Abstract
The cricket paralysis virus intergenic region internal ribosomal entry site (CrPV IGR IRES) can assemble translation initiation complexes by binding to 40S subunits without Met-tRNAMeti and initiation factors (eIFs) and then by joining directly with 60S subunits, yielding elongation-competent 80S ribosomes. Here, we report that eIF1, eIF1A and eIF3 do not significantly influence IRES/40S subunit binding but strongly inhibit subunit joining and the first elongation cycle. The IRES can avoid their inhibitory effect by its ability to bind directly to 80S ribosomes. The IRES's ability to bind to 40S subunits simultaneously with eIF1 allowed us to use directed hydroxyl radical cleavage to map its position relative to the known position of eIF1. A connecting loop in the IRES's pseudoknot (PK) III domain, part of PK II and the entire domain containing PK I are solvent-exposed and occupy the E site and regions of the P site that are usually occupied by Met-tRNAMeti.
Keywords: cricket paralysis virus, IRES, ribosome, translation initiation
Introduction
Translation initiation on capped eukaryotic mRNAs is mediated by at least nine initiation factors (eIFs; Pestova et al, 2001). First, eIF3, which has ribosomal dissociation/antiassociation activity, and a ternary complex of eIF2, guanosine triphosphate (GTP) and initiator tRNA (Met-tRNAiMet) bind to a 40S ribosomal subunit to form a 43S complex. This step is enhanced by eIF1A. By unwinding the secondary structure in the 5′-terminal region of mRNA, eIFs 4F, 4A and 4B promote attachment of 43S complexes, which then scan to the AUG initiation codon to form 48S initiation complexes. Scanning and AUG codon recognition require eIF1, which binds to 40S subunits on the interface surface of the platform near the P (peptidyl) site (Lomakin et al, 2003). Finally, eIF5, which activates hydrolysis of eIF2-bound GTP, and eIF5B mediate factor displacement and joining of 48S complexes to 60S subunits.
The three known mechanisms of viral internal ribosomal entry site (IRES)-mediated initiation have simpler factor requirements than cap-mediated initiation, and are all based on specific noncanonical interactions with canonical components of the translation apparatus (Pestova et al, 2001). The simplest mechanism is used by the ∼190-nucleotide (nt)-long intergenic region (IGR) IRESs of dicistroviruses such as cricket paralysis virus (CrPV) and Plautia stali intestine virus (PSIV), which contain three pseudoknots (PKs; Fig 1). 40S subunits bind IGR IRESs without factors and Met-tRNAMeti, so that a CCU triplet in PK I occupies the P site (Fig 1), and then bind directly to 60S subunits to form 80S ribosomes. Translation begins after Ala-tRNAAla bound by elongation factor (EF) 1 has entered the A (aminoacyl) site and been translocated to the P site by EF2 (Pestova & Hellen, 2003). The first translocation event is unusual because it occurs in the absence of deacylated tRNA in the P site, which is instead occupied by part of the IRES.
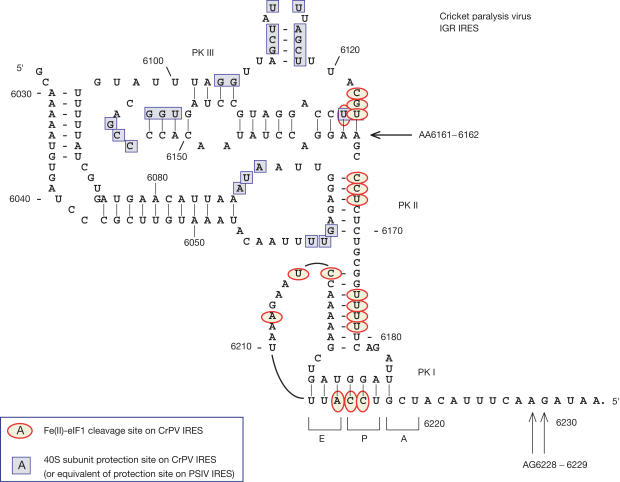
Figure 1.
