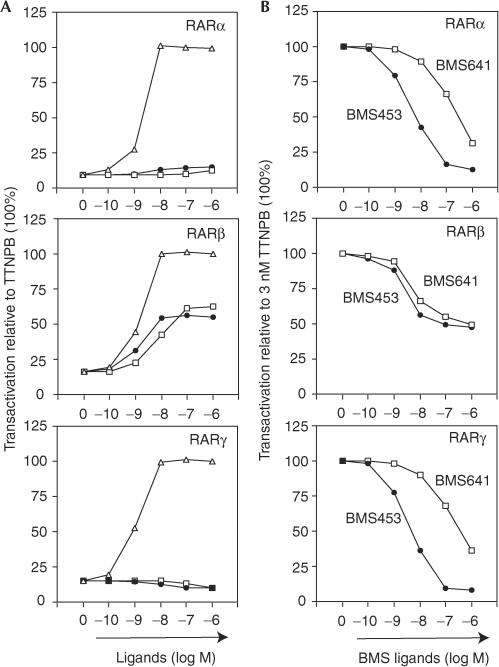
The characterization of retinoids has revealed not only pan- and RAR-selective agonists and antagonists, but also a class of compounds that can act as agonists with one and antagonists with another RAR, such as BMS453 (Chen et al, 1995). The structural basis of this phenomenon has remained elusive. Competition curves derived from challenging 3 nM TTNPB with increasing BMS453 concentrations reveal that BMS453 is a potent antagonist of TTNPB-induced transcription for Gal-RARα and Gal-RARγ, whereas it acts as a ‘mixed' agonist/antagonist (i.e. a weaker agonist than TTNPB) for RARβ (Fig 2A,B; structures are given in Fig 4A).
Figure 2.
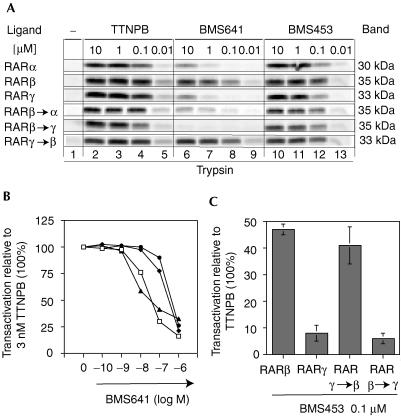
Agonist–antagonist potential and RARβ selectivity of BMS453 and BMS641. (A) HeLa cells were co-transfected with reporter (17m)5x-G-luc and Gal-RARα, Gal-RARβ or Gal-RARγ, as indicated. Cells were incubated with increasing concentrations of TTNPB (open triangles), BMS453 (closed circles) or BMS641 (open squares). (B) Transient transactivation assays as in (A) to assess antagonistic activities of BMS453 (closed circles) or BMS641 (open squares). The reporter was activated with 3 nM TTNPB (100%) and increasing concentrations of the synthetic retinoids were added.
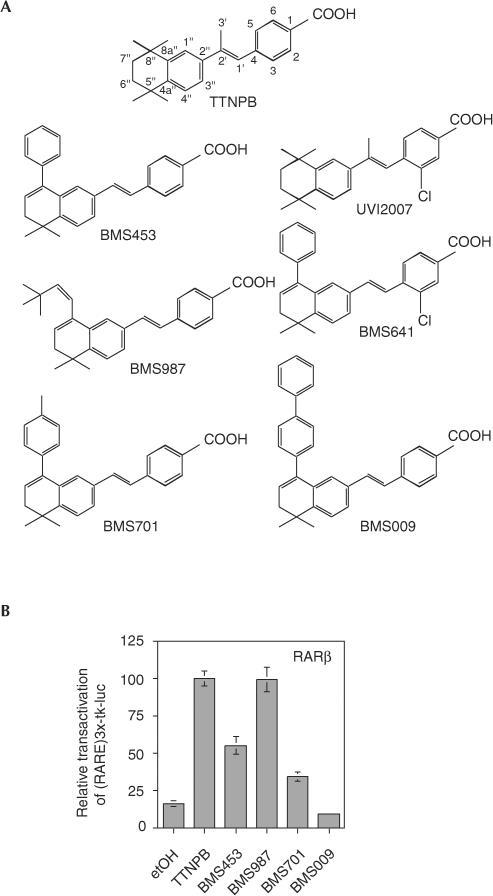
Figure 4.
