Abstract
FtsZ is a filament-forming protein that assembles into a ring at the division site of prokaryotic cells. As FtsZ and tubulin share several biochemical and structural similarities, FtsZ is regarded as the ancestor of tubulin. Chloroplasts—the descendants of endosymbiotic bacteria within plant cells—also harbour FtsZ. In contrast to eubacteria, plants have several different FtsZ isoforms. So far, these isoforms have only been implicated with filamentous structures, rings and networks, inside chloroplasts. Here, we demonstrate that a novel FtsZ isoform in the moss Physcomitrella patens is located not only in chloroplasts but also in the cytoplasm, assembling into rings in both cell compartments. These findings comprise the first report on cytosolic localization of a eukaryotic FtsZ isoform, and indicate that this protein might connect cell and organelle division at least in moss.
Keywords: ancestral tubulin, cell division, organelle division, moss, Physcomitrella patens
Introduction
The protein FtsZ is nearly ubiquitously present in all recent prokaryotes and has an important function in the division process of these organisms (Errington et al, 2003; Pinho & Errington, 2003; Vicente & Löwe, 2003). Before the onset of cytokinesis, FtsZ polymerizes into a ring on the inside of the cytoplasmic membrane (Bi & Lutkenhaus, 1991). After ring formation, additional proteins are recruited to this scaffold in a specific order to complete cell division (Lutkenhaus & Addinall, 1997; Errington et al, 2003). During evolution, chloroplasts arose from free-living cyanobacteria (McFadden, 1999), and, among other genes, ftsZ was transferred to the eukaryotic nucleus (Martin et al, 2002). Nowadays, plants harbour two distinct nuclear-encoded FtsZ families with several members (Kiessling et al, 2000; Osteryoung & McAndrew, 2001; Rensing et al, 2004), indicating morphological and/or functional differences between these proteins in a given species (Reski, 2002). However, all eukaryotic FtsZ proteins analysed so far were found exclusively within plastids (Osteryoung et al, 1998; Strepp et al, 1998; Kiessling et al, 2000; Mori et al, 2001; Vitha et al, 2001; Kuroiwa et al, 2002; Osteryoung & Nunnari, 2003) in addition to mitochondrial FtsZ detected only in non-green algae (Beech et al, 2000; Takahara et al, 2000).
As FtsZ and tubulin share several biochemical features (de Boer et al, 1992; Erickson et al, 1996; Löwe & Amos, 1999) and comprise similar three-dimensional structures (Erickson, 1998; Löwe & Amos, 1998), FtsZ is regarded as the ancestor of tubulin. Moreover, these proteins are involved in the division process of prokaryotes (FtsZ) and eukaryotic cells (tubulin). Here, we demonstrate that an FtsZ isoform in Physcomitrella patens is targeted not only to chloroplasts but also to the cytoplasm assembling into rings in both cell compartments, resulting in the first report on cytosolic location of a eukaryotic FtsZ isoform. As this dual-targeted FtsZ appears to act not only in plastid division but also in cell division, we speculate that this protein may be a molecular link between cell and organelle division, at least in the moss Physcomitrella.
Results And Discussion
The moss P. patens harbours four FtsZ isoforms in two families, FtsZ1 and FtsZ2. Here, we focus on a novel FtsZ1 family member (Fig 1, Physcomitrella 1_2), which is divergent from other plant FtsZ isoforms in terms of amino-acid (aa) composition and genomic organization (Rensing et al, 2004). Subcellular localization of this isoform was analysed by confocal laserscanning microscopy using different green fluorescent protein (GFP) fusions (Fig 2). In transiently transfected moss cells (protoplasts), full-length FtsZ1-2 assembled into ring-like structures not only in plastids but also in the cytoplasm (Fig 2A,B). Dual targeting of this isoform to chloroplasts and the cytoplasm occured irrespective of expression levels: low protein amounts (indicated by low GFP fluorescence) assembled into single rings (Fig 2A), whereas high protein amounts (intense fluorescence) polymerized into multiple rings in both cell compartments (Fig 2B).
