Abstract
Biolog technology was applied to measure the metabolic similarity of plaque biofilm microcosms, which model the complex properties of dental plaque in vivo. The choice of Biolog plate, incubation time, and incubation conditions strongly influenced utilization profiles. For plaque biofilm microcosms, Biolog GP2 plates incubated anaerobically in an H2-free atmosphere gave the clearest profile. To test the application of the Biolog GP2 assay, plaque microcosms were developed under different nutrient conditions in which the frequency of sucrose application was varied. Cluster analysis of Biolog GP2 data from 10 microcosm biofilms correlated with sucrose frequency. Aciduric bacteria (Streptococcus mutans plus lactobacilli) predominated in the plaques receiving high-frequency sucrose applications. Agreement between the Biolog GP2 groupings with nutrient and compositional changes suggests that Biolog analysis is a valuable technique for analyzing the metabolic similarity of dental plaque biofilm microcosms and other high-nutrient or predominantly anaerobic ecosystems.
Dental plaque is a heterogeneous naturally occurring biofilm of up to 600 microbial species (1, 12). The major dental diseases, caries (tooth decay) and periodontitis (gum disease), are caused by shifts in the composition and behavior of plaque bacterial communities (9, 10). Plaque bacteria are predominantly anaerobes underlying layers of aerobic, microaerophilic, and facultative anaerobes. Investigation of natural plaque is limited by its heterogeneity, the small quantities available, limited access, variable and uncontrollable oral environments, and ethical issues (14). To facilitate laboratory investigations into plaque development, composition, and physiology, we have developed a laboratory “artificial mouth” culture system for plaque biofilm microcosms evolved in vitro from the natural oral microbiota (14-16). Plaque biofilm microcosms seem to retain the complexity of natural plaques, with a biodiverse microbiota and heterogeneous structure (14, 16). This has enabled studies of plaque-modifying factors (16-19, 21). A major limitation for such studies is that simple quantitative comparisons of the degree of functional relatedness of such complex and heterogeneous plaque biofilms has been lacking.
Pattern analysis of carbon source catabolism with the Biolog assay (Biolog, Inc., Hayward, Calif.) to give a community-level physiological profile (CLPP), has proven useful as a simple and rapid method to compare microbial communities from low- to moderate-nutrient habitats, such as soil, water, wastewater, contaminated sites, and activated sludge (3, 6, 8). The Biolog assay for community analysis involves outgrowth of the whole ecosystem microbiota on multiple carbon substrates, each contained in a separate well with tetrazolium violet and a minimal amount of proprietary growth medium. Substrate catabolism is measured as the reduction of tetrazolium violet to a colored formazan, evaluated as an optical density (OD) greater than that in a control well that contains no substrate. This control well, however, responds to metabolism of substrates endogenous to the ecosystem. The pattern of substrate catabolism forms a metabolic fingerprint for that community. Communities from similar or disparate environments may be differentiated by cluster and dimensioning techniques.
The objective of this research was to develop a quantitative measurement of differences in the community structure of plaque biofilm microcosms by using carbon source utilization patterns in order to characterize the response to experimental modification. Dental plaques inhabit an environment high in oral fluid nutrients, requiring breakdown of complex macromolecules and, periodically, nutrients from ingested food. This generates significant levels of endogenous storage substrates within the biofilm. For application of Biolog CLPP analysis to dental plaque microcosms, we investigated a range of Biolog microplates (GP/GP2 and GN/GN2) and experimental conditions, including anaerobic incubation. Originally designed to identify gram-positive (GP/GP2) and gram-negative (GN/GN2) bacterial isolates, they contain 95 different carbon substrates and a low concentration of proprietary growth medium. GP/GP2 composition is dominated by carbohydrates (n = 41) and carboxylic acids (n = 16), GN/GN2 plates comprise predominantly carbohydrate (n = 28), carboxylic acids (n = 24), and amino acids (n = 20). Only the basal growth medium was changed between GP/GP2 and GN/GN2 plates.