RARβ agonist potential of a series of 8″substituted TTNPB-like retinoids. (A) Chemical structures of the retinoids used in this study. (B) Transactivation assays. HeLa cells were co-transfected with the reporter (RARE)3x-tk-luc and RARβ in the presence of the indicated BMS compounds at 0.1 μM.
To provide a structural rationale explaining the different transcriptional activities of BMS453 on binding to the three RARs, we performed docking experiments and compared the corresponding fitness scores for the RARβ and RARγ LBDs using GOLD. The validation of the GOLD energy function for retinoid docking is described in the supplementary information online.
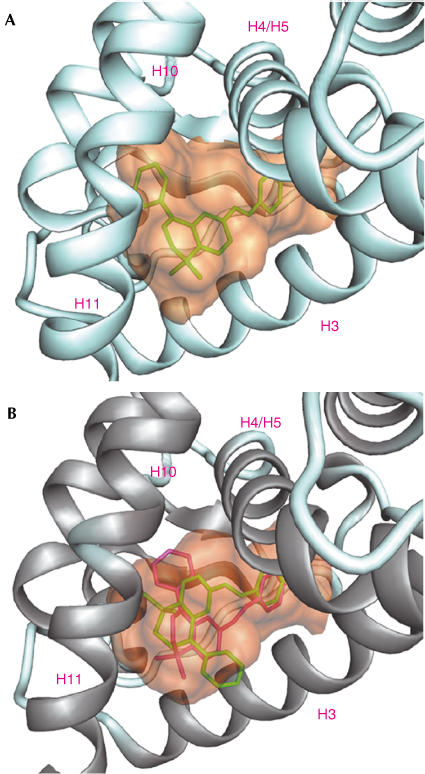
Docking of BMS453 into the RARβ LBD gave similar solutions, with favourable scores; the ligand was completely buried in the LBP with the phenyl oriented towards H5 (Fig 3A shows one solution of VOIDOO). In contrast, any of the proposed solutions for hRARγ had low fitness scores, indicating substantial docking problems. Indeed, inspection of the two predicted types of ligand conformations (Fig 3B, red and green) revealed steric clashes with I412 of H12 (in the agonist conformation) and A234 of H3 for the ‘green' solution, and with M272 of H5 for the ‘red' solution. These distinct docking results reflect the smaller size of the RARγ LBP and the additional cavity in the RARβ LBD, which accommodates the bulky ‘head' of the molecule with its phenyl moiety.
Figure 3.
Docking of BMS453 into the RARβ (A) and RARγ (B) LBP. The ligand cavities were calculated by VOIDOO and MSMS using a probe radius of 1.4 Å. DINO was used for illustration. (A) BMS453 in green is perfectly buried within the RARβ LBP with its phenyl ring oriented towards H12. (B) The two predicted orientations (green and red) could not be accommodated in the RARγ cavity.
Removing H12 from the RARγ LBD structure allowed to manually dock BMS453 readily in the LBP, thus suggesting that this ligand could act as an RARγ antagonist as its binding is incompatible with an agonist conformation. Indeed, transactivation assays (Fig 2B, bottom) confirmed this notion. Together, the above considerations provide a structural rationale accounting for the RARβ agonist–RARγ antagonist activities of BMS453.
Converting an RARβ agonist into an antagonist
Inspired by the structural analysis of the agonist–antagonist switch of BMS453, we predicted that increasing the bulkiness of the hydrophobic group of BMS453-like molecules would generate RARβ ligands that sterically interfere with the agonist positioning of H12 and thus turn into RARβ antagonists. Indeed, studying the agonist–antagonist characteristics of a series of 8″ substituted (E)-4-[2-(5,6,7,8-tetrahydro-5,5,8,8-tetramethylnaphthalene-2-yl)vinyl]-benzoic acid derivatives (Fig 4A) revealed that a (Z)-3,3-dimethylbut-2-en-1-yl substitution yielded a full RARβ agonist (BMS987 in Fig 4B) virtually indistinguishable from TTNPB. BMS453 has a bulkier phenyl moiety in 8″ position and its weaker activity suggests that the phenyl may cause some weak interference with H12 positioning and coactivator recruitment. Increasing the bulkiness at 8″ position by introducing a p-tolyl group (BMS701) further reduced the agonist activity of the ligand, and a p-biphenyl substitution yielded a high-affinity antagonist (BMS009 in Fig 4). All ligands of this series show similar affinity for RARβ (Kd, 2–9 nM) excluding that different affinities could account for distinct transcriptional activities.