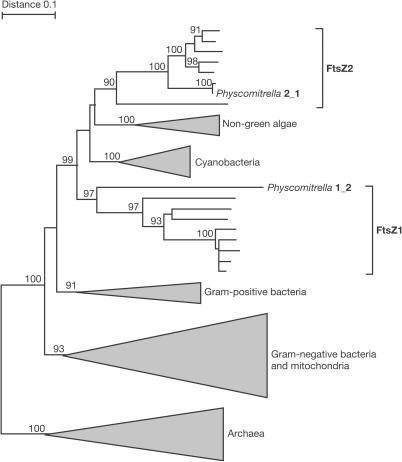
Figure 1.
Phylogeny of FtsZ demonstrating the position of both Physcomitrella isoforms analysed here. Numbers at the branches represent bootstrap values above 90% (1,000 replicates).
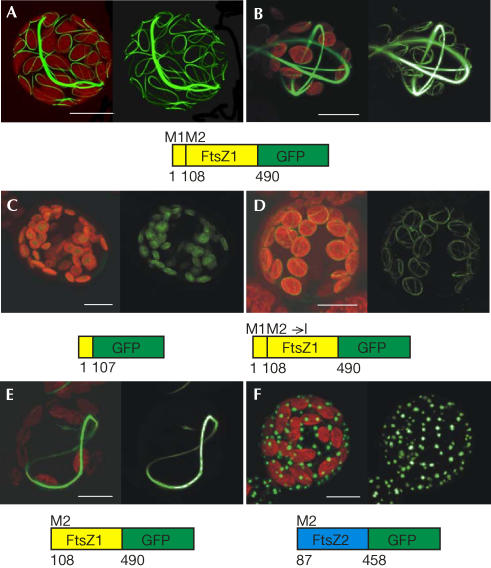
Figure 2.
Dual targeting of FtsZ1-2–GFP to chloroplasts and the cytoplasm in Physcomitrella. Isolated moss cells (protoplasts) were analysed by confocal microscopy 2 days after transfection. Merge of GFP and chlorophyll signal (left side of each part), GFP alone (right side of each part); projections from ∼20 sections of 1 μm depth are shown. (A,B) Full-length FtsZ1-2–GFP (aa 1–490) polymerizes into ring-like structures in chloroplasts and the cytosol at low (A) and high (B) levels. Plastidic targeting of aa 1–107 (C) and the full-length protein with M108 to I mutation (D). Cytosolic targeting of FtsZ1-2–GFP, starting at the second methionine (M2, aa 108–490; E) and artificial cytosolic targeting of FtsZ2-1–GFP, starting from Met 87 (M2, aa 87–458; F). Scale bars, 10 μm.
In many eukaryotes—including Physcomitrella—dual targeting of nuclear-encoded proteins has been observed (Small et al, 1998; Hedtke et al, 2000; Richter et al, 2002); however, for FtsZ proteins, this has not been described so far. Dual targeting of a protein within a cell can result from a differential use of two translation initiation sites within a single mRNA (Small et al, 1998). As the FtsZ isoform described here contains methionines at aa positions 1 and 108, these methionines may represent the amino-termini of the plastidic and cytosolic form of FtsZ1-2, respectively. To test this assumption, different GFP fusions were created (Fig 2C–F). As aa 1–107 conferred plastidic localization to GFP (Fig 2C), they contain the chloroplast-targeting signal consistent with the predicted cleavage site at aa 69 obtained through TargetP (Nielsen et al, 1997; Emanuelsson et al, 2000). Mutation of the second putative initiation codon in the background of the full-length complementary DNA (cDNA) resulted in an exclusive localization of the corresponding fusion protein inside chloroplasts (Fig 2D). In contrast, a truncated form starting at methionine 108 assembled into rings solely in the cytoplasm (Fig 2E). Thus, methionine 108 represents the N-terminus of cytosolic FtsZ1-2.
By comparison, a member of the second FtsZ family (Fig 1, Physcomitrella 2_1) was analysed. Previously, this FtsZ2 isoform has been detected exclusively inside plastids, where it assembles into networks (Kiessling et al, 2000). By deleting aa 1–86, FtsZ2-1 was artificially targeted to the cytosol, where it aggregated into dots (Fig 2F). Thus, targeting to the ‘wrong' compartment (FtsZ2 to the cytosol) led to a nonspecific accumulation of the fusion protein. Therefore, plant FtsZ proteins—like their bacterial homologues—might require specific cofactors for assembly, which seem to be absent in the cytoplasm for FtsZ2-1 and present for FtsZ1-2, as described here.