Plaque biofilms were cultured from the plaque-enriched saliva of one person who had abstained from oral hygiene for 24 h. Plaques were generally grown for 24 days at 35°C in a multiplaque artificial mouth (MAM) culture system on 25-mm-diameter Thermanox coverslips (Nunc, Inc., Naperville, Ill.) as previously described (14-17, 21). A chemically defined simulated analogue of saliva, defined medium with mucin (DMM) (21), was supplied continuously at a rate of 2.5 ml/h/plaque with periodic applications of 10% (wt/vol) sucrose (at 8-h intervals as standard, with 1.5 ml delivered over 6 min). To induce increasingly frequent acidification in the biofilm, sucrose was also applied at 6- and 4-h intervals. As plaque biofilm development occurs on the coverslips, plaque bacteria slough off, accumulate in the base of the MAM chamber, and continue growing as a biofilm. This plaque, termed “base-plaque,” was used for optimization experiments.
Experimental biofilm plaque (and base-plaque) was sampled following 2 to 3 weeks of growth. For each sample, an 8.0-mg/ml suspension was prepared by homogenization (Ika-UltraTurrax, Janke and Kunkel GmbH & Co., Staufen, Germany) of approximately 0.4 g (wet weight) of plaque in 50 ml of sterile deionized water for 90 s. Portions were taken for Biolog and other analyses. Each plaque suspension was inoculated (140 μl per well) into Biolog microplates.
The OD at 595 nm (OD595) in each well was recorded periodically, for up to 4 days, with a microplate reader and associated software (Benchmark; Bio-Rad Laboratories, Calif.). Biolog microplate data were normalized by average well color development (AWCD) as described by Garland and Mills (6): AWCD = [Σ(C − R)]/n, where C is the color production within each well (OD measurement), R is the OD value of the no-substrate control well of each plate, and n is the number of substrates (n = 95). Each blank-corrected well was then divided by the plate AWCD: (C − R)/AWCD (4, 6). To normalize the analysis, a standard AWCD of 0.5 was adopted (5). Statistical processing was performed with SAS release 8.0 (SAS Institute, Inc., Cary, N.C.). Cluster analysis by the unweighted pair-group method with arithmetic averages (UPGMA), was performed on AWCD-transformed data and evaluated as well by principal component analysis. A positive substrate response was taken to be an OD595 25% greater than that of the control well (no added substrate).
To examine the composition of microcosm plaque, serial dilutions (1:50 and then 1:100) in 1% peptone (Difco Laboratories, Detroit, Mich.) of the homogenized plaque samples prepared for Biolog assays were plated in triplicate (50-μl volumes) by using a spiral plater (model D; Spiral Biotech, Inc., Norwood, Mass.) (13, 14). The following media were used: TSBYK agar plus 5% defibrinated sheep blood (TSBYK+B) to obtain total anaerobic and aerobic CFU; MS agar (Difco) for total Streptococcus spp.; MS agar plus bacitracin (1.45 μg/ml) (MSB) for Streptococcus mutans; and Rogosa SL agar (Difco) for lactobacilli. TSBYK+B agar contains 15 g of Trypticase soy broth per liter, 18.5 g of brain heart infusion broth per liter, 1% yeast extract, 1.5% agar (all Difco), 5 mg of hemin per liter, autoclaved, and supplemented with vitamin K (4 μg/ml) and 5% defibrinated sheep blood (20). TSBYK+B-anaerobic, MSB, and Rogosa plates were incubated anaerobically for 3 days at 35°C in an atmosphere of 80% N2, 10% CO2, and 10% H2. TSBYK+B-aerobic and MS agar plates were incubated aerobically in an atmosphere of 10% CO2 in air at 35°C for 2 days. Values were expressed as ratios to the total anaerobic counts for aciduric bacteria (S. mutans plus lactobacilli) and for non-mutans total streptococci (MS agar CFU − S. mutans and lactobacillus CFU).