Structure of the CrPV IGR IRES (Kanamori & Nakashima, 2001; Jan & Sarnow, 2002) showing toe-prints (black arrows) due to binding of 40S subunits and 80S ribosomes (Wilson et al, 2000) and sites of cleavage (red circles) induced by 40Ssubunit-bound Fe(II)-eIF1 derivatives (this work). Sites protected by 40S subunits from chemical and enzymatic cleavage (blue squares) in CrPV and PSIV IRESs (Jan & Sarnow, 2002; Nishiyama et al, 2003) have been combined.
We now report that although eIF1 can bind to the 40S subunit simultaneously with the IRES, it did not affect initiation, whereas eIFs 1, 1A and 3 together had little effect on IRES–40S subunit binding but strongly inhibited subunit joining and the first elongation step. The IRES could avoid this inhibitory effect by its ability to bind directly to 80S ribosomes. An understanding of how the IRES can bind 40S subunits, mimic the AUG codon/Met-tRNAMeti anticodon base pairs in the P site, set the reading frame for translation and promote translocation requires a knowledge of how it interacts with the ribosome. We therefore mapped its position on the 40S subunit in IRES–eIF1–40S subunit complexes using directed hydroxyl radical cleavage. The entire PK I domain, part of PK II and a loop in the PK III domain were found to be solvent-exposed. They occupy the E site, and parts of the P site that are usually occupied by Met-tRNAMeti.
Results
Factor-dependent IRES–40S and IRES/80S assembly
The influence of eIFs on the IRES–40S subunit binding was studied by sucrose density gradient centrifugation and toe-print analysis. Toe-printing involves extension by reverse transcriptase (RT) of a primer annealed to mRNA, to which a 40S subunit is also bound. 48S complexes arrest RT nt 15–17 downstream of the initiation codon. Binding of a 40S subunit to the IRES yields two sets of toe-prints (Fig 1). Those at AG6228–9 correspond to the leading edge of the 40S subunit, nt 15–16 from the CCU codon in the P site. The set at AA6161–2 is indicative of a second interaction of 40S subunits with the IRES involving the PK III domain. In centrifugation experiments, eIFs 1, 1A and 3 did not affect IRES–40S subunit binding (Fig 2A). In toe-printing experiments, eIF1 enhanced the AG6228–9 toe-prints by 50% and strongly reduced those at AA6161–2; eIF1A had no effect and did not augment the activity of eIF1 (Fig 2G, lanes 3–5). eIF3 did not affect IRES–40S complex formation in centrifugation assays (Fig 2A) but reduced both sets of toe-prints by ∼35% when added either to preformed IRES–40S subunit complexes or to 40S subunits before the IRES (Fig 2G, lanes 6 and 14). eIF1A did not alter this effect of eIF3, but inclusion of eIF1 or eIFs 1 and 1A with eIF3 enhanced toe-prints at AG6228–9 and reduced those at AA6161–2, as in other reaction mixtures containing eIF1 (Fig 2G, lanes 7–9).
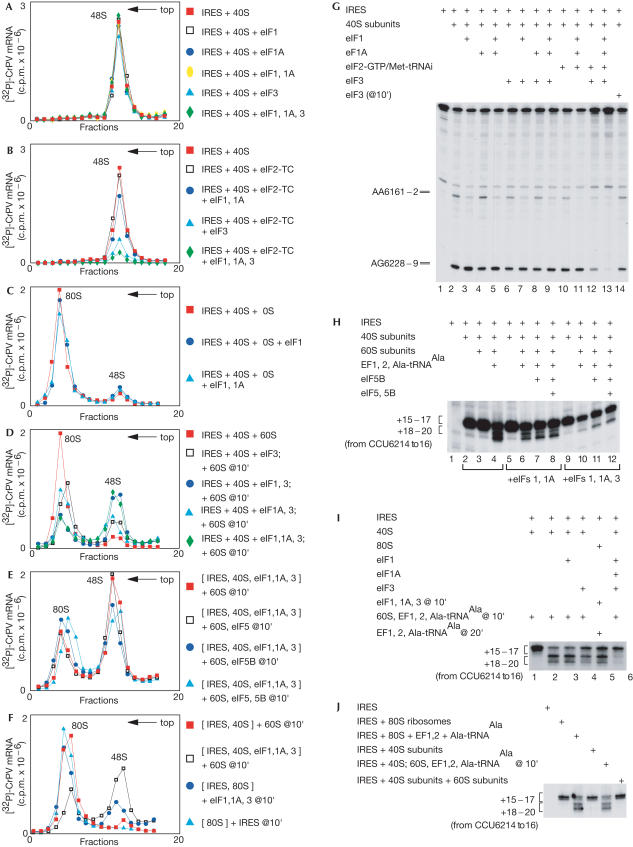
Figure 2.