To visualize endogenous FtsZ1-2 in nontransgenic moss, a polyclonal antibody was raised against the carboxy-terminal aa 441–490, where similarity with other FtsZ isoforms is low (∼27%) and even lower with tubulins, including Physcomitrella tubulins (Jost et al, 2004). In protein extracts of Physcomitrella, anti-FtsZ detected specific bands of approximately 40,000 and 46,000/47,000 Da (Fig 3A). The sizes of the bands support our findings on dual localization of FtsZ1-2: the predicted masses for cytosolic (40,500 Da) and plastidic (44,600 Da) FtsZ1-2 correspond with the sizes of the bands detected through immunoblotting. In addition, the 46,000/47,000 Da double band may hint at post-translational modifications and a faint band of approximately 75,000 Da at dimerization of cytosolic FtsZ. Crossreactivity with tubulin was excluded by use of an antitubulin antibody, which detected proteins of ∼51,000 Da (Fig 3A).
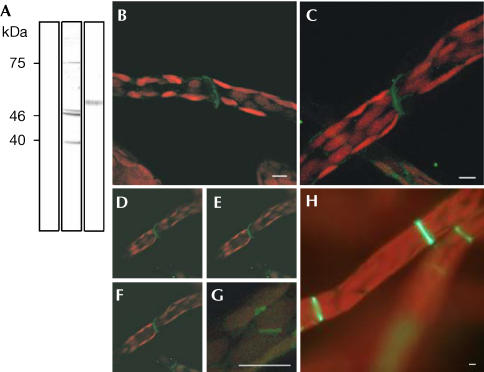
Figure 3.
FtsZ1-2 assembles into annular–discoidal structures at the interface of moss protonema cells. (A) Western blot of Physcomitrella protein extract, probed with preimmune serum (lane 1), anti-FtsZ1-2 (lane 2) and anti-tubulin (lane 3). Signals at ∼40,000 and ∼46,000/47,000 Da represent the expected sizes of cytosolic and plastidic FtsZ1-2 (lane 2). Anti-tubulin detected proteins at ∼51,000 Da (lane 3). (B–G) Immunocytochemistry using Physcomitrella protonema, a moss cell filament. Merge of FITC signal (green, anti-FtsZ1-2) and chlorophyll (red). (B,C,G) Projections from ∼20 sections of 1 μm depth are shown. (D–F) Optical sections from top to the middle of (C) at 1 μm intervals. Anti-FtsZ detects an annular–discoidal structure at the interface of moss cells in protonemal filaments (B–F) and the division site of chloroplasts (G). Stably FtsZ1-2-overexpressing plants confirm FtsZ localization obtained after immunocytochemistry of nontransgenic plants (H, compare with B–F). Scale bars, 5 μm.
As protoplasts used for transfections were isolated from Physcomitrella protonema, a filamentous tissue growing by subsequent division of apical cells, immunocytochemistry was performed using this tissue. Anti-FtsZ not only labelled the division site of chloroplasts (Fig 3G) but also visualized a cytoplasmic signal allocating cytosolic localization (Fig 3B–F). Prominent annular to discoidal structures were observed at the interface of subsequent protonema cells (Fig 3B–F).
Thus, two independent experiments proved the cytoplasmic localization of FtsZ in Physcomitrella. As, however, transgenic studies have been performed by transiently transfecting protoplasts, we additionally analysed intracellular localization of this FtsZ isoform in stably transformed differentiated protonema tissue. A total of 13 independent transgenic lines showed enlarged plastids as well as reduced growth when compared with wild type (WT), indicating that stable overexpression of this FtsZ isoform interferes with plastid division as well as with cell division. Moreover, these experiments perfectly confirmed the results obtained with immunocytochemistry of nontransgenic WT tissue, as FtsZ–GFP assembled to prominent annular to discoidal structures at the interface of subsequent protonema cells also in stably overexpressing moss plants (Fig 3H). The use of GFP fusions for unravelling intracellular protein sorting in Physcomitrella has recently been validated not only for protein import into plastids and mitochondria (Kiessling et al, 2000; Richter et al, 2002) but also for protein sorting through the secretory pathway to the vacuole (Schaaf et al, 2004).