Identification of an RARβ-selective agonist
By screening a panel of synthetic retinoids with an in vivo reporter cell system (Chen et al, 1995), we identified BMS641, a retinoid that exclusively activated RARβ in reporter cells. Limited proteolysis with trypsin (Fig 5A, middle panel) revealed BMS641-dependent protection of RARβ similar to that seen with the panRAR agonist TTNPB (left panel), but no protection of RARα or RARγ. Indeed, direct binding assays revealed a higher affinity for RARβ (Kd, 2.5 nM) that was 100 times higher than that for RARα (Kd, 225 nM) or RARγ (Kd, 223 nM). In contrast, BMS453 induced a digestion pattern very similar to that of TTNPB, indicating that BMS453 can bind to all three RARs with similar affinities. Interestingly, BMS641 is a 3-chloro derivative of BMS453 implying that a halogen in position 3 is a crucial determinant for generating RARβ binding selectivity.
Figure 5.
The three divergent LBP residues determine the RARβ selectivity of BMS641 and the isotype-dependent potential of BMS453. (A) Partial proteolysis maps of in vitro-translated RARs in the presence or absence of increasing concentrations of either TTNPB, BMS453 or BMS641, as indicated. (B) Dose–response curves to assess the binding affinity of BMS641 relative to TTNPB in RAR pocket mutants. HeLa cells were co-transfected with (RARE)3x-tk-luc and either RARβ (closed triangles), RARγ (open circles), RARγ → β (RARγ(M272I, A397V); open squares) or RARβ → γ (RARβ(I263M, V388A); closed diamonds) and reporter gene transcription was induced with 3 nM TTNPB (100%). (C) BMS453-induced luciferase activity in HeLa cells co-transfected with (RARE)3x-tk-luc and the indicated receptors.
Transient transactivation (Fig 2) reflected ligand binding. BMS641 did not antagonize TTNPB-induced transcription for RARα and RARγ, in keeping with its low affinity for these receptors. In contrast, a ‘mixed' agonistic/antagonistic activity was seen for RARβ, as competition curves plateaued at about 50% of the TTNPB-induced activity. Virtually identical inhibition was obtained with BMS453, in keeping with similar relative binding and agonistic activities of BMS453 and BMS641 for RARβ.
Three residues dictate RARβ selectivity
We previously demonstrated that swapping of three divergent residues in the LBPs of RARs suffices to modulate isotype-selective binding (Gehin et al, 1999). To assess whether these residues are also important for RARβselective binding of BMS641, partial proteolysis maps were established (Fig 5A). Indeed, converting the single divergent residue of RARβ into that of RARα (mutant RARβ → α; RARβA225S) or two divergent residues into those of RARγ (mutant RARβ → γ; RARβ(I263M, V388A)) resulted in a loss of BMS641-dependent protection, whereas a gain of protection was observed with RARγ if its LBP was changed to that of RARβ (mutant RARγ → β; RARγ(M272I, A397V)). Importantly, identical maps were obtained with TTNPB or BMS453 (Fig 5A). Transactivation experiments supported these results (Fig 5B) and showed that these residues also determine the agonistic/antagonistic profile of ligands with this chemotype (Fig 5C). Indeed, in the context of the mutant RARγ → β BMS453 acts as a partial agonist (∼40% of TTNPB), but when the LBD is converted into that of RARγ (mutant RARβ → γ) BMS453 shows a strong antagonistic activity as with wild-type RARγ (less than 5% of the TTNPB activity). These results underscore the importance of the divergent residues and suggest that a 3-chloro substitution in TTNPB-like retinoids may sense the RARβ pattern.
A 3-chloro substitution is RARα discriminatory
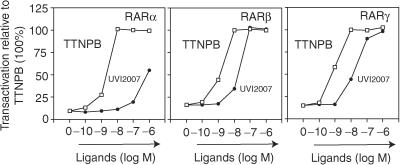
To study whether a 3-chloro substitution on its own would generate RARβ selectivity, we introduced it into the pan-agonist TTNPB, generating UVI2007 (Figure 4A; see supplementary section online for synthesis). Transcriptional (Fig 6) and proteolysis (not shown) assays demonstrated that UVI2007 shows a lower relative binding affinity to all three RAR isotypes. Notably, the decrease in the affinity is much more pronounced for RARα than for RARβ or γ, revealing an RARα discriminatory effect of the 3-chloro group. This is most likely because of a steric interference originating from RARαS232, which is bulkier than RARβA225. In contrast to BMS641, no significant difference was seen for UVI2007 binding to RARβ and RARγ (Fig 6), indicating that on its own a chlorine in position 3 is insufficient to discriminate between these isotypes.
Figure 6.