Influence of initiation factors on ribosomal binding to the IRES. Sucrose density gradient analysis of the influence of eIFs (as indicated) on (A,B) IRES–40S subunit binding and (C–F) joining of 60S subunits to IRES–40S–eIF complexes. The positions of 48S and 80S complexes are indicated. Fractions from the upper parts of gradients have been omitted for clarity. (G) Toe-print analysis of 40S subunit binding to IRES RNA in the presence of eIFs as indicated. The 40S-dependent toe-prints are at AA6161–2 and AG6228–9. (H–J) Toe-print analysis of ribosome translocation on the IRES in reactions containing components as indicated. The toe-prints seen on binding of 40S subunits or 80S ribosomes and following ribosomal translocation are indicated to the left.
The eIF2 ternary complex alone did not influence IRES–40S binding (Fig 2B,G, lane 10), but with eIFs 1 and 1A reduced binding by 10–15% in centrifugation assays (Fig 2B) and strongly reduced the AA6161–2 toe-prints without altering those at AG6228–9 (Fig 2G, lane 11). eIF2 ternary complex and eIF3 or eIFs 3, 1 and 1A reduced IRES–40S subunit binding by ∼75 and ∼95%, respectively, in both assays (Fig 2B,G, lanes 12 and 13).
The inability of eIFs 1, 1A and 3 to affect IRES–40S subunit binding assayed by centrifugation shows that they do not compete directly with the IRES for binding to 40S subunits. The simultaneous enhancement of toe-prints at AG6228–9 and weakening at AA6161–2 by eIF1 indicate that it stabilizes binding to the 40S subunit of the IRES around the P-site CCU codon, so that RT can less easily displace this part of the bound IRES. Conversely, weakening of both sets of toe-prints by eIF3 implies that it destabilizes IRES–40S subunit complexes sufficiently to enhance RT readthrough. Levels of inhibition of IRES–40S subunit binding by the eIF2–ternary complex with combinations of eIFs 1, 1A and 3 correlated with their enhancement of eIF2–ternary complex binding to 40S subunits.
Next, we investigated whether factors that do not prevent IRES–40S subunit binding could nonetheless influence 80S ribosome assembly on the IRES. eIF1 alone or with eIF1A had no effect, but eIF3 alone inhibited assembly by ∼25% and with eIF1, by ∼70% (Fig 2C,D). eIF1A did not exacerbate inhibition by eIF3 or eIF3/eIF1 (Fig 2D). During cap-dependent initiation, eIFs 5 and 5B mediate the release of eIFs 1, 1A and 3 from 40S subunits during subunit joining. The inhibition by eIFs 1, 1A and 3 was relieved only twofold by eIF5B; eIF5 had no effect and did not enhance the activity of eIF5B (Fig 2E). IRES–40S subunit complexes bound to 60S subunits in the presence of eIFs 1 and 1A, but their elongation was reduced by 30–40% (Fig 2H). Inclusion of eIF3 reduced elongation up to eightfold (Fig 2H). eIF5B alone or with eIF5 doubled elongation in the presence of eIFs 1 and 1A or eIFs 1, 1A and 3, just as they doubled IRES–80S complex formation in the presence of these factors (Fig 2H, lanes 6–8 and 10–12). eIF3 alone inhibited elongation by ∼25%; eIF1 alone had no effect (Fig 2I, lanes 3 and 4).