As transgenics stably overexpressing cytosolic FtsZ showed reduced growth, a possible role of this protein in moss cell division was assessed in transient transfection assays. Such assays are suitable to monitor the effects of different FtsZ levels on division processes. In bacteria, slightly enhanced FtsZ concentrations resulted in minicells, whereas higher FtsZ levels blocked cell division (Ward & Lutkenhaus, 1985). In plants, plastidic FtsZ affected chloroplast division in a similar, dose-dependent manner (Kiessling et al, 2000).
The experiments described here demonstrate that a high level of cytosolic FtsZ1-2–GFP impeded 30% of the protoplasts in cell division (Fig 4A,B). Whereas nontransfected Physcomitrella cells managed to divide at the latest 9 days after isolation, cells expressing high amounts of FtsZ1-2–GFP were affected in cell division: more than 30% of all transfected cells were impeded in cell division, showing ring-like structures at the future division site (Fig 4A,B). By comparison, protoplasts harbouring only low FtsZ1-2–GFP levels managed to divide but presented a block in plastid division: they contained only one or a few huge plastids rather than about 50 small, WT-like chloroplasts (Fig 4C).
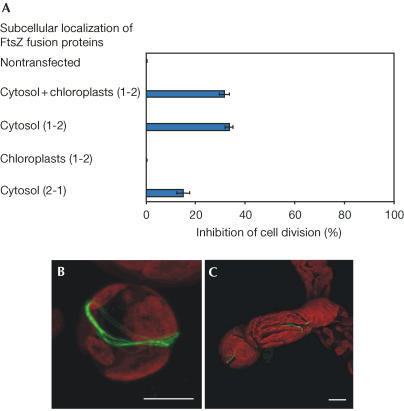
Figure 4.
Overexpression of cytosolic FtsZ interferes with cell division. (A) Cells transfected with different ftsZ–GFP vectors were monitored (200 per assay) for lack of division 9 days after transfection. For construct description, see Fig 2A,B (dual targeting of full-length FtsZ1-2 to the cytosol and chloroplasts), E (targeting of truncated FtsZ1-2 to the cytosol), D (targeting of mutated FtsZ1-2 to chloroplasts) and F (targeting of truncated FtsZ2-1 to the cytosol). Nontransfected cells served as an additional control. Error bars represent the standard deviation from the mean of three independent assays. (B,C) Cells 12 days after transfection with ftsZ1-2–GFP, demonstrating a blocked cell (B) and chloroplast division (C). Scale bars, 10 μm.
As controls, the effects of further FtsZ–GFP fusions were monitored. Cytosolic FtsZ1-2 (for localization, refer to Fig 2E) also impeded ∼30% of the protoplasts in cell division (Fig 4A). By comparison, high levels of plastidic FtsZ1-2 (for localization, refer to Fig 2D) only blocked organelle division (data not shown), whereas cell division was not affected (Fig 4A). In addition, some cells comprising high amounts of FtsZ2-1 targeted to the cytosol (for localization, refer to Fig 2F) were also impeded in cell division (Fig 4A). This might indicate that components required for cell division were accumulated by this fusion. However, the slight interference with cell division could also reflect the fact that overexpression of proteins in general is deleterious for cells. Nevertheless, the effects of this originally plastidic isoform were less severe compared with cytosolic FtsZ1-2 and, moreover, plastidic FtsZ1-2 did not interfere with cell division (Fig 4A).
A unique feature of Physcomitrella is the high rate of homologous recombination in its nuclear DNA (Schaefer & Zryd, 1997), making gene-knockout strategies as efficient as in yeast (Strepp et al, 1998). However, as mosses are haploid plants, knockouts in essential genes are not possible, and, consequently, such attempts will generate transgenic plants by means of illegitimate recombination, with the essential gene still being functional (Hohe et al, 2004). Our recent attempts to generate knockouts in ftsZ1-2 and subsequently analyse the effects of this loss-of-function yielded nine transgenics so far, none of which was affected in this gene (data not shown). Previously, targeting of ftsZ2-1 encoding a plastidal FtsZ isoform yielded 14% (seven out of 51) knockouts among transgenics (Strepp et al, 1998) although that targeting construct was based on a suboptimal partial cDNA sequence. In contrast the nine transgenics produced in this study were generated by constructs based on long stretches of genomic DNA, which greatly enhances targeting efficiencies in nonessential genes up to 90% (Hohe et al, 2004). Thus, these nine plants, all unaffected in ftsZ1-2, may be taken as further, however indirect, evidence for an essential role of the FtsZ1-2 protein in moss cell division.