A 3-chloro substitution discriminates RARα from RARβ/γ binding. HeLa cells were transfected as in Fig 2A. Dose–response curves for TTNPB (open squares) or UVI 2007 (closed circles).
Structural features contributing to RARβ selectivity
From this and previous studies (reviewed in Bourguet et al, 2000a), the following structural features can be deduced for the synthesis of RAR isotypeselective retinoids: (i) the three divergent residues and the different shape of the RARβ LBP reported here are the most important discriminatory elements; (ii) existing isotype-selective ligands dissociate mostly RARα from RARβ/γ by exploiting the presence of RARαS232, which can establish hydrogen bonds with suitable ligands; this was predicted for ligands harbouring an amino group, such as Am580 or BMS753 (Gehin et al, 1999); (iii) to separate RARβ from RARα binding, a 3-chloro substitution that appears to create steric hindrance within the RARα pocket can be introduced in TTNPB-like retinoids; (iv) to separate RARβ from RARγ binding, two structural aspects can be exploited, both of which are a consequence of the replacement of RARβI263 by RARγM272. First, the side-chain orientation of I263 opens a cavity that is closed in RARγ; ligands that require such a cavity to accommodate a bulky substituent acquire βselectivity. Our functional analyses have shown that a 3-chloro (in UVI2007) or an 8″-phenyl (in BMS453) on its own cannot discriminate between RARβ and RARγ; however, the combination of both substitutions (in BMS641) results, despite an overall lower affinity, in the acquisition of β-selective binding that originates from a marked loss of RARγ affinity. This is highly suggestive of a structural model in which the 3-chloro substitution positions the ligand in the RARγ pocket such that the 8″-phenyl clashes with M272 (but not the corresponding I263 of RARβ). Second, the presence of M272 allows establishing hydrogen bonds to a suitable ligand, for example, BMS270394 (Klaholz et al, 2000a), to augment γ affinity and selectivity; (v) in all cases, antagonists are characterized by the presence of substitutions that interfere with the holo-conformation of H12.
Speculation
The novel structure of the RARβ LBD allows, for the first time to our knowledge, the establishment of a comprehensive set of guidelines for the rational drug design of RAR-selective retinoids. The availability of agonists and antagonists selective for each of the RARs will allow one to dissect the various activities exerted by the three RARs, which are the basis for the well-established cancer therapeutic and cancer preventive activity of retinoids (Altucci & Gronemeyer, 2001; Sun & Lotan, 2002), assess the role of the suspect tumour suppressor RARβ and will aid in generating novel retinoids with reduced side effects.
Methods
Software. The software used in this study is described in the supplementary section.
Ligands and plasmids. For ligands, see the supplementary section online and supplementary Table 2 online. pSG5-based RAR and GAL-RAR expression vectors and reporter genes were described previously (Nagpal et al, 1993; Chen et al, 1995; Gehin et al, 1999). For docking experiments, ligands with all hydrogen atoms were built with QUANTA-CHARMM.
Protein purification, crystallization and data collection. The His-tagged hRARβ LBD (173–409) was purified to 98% homogeneity and crystallized as described in the supplementary section online. X-ray diffraction data were collected (Synchrotron Light Source, Zürich, Switzerland) and processed with DENZO and SCALEPACK from the HKL2000 package.
Structure refinement. Structure determination and refinement was carried out with CNS solve package using the hRARγ LBD complex (PDB ID, 3LBD) as a search model. Data between 20 and 2.1 Å resolution were included in the refinement process (supplementary section online and supplementary Table 1 online).
Cell culture and transient transfections. HeLa cells, cultured in Dulbecco's modified Eagle's medium–5% fetal calf serum, were transfected as described (Vivat et al, 1997).
Limited proteolytic digestion. For limited proteolytic digestions (Vivat et al, 1997), in vitro-made 35S-labelled human RARs (TNT kit, Promega) were used. Briefly, after incubating on ice for 1 h with ligand, receptor proteins were digested at 25°C for 10 min with 100 μg/ml (RARα, RARβ) or 50 μg/ml (RARγ) trypsin.
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v5/n9/extref/7400235s1.pdf).
Supplementary Material
Supplementary Section
Acknowledgments
We are grateful to D. Moras for supporting this project. We thank A. Pornon for excellent technical assistance, T. Lerouge and P. Balaguer for plasmids, A. Mitschler for data collection, S. Sasorith and C. Loch for advice in docking analysis, and the beamline staff at SLS (Zürich) for technical assistance. S.K. received fellowships from the MRT and the ARC, and P.G. and E.P. are supported from EU grants. This work was supported by grants from the EU (QLG1-CT2000-01935, QLK3-CT2002-02029, HPRN-CT2002-00268), the Spanish MCYT (SAF2001-3288), the Fondation de France, the AICR, the ARC, the ULP, the INSERM, the CNRS and Bristol-Myers Squibb.