Although the concentrations of eIFs and free 40S subunits in insect cells are not known, these data raise the question as to how the IRES can function efficiently if eIFs are abundant. Initiation on the IRES is normally inefficient (Thompson et al, 2001) but is activated in infected insect cells and is maximal following the CrPV-induced shut-off of host-cell protein synthesis (Moore et al, 1985). Shut-off in picornavirus-infected animal cells is accompanied by polysome disaggregation and 80S monosome accumulation (e.g. Penman et al, 1963). These reports refer to yeast, insect and animal cells, and the IRES is active in all of them. A potentially general mechanism for the IRES to bypass the impediment to initiation caused by eIFs would be to bind directly to 80S ribosomes; if 80S ribosomes accumulate, this could explain activation of the IRES in virus-infected insect cells. The IRES could indeed bind directly to purified, preassembled 80S ribosomes and undergo elongation (Fig 2J). It did not destabilize 80S ribosomes as monitored by optical density (data not shown) or by formation of 40S–IRES complexes (Fig 1F). Addition of eIFs 1, 1A and 3 to 80S–IRES complexes led to 25% dissociation (Fig 2F), but the amount of 40S–IRES complex formed was much less than when 60S subunits were added to 40S subunits that had been pre-incubated with eIFs 1, 1A and 3 (Fig 2D). Incubation of IRES–80S ribosome complex with eIFs 1, 1A and 3 also reduced elongation by 20%, which was still much higher than when 40S subunits were pre-incubated with eIFs 1, 1A and 3 (Fig 2I, lanes 2, 5 and 6).
Simultaneous binding of IRES and eIF1 to 40S subunits
eIF1 did not affect IRES–40S subunit binding (Fig 2G, lane 3), and the IRES and eIF1 bound to 40S subunits simultaneously in pull-down assays (Fig 3A, lane 5). Residues 29–113 of eIF1 form a tightly folded domain with two α-helices on one side of a βsheet (Fletcher et al, 1999; Fig 4A). We determined the position of this functional domain of eIF1 on 40S subunits by directed hydroxyl radical cleavage (Lomakin et al, 2003), which uses locally generated hydroxyl radicals to cleave RNA (in this case, 18S rRNA) near Fe(II) that is sitespecifically tethered by the linker 1-(p-bromoacetamidobenzyl)-EDTA (BABE) to a unique cysteine residue on the surface of an RNA-bound protein (in this case, eIF1). Hydroxyl radicals are generated near the Fe(II) by ascorbic acid/H2O2 treatment and cleave the backbone of adjacent 18S rRNA at sites that are identified by primer extension. The small radius of action of hydroxyl radicals (∼20 Å) allows precise localization of protein. eIF1 binds to the interface surface of the 40S subunit near the P site between the platform and initiator tRNA. To investigate whether the IRES affects eIF1–40S subunit binding, we used the same eIF1 mutants with single surface-exposed cysteine residues that were employed to localize eIF1 on the 40S subunit (Lomakin et al, 2003) to study cleavage of 18S rRNA in IRES–40S–eIF1 complexes. Cleavage of 18S rRNA in helices 23 (from position 75), 24 (from positions 38, 42, 57 and 75) and 44 (from positions 38 and 42) was unaffected by the bound IRES (Fig 3B–D; Lomakin et al, 2003). Thus, the IRES did not alter the position of eIF1 on the 40S subunit. eIF1, in turn, did not impair the assembly of IRES–40S subunit complexes or elongation-competent 80S ribosomes, and strengthened only toe-prints (AG6228–9) that correspond to the leading edge of correctly bound 40S subunits. It therefore did not alter the functional interaction of IRES with 40S subunits. We conclude that the IRES and eIF1 are both properly positioned on the 40S subunit in IRES–eIF1–40S subunit complexes and that they could be used to study the position of the IRES on 40S subunits. Although our data suggest that there is an alternative initiation mechanism in which the IRES binds to 80S ribosomes directly, we have no evidence to suggest that the interaction of the IRES with 80S ribosomes differs much from its interaction with 40S subunits: IGR IRESs do not bind to 60S subunits (Nishiyama et al, 2003), which implies that the major determinants of interaction with 80S ribosomes are on the 40S subunit.