Taken together, these results point to a role of cytosolic FtsZ1-2 in moss cell division. Similar to FtsZ in bacteria and chloroplasts, FtsZ1-2 could be part of the cell division machinery, the proper function of which is dependent on a fixed ratio of several components. Thus, this Physcomitrella FtsZ isoform may function not only in organelle division but also in eukaryotic cell division.
As plant FtsZ proteins seem to originate from the ancestors of chloroplasts, these proteins are located in plastids and are involved in the division of these organelles. Here, we have demonstrated that an FtsZ isoform of Physcomitrella (PpFtsZ1-2) is dual targeted to plastids and the cytosol. Both forms, plastidic and cytosolic FtsZ, are encoded by a single gene. Thus, this novel FtsZ may represent a molecular link of both division processes. Such a link is pivotal to coordinate organelle and cell division and thus ensures even distribution of chloroplasts during cytokinesis.
Methods
Plant material. P. patens (Hedw.) B.S.G. was cultivated as described previously (Strepp et al, 1998).
Isolation of ftsZ1-2. A partial cDNA clone was identified by a BLAST search of P. patens EST data (Rensing et al, 2002). The missing 500 bp of the 5′ end were obtained by 5′ rapid amplification of cloned ends (RACE)-PCR with the FirstChoiceTM RLM-RACE Kit (Ambion, USA).
Phylogenetic tree reconstruction. A structure-based alignment of FtsZ peptide sequences was calculated using CLUSTAL W 1.83 (http://www.ebi.ac.uk) and the Methanococcus jannaschii 2.8a structure (1FSZ) as template. After removal of the nonconserved N- and C-terminal regions, phylogenetic trees were constructed using quartet puzzling (TREE-PUZZLE 5.1, http://www.tree-puzzle.de) with eight γ-distributed rates and data-derived frequencies as well as neighbour-joining (TREECON 1.3, http://www.psb.rug.ac.be/bioinformatics/psb/current_projects_soft.htm) using Tajima and Nei parameters (with InDels). No major differences were apparent in both reconstructed phylogenies.
Sequence analysis. To assess subcellular localization of FtsZ1-2, the sequence was analysed with TargetP (http://www.cbs.dtu.dk/services/TargetP/; Nielsen et al, 1997; Emanuelsson et al, 2000). A plastid-targeting probability of 0.545 and a putative cleavage site at aa 69 were predicted. The relative molecular masses of the plastidic (40505) and cytosolic (44556) forms were predicted using ExPASy (http://www.expasy.org/tools/pi_tool.html).
Cloning of FtsZ–GFP fusions, transfection assays, generation of stable plants and microscopical inspection. For construction of full-length FtsZ1-2–GFP (aa 1–490), the coding region of ftsZ1-2 was PCR amplified with primers F1 (5′-TTTGGATCCATGATCACGTGTAGGGTTTGGG-3′) and R1 (5′-TTTAGATCTCCTGCCACTGCCACGCCTTC-3′) introducing BamHI and BglII restriction sites 5′ and 3′ of the cDNA, respectively. The PCR product was cleaved and ligated to the BamHI–BglII-digested GFP-reporter plasmid mAV4 (Kircher et al, 1999). The plasmid coding for aa 1–107 (Fig 2C) was constructed in a similar way using primers F1 and R2 (5′-TTTGCTAGCCGCCGTGGAGAAAGTGTGAGGATC-3′), adding BamHI and NheI restriction sites to the PCR product. For mutation of the second in-frame methionine (Fig 2D), the coding sequence for aa 108–490 was PCR amplified using primers F2 (5′-TTTGCTAGCATCGACTCCTTAGCTATTAAAGCAG-3′) and R1, by introducing an ATG → ATC mutation and NheI and BglII restriction sites, respectively. This PCR product was inserted by means of NheI and BglII into the plasmid