References
- Altucci L, Gronemeyer H (2001) The promise of retinoids to fight against cancer. Nat Rev Cancer 1: 181–193 [DOI] [PubMed] [Google Scholar]
- Bourguet W, Germain P, Gronemeyer H (2000a) Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci 21: 381–388 [DOI] [PubMed] [Google Scholar]
- Bourguet W, Vivat V, Wurtz JM, Chambon P, Gronemeyer H, Moras D (2000b) Crystal structure of a heterodimeric complex of RAR and RXR ligand-binding domains. Mol Cell 5: 289–298 [DOI] [PubMed] [Google Scholar]
- Chen JY, Penco S, Ostrowski J, Balaguer P, Pons M, Starrett JE, Reczek P, Chambon P, Gronemeyer H (1995) RARspecific agonist/antagonists which dissociate transactivation and AP1 transrepression inhibit anchorage-independent cell proliferation. EMBO J 14: 1187–1197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehin M, Wurtz JM, Losson R, Chambon P, Moras D, Gronemeyer H (1999) Structural basis for engineering of retinoic acid receptor isotypeselective agonists and antagonists. Chem Biol 6: 519–529 [DOI] [PubMed] [Google Scholar]
- Germain P, Iyer J, Zechel C, Gronemeyer H (2002) Co-regulator recruitment and the mechanism of retinoic acid receptor synergy. Nature 415: 187–192 [DOI] [PubMed] [Google Scholar]
- Gronemeyer H, Gustafsson JA, Laudet V (2004) Prospects for modulation of the nuclear receptor superfamily. Nat Rev Drug Disc (in press) [DOI] [PubMed] [Google Scholar]
- Klaholz BP, Renaud JP, Mitschler A, Zusi C, Chambon P, Gronemeyer H, Moras D (1998) Conformational adaptation of agonists to the human nuclear receptor RARγ. Nat Struct Biol 5: 199–202 [DOI] [PubMed] [Google Scholar]
- Klaholz BP, Mitschler A, Belema M, Zusi C, Moras D (2000a) Enantiomer discrimination illustrated by high-resolution crystal structures of the human nuclear receptor hRARγ. Proc Natl Acad Sci USA 97: 6322–6327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaholz BP, Mitschler A, Moras D (2000b) Structural basis for isotype selectivity of the human retinoic acid nuclear receptor. J Mol Biol 302: 155–170 [DOI] [PubMed] [Google Scholar]
- Laudet V, Gronemeyer H (2002) The Nuclear Receptor Facts Book. San Diego, CA: Academic Press [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F (2003) Estrogen receptor-α directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell 115: 751–763 [DOI] [PubMed] [Google Scholar]
- Nagpal S, Friant S, Nakshatri H, Chambon P (1993) RARs and RXRs: evidence for two autonomous transactivation functions (AF-1 and AF-2) and heterodimerization in vivo. EMBO J 12: 2349–2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaud JP, Rochel N, Ruff M, Vivat V, Chambon P, Gronemeyer H, Moras D (1995) Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature 378: 681–689 [DOI] [PubMed] [Google Scholar]
- Sirchia SM, Ren M, Pili R, Sironi E, Somenzi G, Ghidoni R, Toma S, Nicolo G, Sacchi N (2002) Endogenous reactivation of the RARβ2 tumor suppressor gene epigenetically silenced in breast cancer. Cancer Res 62: 2455–2461 [PubMed] [Google Scholar]
- Sun SY, Lotan R (2002) Retinoids and their receptors in cancer development and chemoprevention. Crit Rev Oncol Hematol 41: 41–55 [DOI] [PubMed] [Google Scholar]
- Vivat V, Zechel C, Wurtz JM, Bourguet W, Kagechika H, Umemiya H, Shudo K, Moras D, Gronemeyer H, Chambon P (1997) A mutation mimicking ligand-induced conformational change yields a constitutive RXR that senses allosteric effects in heterodimers. EMBO J 16: 5697–5709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widschwendter M, Berger J, Muller HM, Zeimet AG, Marth C (2001) Epigenetic downregulation of the retinoic acid receptor-β2 gene in breast cancer. J Mammary Gland Biol Neoplasia 6: 193–201 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Section