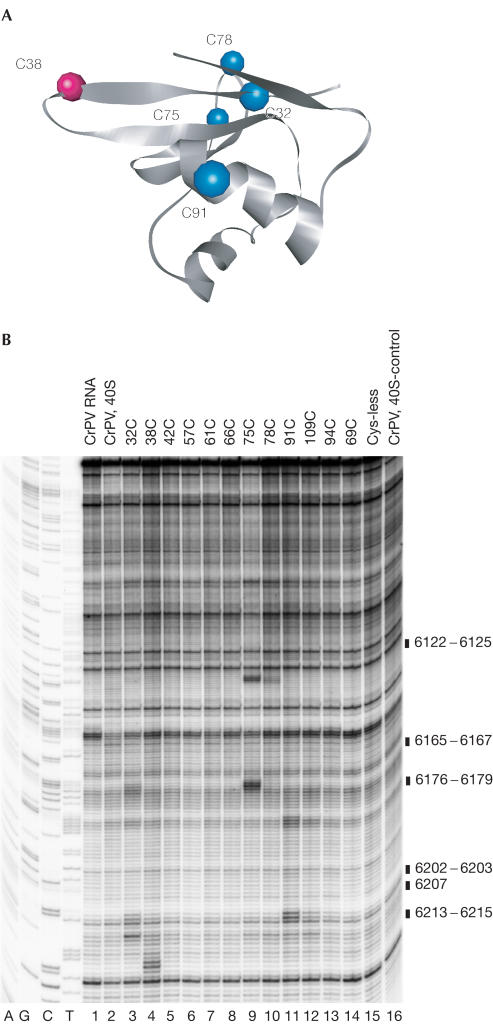
Figure 3.
Formation of eIF1–40S subunit–IRES complexes. (A) Binding of 40S subunits and the IRES to T7-Tag antibody agarose-immobilized eIF1. Ribosomal protein S6, eIF1 and the IRES were visualized by western blotting, Coomassie staining and primer extension, respectively. (B–D) Primer extension analysis of directed hydroxyl radical cleavage of 18S rRNA helices 23, 24 and 44, respectively, in 40S–eIF1–IRES complexes from Fe(II) tethered to positions on the surface of eIF1, as indicated. Reaction mixtures marked ‘40S, IRES' did not contain eIF1, those marked ‘40S' lacked eIF1 and the IRES, and those marked ‘Cys-less' contained the cysteine-less eIF1 mutant. Lanes A, G, C and T depict 18S rRNA sequence obtained using the same primer. The positions of cleaved nucleotides are indicated to the right of each panel.
Figure 4.
Directed hydroxyl radical cleavage of the CrPV IRES bound to 40S subunits. (A) Ribbon diagram of the structured domain of human eIF1. Coloured spheres indicate positions of cysteines on the surface of eIF1 from which hydroxyl radicals cleaved the IRES. (B) Primer extension analysis of directed hydroxyl radical cleavage of the IRES in eIF1–40S–IRES complexes from Fe(II) tethered to surface positions on eIF1, as indicated (lanes 3–14). Control reactions were carried out without eIF1 (lane 16) or with cysteine-less eIF1 mutant (lane 15). Hydroxyl radical cleavage was not induced in reactions shown in lanes 1 and 2. The positions of cleaved nucleotides are indicated to the right of the panel.
Hydroxyl radical probing of 40S-bound CrPV IRES
The ability of eIF1 and the IRES to bind simultaneously to 40S subunits and the fact that eIF1 binds near the P site indicated that directed cleavage of the IRES by hydroxyl radicals generated on the surface of eIF1 could be used to localize the position of the IRES on the 40S subunit relative to eIF1 in IRES–40S–eIF1 complexes. To do so, we used the same eIF1 cysteine mutants.
Hydroxyl radicals generated from five unique cysteines on the eIF1 surface (shown in colour in Fig 4A) cleaved the 40S subunit-bound IRES specifically (Fig 4B). Hydroxyl radicals generated from Cys 38 (which faces the base-paired codon/anticodon in 40S–eIF1 complexes; Fig 5) cleaved the IRES at nt 6213–5 overlapping the triplet base pairs in PK I located in P and E sites (Fig 4B, lane 4). Hydroxyl radicals generated from Cys 91 located on the Asite side of eIF1 (Fig 5) cleaved strongly at nt 6202–3 in the connecting loop and weakly at nt 6176–9 in the adjacent helix of PK I (Fig 4B, lane 11). Hydroxyl radicals generated from Cys 75 located on the Esite side of eIF1 (Fig 5) cleaved strongly at nt 6165–7 in PK II and at nt 6122–5 in an internal loop of PK III (Fig 4B, lane 9). Hydroxyl radicals generated from Cys 32 located near the D-loop of initiator tRNA (Fig 5) cleaved strongly at nt 6202–3 and nt 6207 in a connecting loop in PK I and more weakly at nt 6165–7 in PK II (Fig 4B, lane 3). Hydroxyl radicals generated from Cys 78 (also located on the Esite side of eIF1; Fig 5) cleaved weakly at nt 6122–5 in an internal loop of PK III (Fig 4B, lane 10). Some parts of the IRES were not cleaved, either because they were outside the range of hydroxyl radicals or were shielded from them by other regions of the IRES.