already containing the aa 1–107. The plasmid starting with the second methionine (aa 108–490) was constructed using primers F3 (5′-TTTGCTAGCATGGACTCCTTAGCTATTAAAGCAG-3′) and R1 adding NheI and BglII restriction sites. The GFP fusion with FtsZ2-1 beginning at aa 87 was generated using primers F4 (5′-TTTGTCGACATGGCATTTGAAGGAAGCGGAG-3′) and R3 (5′-TTTAGATCTATGACGTGTCTGGCCTCGCTTC-3′), introducing SalI and BglII restriction sites. PCR products were cleaved and ligated to mAV4 digested with the corresponding enzymes. DNA for transfection was prepared using the QIAfilter Plasmid Maxi Kit (Qiagen, Germany). Before transient transfection, single cells (protoplasts) were isolated from protonema and, subsequently, 3 × 105 protoplasts were transiently transfected with 50 μg of circular plasmid DNA. Isolation and regeneration of protoplasts was carried out as described previously (Rother et al, 1994). Six days after transfection, protoplasts were transferred from the regeneration medium (Rother et al, 1994) to Knop medium (Strepp et al, 1998). Localization of FtsZ–GFP fusions was performed as described previously (Kiessling et al, 2000). For the generation of stably transformed moss plants, 1.2 × 106 protoplasts were co-transfected with 50 μg of the PstI–EcoRI-digested FtsZ1-2–GFP construct and 25 μg of a plasmid containing the 35S promoter-driven nptII gene (Töpfer et al, 1988). After 8 days in the regeneration medium, the protoplast suspension was transferred to cellophane-covered solidified Knop medium (1 ml per 9 cm Petri dish). After another 3 days, the cellophane discs with the regenerated protoplasts were transferred to Knop medium supplemented with 25 μg G418 per millilitre (Promega, Germany) for 3 weeks. Single plants were placed on Knop medium without the antibiotic for a release period of 3 weeks. Moss protonemata were scanned for GFP fluorescence using an Olympus BX41microscope.
Antibodies and western blot analysis. Polyclonal antibodies against aa 441–490 of FtsZ1 were obtained by genetic immunization of rats (for further details, see http://www.genovac.com). For western blot analysis, 1 g protonema was ground in liquid nitrogen, homogenized in 100 μl Tris–HCl (0.1 M, pH 8.0) and centrifuged (15 min, 14,000g). Before electrophoresis, 20 μg of protein was dissolved (1:2) in SDS sample buffer (62.5 mM Tris–HCl, pH 6.8, 100 mM dithiothreitol, 2% (w/v) SDS, 50% (v/v) glycerine, 0.01% (w/v) bromophenol blue), heated to 90°C for 3 min and centrifuged (3 min, 14,000g). Proteins were separated by standard SDS–PAGE on 12.5% (w/v) polyacrylamide gels and transferred to nitrocellulose membranes (0.2 μm; Schleicher & Schuell, Germany). Membranes were blocked for 1 h in phosphate-buffered saline (PBS)-T (0.5% (v/v) Tween 20) including 5% (w/v) skimmed milk powder (Fluka, Switzerland). After three washes (1 × 15 min, 2 × 5 min) in PBS-T, membranes were incubated for 1 h in PBS-T containing preimmune serum, anti-FtsZ serum or anti-tubulin (monoclonal antibody against aa 414–422 of yeast α-tubulin; Abcam, UK), respectively, at dilutions of 1:1,000. Subsequently, membranes were washed three times (1 × 15 min, 2 × 5 min) in PBS-T and incubated for 1 h with peroxidase-linked goat anti-rat IgG antibody (GE Healthcare, Germany) at 1:2,500 dilution in PBS-T. After three washes (1 × 15 min, 2 × 5 min) in PBS-T, membranes were developed using the chemiluminescent ECL western blotting detection reagents RPN 2105 (GE Healthcare) and the signal was recorded on HyperfilmTM ECLTM (GE Healthcare).
Immunocytochemistry Protonema was fixed for 5 min in F-MSB (5% (v/v) DMSO, 100 mM PIPES, 10 mM EGTA, 5 mM MgSO4, pH 6.8) containing 1.25% (v/v) glutaraldehyde. The tissue was subsequently washed in W-MSB (33 mM PIPES, 3.3 mM EGTA, 1.7 mM MgSO4, pH 6.8) and fixed for 40 min in F-MSB containing 2% (w/v) paraformaldehyde. After three washes in W-MSB (1 × 1 min, 2 × 5 min), the protonema was incubated in W-MSB containing 0.5% (w/v) NaBH4 for 15 min. After three washes in W-MSB, cell walls were digested by incubation in MSB (100 mM PIPES, 10 mM EGTA, 5 mM MgSO4, pH 5.6) containing 1% (w/v) cellulase (Serva, Germany), 1% (w/v) pectinase (Fluka), and 2% (w/v) driselase (Sigma-Aldrich, Germany). After a single washing step in MSB (pH 6.8), the protonema was rinsed in W-MSB three times (1 × 1 min, 2 × 5 min). To partly extract chlorophyll and unbound particles, the tissue was incubated in E-MSB (5% (v/v) DMSO, 0.5% (v/v) Triton X-100 in MSB, pH 6.8) at 25°C for 1 h. The protonema was subsequently washed three times in W-MSB and incubated with primary antibodies at dilutions of 1:50 in PBS at 37°C for 1 h. After three washes in W-MSB, the tissue was incubated at 37°C with an FITC-conjugated rabbit anti-rat IgG antibody (Sigma-Aldrich) at dilutions of 1:150 in PBS for 1 h. After three washes, the protonema was stored at 4°C in PBS overnight. Controls were treated in the same way, except that the primary antibody was omitted or that the primary antibody was replaced by preimmune serum. FITC and chlorophyll fluorescence was visualized by confocal microscopy using the same filter set that had been used for detection of GFP and chlorophyll.
Accession numbers. The PpftsZ1-2 sequence has been submitted to the EMBL database (accession number AJ428994). Accession number of PpftsZ2-1 is AJ249138.
Acknowledgments
Funding by Deutsche Forschungsgemeinschaft (Re 837/4 and SFB388 project C14) is gratefully acknowledged.
References
- Beech PL et al. (2000) Mitochondrial FtsZ in a chromophyte alga. Science 287: 1276–1279 [DOI] [PubMed] [Google Scholar]
- Bi EF, Lutkenhaus J (1991) FtsZ ring structure associated with division in Escherichia coli. Nature 354: 161–164 [DOI] [PubMed] [Google Scholar]
- de Boer P, Crossley R, Rothfield L (1992) The essential bacterial cell-division protein FtsZ is a GTPase. Nature 359: 254–256 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Erickson HP (1998) Atomic structures of tubulin and FtsZ. Trends Cell Biol 8: 133–137 [DOI] [PubMed] [Google Scholar]
- Erickson HP, Taylor DW, Taylor KA, Bramhill D (1996) Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA 93: 519–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Errington J, Daniel RA, Scheffers DJ (2003) Cytokinesis in bacteria. Microbiol Mol Biol Rev 67: 52–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedtke B, Börner T, Weihe A (2000) One RNA polymerase serving two genomes. EMBO Rep 1: 435–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohe A, Egener T, Lucht JM, Holtorf H, Reinhard C, Reski R (2004) An improved and highly standardised transformation procedure allows efficient production of single and multiple targeted gene knockouts in a moss, Physcomitrella patens. Curr Genet 44: 339–347 [DOI] [PubMed] [Google Scholar]
- Jost W, Baur A, Nick P, Reski R, Gorr G (2004) A large plant β-tubulin family with minimal C-terminal variation but differences in expression. Gene (in press) [DOI] [PubMed] [Google Scholar]
- Kiessling J, Kruse S, Rensing SA, Harter K, Decker EL, Reski R (2000) Visualization of a cytoskeleton-like FtsZ network in chloroplasts. J Cell Biol 151: 945–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kircher S, Wellmer F, Nick P, Rügner A, Schäfer E, Harter K (1999) Nuclear import of the parsley bZIP transcription factor CPRF2 is regulated by phytochrome photoreceptors. J Cell Biol 144: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroiwa H, Mori T, Takahara M, Miyagishima S, Kuroiwa T (2002) Chloroplast division machinery as revealed by immunofluorescence and electron microscopy. Planta 215: 185–190 [DOI] [PubMed] [Google Scholar]
- Löwe J, Amos LA (1998) Crystal structure of the bacterial cell-division protein FtsZ. Nature 391: 203–206 [DOI] [PubMed] [Google Scholar]