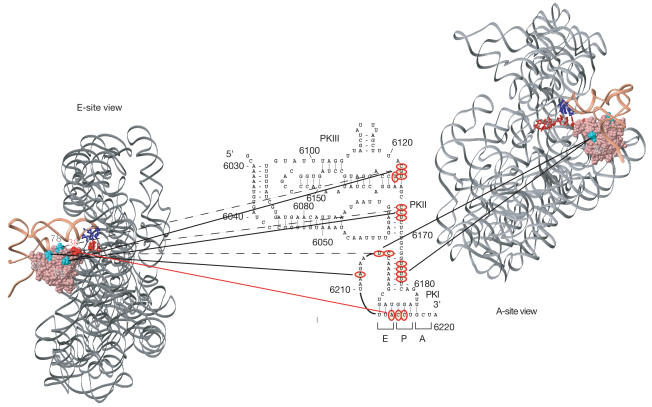
Figure 5.
Orientation of the CrPV IRES bound to a 40S subunit relative to eIF1. E- and Asite views of the position of eIF1 (pink) on the small ribosomal subunit (grey) relative to initiator tRNA (coral) and A- and P-site mRNA (red) were modelled as described by Lomakin et al (2003). eIF1 residues from which hydroxyl radicals cleaved the IRES are shown in turquoise (32, 78, 75 and 91) and red (38). The anticodon bases of Psite tRNA are dark blue. The strong sites of hydroxyl radical cleavages in the IRES (red circles) are connected by solid lines to the positions on eIF1 from which they occurred. Weaker cleavage sites are connected by dashed lines.
These data confirm that the CCU triplet is located in the P site. The connecting loop of PK III is probably situated in the E site near residue 75, and between it and residue 78 (Fig 5). PK II is also in the E site, between residues 32 and 75. The connecting loop of PK I is on top of eIF1 and may occupy the position of the D-loop of initiator tRNA. The stem of PK I is probably positioned in front or on top of eIF1 towards the A site.
These data indicate that the region of the IRES that is accessible to hydroxyl radicals is folded compactly and located mostly in the P and E sites. Parts of the IRES and initiator tRNA occupy overlapping sites on the 40S subunit, which may account for the competition between Met-tRNAMeti–eIF2 and the IRES for 40S subunits.