- Löwe J, Amos LA (1999) Tubulin-like protofilaments in Ca2+-induced FtsZ sheets. EMBO J 18: 2364–2371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus L, Addinall SG (1997) Bacterial cell division and the Z ring. Annu Rev Biochem 66: 93–116 [DOI] [PubMed] [Google Scholar]
- Martin W et al. (2002) Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc Natl Acad Sci USA 99: 12246–12251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden GI (1999) Endosymbiosis and evolution of the plant cell. Curr Opin Plant Biol 2: 513–519 [DOI] [PubMed] [Google Scholar]
- Mori T, Kuroiwa H, Takahara M, Miyagishima S, Kuroiwa T (2001) Visualization of an FtsZ ring in chloroplasts of Lilium longiflorum leaves. Plant Cell Physiol 42: 555–559 [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G (1997) Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Prot Eng 10: 1–6 [DOI] [PubMed] [Google Scholar]
- Osteryoung KW, McAndrew RS (2001) The plastid division machine. Annu Rev Plant Physiol Plant Mol Biol 52: 315–333 [DOI] [PubMed] [Google Scholar]
- Osteryoung KW, Nunnari J (2003) The division of endosymbiotic organelles. Science 302: 1698–1704 [DOI] [PubMed] [Google Scholar]
- Osteryoung KW, Stokes KD, Rutherford SM, Percival AL, Lee WY (1998) Chloroplast division in higher plants requires members of two functionally divergent gene families with homology to bacterial ftsZ. Plant Cell 10: 1991–2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinho MG, Errington J (2003) Dispersed mode of Staphylococcus aureus cell wall synthesis in the absence of the division machinery. Mol Microbiol 50: 871–881 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Rombauts S, Peer Y, Reski R (2002) Moss transcriptome and beyond. Trends Plant Sci 7: 535–553 [DOI] [PubMed] [Google Scholar]
- Rensing SA, Kiessling J, Reski R, Decker EL (2004) Diversification of ftsZ during early land plant evolution. J Mol Evol 58: 154–162 [DOI] [PubMed] [Google Scholar]
- Reski R (2002) Rings and networks: the amazing compelxity of FtsZ in chloroplasts. Trends Plant Sci 7: 103–105 [DOI] [PubMed] [Google Scholar]
- Richter U, Kiessling J, Hedtke B, Decker E, Reski R, Börner T, Weihe A (2002) Two RpoT genes of Physcomitrella patens encode phage-type RNA polymerases with dual targeting to mitochondria and plastids. Gene 290: 95–105 [DOI] [PubMed] [Google Scholar]
- Rother S, Hadeler B, Orsini JM, Abel WO, Reski R (1994) Fate of a mutant macrochloroplast in somatic hybrids. J Plant Physiol 143: 72–77 [Google Scholar]
- Schaaf A, Reski R, Decker EL (2004) A novel aspartic proteinase is targeted to the secretory pathway and to the vacuole in the moss, Physcomitrella patens. Eur J Cell Biol 83: 145–152 [DOI] [PubMed] [Google Scholar]
- Schaefer D, Zryd JP (1997) Efficient gene targeting in the moss Physcomitrella patens. Plant J 11: 1195–1206 [DOI] [PubMed] [Google Scholar]
- Small I, Wintz H, Akashi K, Mireau H (1998) Two birds with one stone: genes that encode products targeted to two or more compartments. Plant Mol Biol 38: 265–277 [PubMed] [Google Scholar]
- Strepp R, Scholz S, Kruse S, Speth V, Reski R (1998) Plant nuclear gene knockout reveals a role in plastid division for the homolog of the bacterial cell division protein FtsZ, an ancestral tubulin. Proc Natl Acad Sci USA 95: 4368–4373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahara M et al. (2000) A putative mitochondrial ftsZ gene is present in the unicellular primitive red alga Cyanidioschyzon merolae. Mol Gen Genet 264: 452–460 [DOI] [PubMed] [Google Scholar]
- Töpfer R, Schell J, Steinbiss HH (1988) Versatile cloning vectors for transient gene expression and direct gene transfer in plant cells. Nucleic Acids Res 16: 8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente M, Löwe J (2003) Ring, helix, sphere and cylinder: the basic geometry of prokaryotic cell division. EMBO Rep 4: 655–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitha S, McAndrew RS, Osteryoung KW (2001) FtsZ ring formation at the chloroplast division site in plants. J Cell Biol 153: 111–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward JE, Lutkenhaus J (1985) Overproduction of FtsZ induces minicell formation in E. coli. Cell 42: 941–949 [DOI] [PubMed] [Google Scholar]