Discussion
The observation that IRES–40S subunit binding was inhibited by eIF2-GTP/Met-tRNAMeti and combinations of eIFs 1, 1A and 3 (Fig 2G) in direct proportion to their activity in promoting binding of eIF2–ternary complex to 40S subunits is consistent with the inverse relationship between IRES activity and levels of active eIF2–ternary complex (Wilson et al, 2000; Thompson et al, 2001). Inhibition by eIF3 of subunit joining on the IRES, particularly with eIF1, is consistent with the known subunit anti-association activity of eIF3. eIF1 is not known to have such activity and, consistently, eIF1 alone did not inhibit subunit joining on the IRES. However, its position on the 40S subunit would block the interaction of the 60S subunit with parts of the 40S subunit that form intersubunit bridges B2b and B2d, as suggested previously (Lomakin et al, 2003). This may be how eIF1 enhances the anti-association activity of eIF3. There are several possible reasons as to why the ability of eIF5 and eIF5B to relieve eIF3's inhibition of 80S formation on the IRES is lower than during cap-dependent initiation. For example, the IRES may interfere with binding of eIF5B to 40S subunits, or eIF5B may function better if aminoacylated tRNA is in the P site. The fact that eIF5 did not stimulate subunit joining on the IRES is consistent with its role in promoting hydrolysis of eIF2-bound GTP, which these complexes do not contain. The ability of eIF5B to promote subunit joining on the IRES in the presence of eIFs 1, 1A and 3 is lower than in cap-dependent initiation, but is nonetheless apparent. It is consistent with our previous suggestion that the role of eIF5B in subunit joining is, at least in part, to promote the release of initiation factors other than eIF2 from 40S subunits (Pestova et al, 2000). The possibility that eIF1A remains bound to 80S ribosomes assembled on the IRES could account for it inhibiting elongation. eIF1A is a homologue of IF1, which binds to the 30S subunit A site so that it sterically blocks binding of tRNA (Carter et al, 2001). If eIF1A occupies the equivalent region on 40S subunits, then its association with 80S ribosomes would block aminoacyl-tRNA entry to the A site and prevent elongation. However, the ability of the IRES to bind directly to 80S ribosomes in a manner that allows translocation (Fig 2D) suggests a mechanism for efficient initiation when free 40S subunit levels are low, and to avoid inhibition by eIFs 1, 1A and 3. Although eIFs 1, 1A and 3 weakly dissociated preassembled IRES–80S complexes, this activity was much lower than their subunit anti-association activity on IRES–40S subunit complexes.
The distribution of cleavage sites on the IRES bound to a 40S subunit (Fig 1) indicates that much of it is solvent-exposed and occupies the ribosomal intersubunit cavity through which tRNAs move during elongation. PK I, the PK III connecting loop and PK II occupy regions that are usually occupied by deacylated tRNA in the E site and by Met-tRNAMeti in the P site. The A site is mostly unoccupied; therefore it can accept an (EF1-GTP/Ala-tRNAAla) complex to decode the first codon of the open reading frame.
The results of hydroxyl radical cleavage complement footprinting data for IRES–40S subunit complexes (Jan & Sarnow, 2002; Nishiyama et al, 2003; Fig 1). Regions that are protected from modification, and that are solvent-exposed and accessible for hydroxyl radical cleavage in IRES–40S subunit complexes do not overlap. Together, these results and data from mutational and biochemical analyses yield an outline of the functional interactions with the ribosome, which enable the IRES to initiate translation.
The toe-prints at AA6161–2 suggest that 40S subunits bind the PK III domain, consistent with the 40S subunit footprint on the IRES being mostly restricted to this domain and adjacent loops in PK II (Fig 1). Moreover, the PK III domain of the PSIV IRES alone binds stably to 40S subunits (Nishiyama et al, 2003), and mutations in the CrPV PK III domain impair binding, whereas those outside it have little effect (Jan & Sarnow, 2002). This domain is probably responsible for IRES binding to 40S subunits, but much of the rest of the IRES is not protected by bound 40S subunits from modification and is accessible to hydroxyl radical cleavage. It is probably exposed and can interact with the 60S subunit. Mutations outside the PK III domain do not greatly reduce IRES–40S subunit binding, but do abrogate IRES function (Kanamori & Nakashima, 2001; Jan & Sarnow, 2002; Nishiyama et al, 2003). This suggests that those regions are involved in later essential functions, such as placement of PK I in the P site or translocation.
Methods
Purification of factors and ribosomal subunits. Aminoacyl-tRNA synthetases, ribosomal subunits, native (eIF2, eIF3, eIF5B, EF1 and EF2) and recombinant (eIF1, eIF1A and eIF5) factors were purified, and aminoacylated Ala-tRNAAla and Met-tRNAiMet were prepared, as described (Pestova et al, 2000; Pestova & Hellen, 2003). Purified single-cysteine eIF1 mutants were modified with BABE (Lomakin et al, 2003).
Assembly and analysis of ribosomal complexes. Ribosomal complexes were assembled by incubating 0.3 μg CrPV nt 5997–6395 RNA for 5 min at 37°C in 40 μl reaction mixtures containing buffer A (20 mM Tris pH 7.5, 100 mM KAc, 2 mM dithiothreitol (DTT), 2.5 mM MgAc), 1 mM ATP, 0.4 mM GTP, 0.25 mM spermidine, 5 pmol 40S subunits and combinations of 15 pmol eIF2, eIF3, eIF1, eIF1A, eIF5, eIF5B, EF1 and EF2, 20 pmol Met-tRNAMeti and Ala-tRNAAla, 7 pmol 60S subunits and 5 pmol preassembled purified 80S ribosomes (see supplementary information online) as indicated in the text, and were analysed either by sucrose density gradient centrifugation or by primer extension using primer #1 complementary to CrPV nt 6341–59 (Pestova & Hellen, 2003).
For pull-down analysis of eIF1–40S–IRES complexes, 5 μg eIF1 was immobilized on 10 μl T7-antibody-agarose by incubation in 50 μl buffer B (buffer A with 10% glycerol and 0.5% Triton X-100) at 25°C for 30 min. Then, 10 pmol 40S subunits and 20 pmol CrPV mRNA were added to immobilized eIF1 in 40 μl buffer B with 2 mg/ml plasmid DNA (to prevent nonspecific binding of CrPV RNA to the matrix), and incubated at 25°C for 30 min. Beads were washed 4 × with 400 μl buffer B. In all, 25% of the washed matrix-bound complex was phenol–chloroform extracted, ethanol-precipitated and analysed by primer extension (to detect CrPV RNA). The rest of the bound material was analysed by western blotting using antibodies against ribosomal protein S6.
Directed hydroxyl radical probing. Complexes were assembled by incubating 1–2 pmol CrPV nt 5997–6395 RNA in 40 μl buffer C (80 mM K+-Hepes (pH 7.6), 100 mM KCl, 2 mM MgCl2 and 10% glycerol) with ∼50 pmol Fe(II)-eIF1 and 10 pmol 40S subunits, as indicated, at 37°C for 5 min and then on ice. Directed hydroxyl radical probing and subsequent analyses of 18S rRNA were carried out as described by Lomakin et al (2003).
Supplementary information is available at EMBO reports online (http://www.nature.com/embor/journal/v5/n9/extref/7400240s1.pdf).
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by NIH grants AI51340 and GM63940 to C.U.T.H. and T.V.P., respectively.
References
- Carter AP et al. (2001) Crystal structure of an initiation factor bound to the 30S ribosomal subunit. Science 291: 498–501 [DOI] [PubMed] [Google Scholar]
- Fletcher CM, Pestova TV, Hellen CUT, Wagner G (1999) Structure and interactions of the translation initiation factor eIF1. EMBO J 18: 2631–2637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jan E, Sarnow P (2002) Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J Mol Biol 324: 889–902 [DOI] [PubMed] [Google Scholar]
- Kanamori Y, Nakashima N (2001) A tertiary structure model of the internal ribosome entry site (IRES) for methionine-independent initiation of translation. RNA 7: 266–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomakin IB, Kolupaeva VG, Marintchev A, Wagner G, Pestova TV (2003) Position of eukaryotic initiation factor eIF1 on the 40S ribosomal subunit determined by directed hydroxyl radical probing. Genes Dev 17: 2786–2797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore NF, Reavy B, King LA (1985) General characteristics, gene organization and expression of small RNA viruses of insects. J Gen Virol 66: 647–659 [Google Scholar]
- Nishiyama T et al. (2003) Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res 31: 2434–2442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penman S, Scherrer K, Becker Y, Darnell JE (1963) Polyribosomes in normal and poliovirus-infected HeLa cells and their relationship to messenger-RNA. Proc Natl Acad Sci USA 49: 654–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Hellen CUT (2003) Factor requirements for translation elongation after initiation at the A site on the cricket paralysis virus internal ribosomal entry site. Genes Dev 17: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV et al. (2000) The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature 403: 332–335 [DOI] [PubMed] [Google Scholar]
- Pestova TV et al. (2001) Molecular events in initiation of translation in eukaryotes. Proc Natl Acad Sci USA 98: 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson SR, Gulyas KD, Sarnow P (2001) Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc Natl Acad Sci USA 98: 12972–12977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JE, Pestova TV, Hellen CUT, Sarnow P (2000) Initiation of protein synthesis from the A site of the ribosome. Cell 102: 511–520 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information