Biolog plate selection and incubation conditions proved crucial for differentiating plaque communities. Because dental plaque and microcosms cultured under standard conditions contain mainly strict and facultatively anaerobic bacteria, we tested the plaque metabolic response under a variety of gas conditions: aerobic, anaerobic (H2-free, Anaero Pouch; Mitsubishi Gas Chemical Co., Inc., Tokyo, Japan), and 10% CO2 in air (capnophilic conditions). An anaerobic atmosphere containing H2 directly reduces the tetrazolium violet. Recent modification by the manufacturer of Biolog GP and GN plates to the new formulations GP2 and GN2 gave different responses with microcosm plaque under the aerobic experimental conditions recommended by the manufacturer and standardized in previous soil ecosystem studies (6). The GP2 plate compared to the original GP plate yielded much lower and less frequent aerobic metabolic responses, while GN and GN2 plates had similar but not identical responses. Nevertheless, early experiments showed that variation in the metabolic responses of microcosm plaque cultured from saliva inocula of different donors was discernible with GP and GN microplates under aerobic incubation conditions (data not shown).
In communications with the manufacturer, it was indicated that a low level of nutrient medium was present in each well and that this differed in composition between the GP and GP2 plates and between the GN and GN2 plates. We observed a significant difference between GP2 and GN2 plates in plaque sample response to the same carbon substrate inoculated with the same plaque sample. O'Connell and Garland (11) have reported differences in the metabolic responses of soil and groundwater samples when tested in GN and GN2 plates. These differences are probably due to modification of the proprietary basal media by the manufacturer (2, 11).
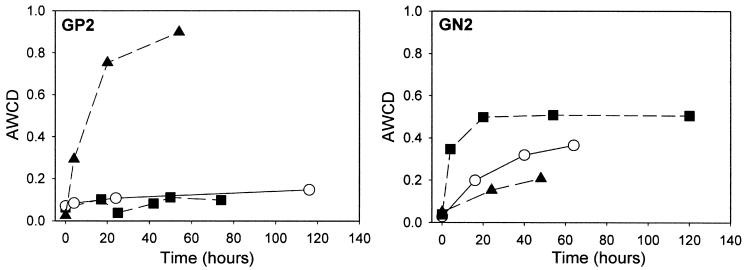
The GP2 plates yielded the greatest metabolic response when incubated under H2-free anaerobic conditions (Fig. 1). After 54 h of anaerobic incubation, the AWCD value was eightfold greater than the AWCD under aerobic conditions and 10% CO2-in-air conditions. For the GN2 plates, aerobic incubation resulted in a better metabolic response than under anaerobic conditions and 10% CO2-in-air conditions, but the level of overall color development was much less than that observed in the anaerobic GP2 plates (Fig. 1). Biolog ECO plates, designed for community analysis and microbial ecological studies, contain three sets of 31 substrates selected for soil community analysis. Tested on microcosm plaques, a capnophilic environment, rather than aerobic conditions prescribed by the manufacturer, was necessary to obtain a reasonable Biolog pattern (data not shown).
FIG. 1.
Comparison of GP2 and GN2 metabolic responses (AWCD) of microcosm plaque under anaerobic (H2 free) (▴), 10% CO2 (○), and aerobic incubation (▪) conditions.
Minimal reaction in the no-substrate control well of the Biolog plates was crucial for obtaining a useful metabolic profile. Rapid development of the control well of some plaque samples poses a major problem for substrate well OD595 measurement, data normalization, and analysis. It indicates the presence of substantial endogenous and storage substrates in the biofilm (GP2 Microplate instructions, Biolog, Inc., Hayward, Calif.). This was a particular problem in GP and GP2 plates under the manufacturer's standard aerobic conditions. To reduce the level of the control well reaction, preincubation of the plaque suspension (30 min, 35°C) and washing by centrifugation were investigated to remove the endogenous substrate. This did not significantly reduce the control well reaction in GP plates, indicating ongoing metabolism of storage, possibly extracellular, substrates. In a set of optimization experiments (data not shown), the control well of the anaerobic (H2-free) GP2 plate increased by 30% over the original OD595 blank value, with 60% of the substrate wells positive after 98 h. In comparison, incubation of GP2 plates in 10% CO2 in air and aerobically showed little reaction in the no-substrate control and substrate wells, with only 25% of the substrate wells positive. The same plaque inoculated into GN2 plates and incubated anaerobically gave rapid control well development and similarly a low number of positive substrate well responses (27%). However, control wells in GN2 plates incubated aerobically or with 10% CO2 in air did not develop color, and both conditions yielded 40% positive substrate wells.
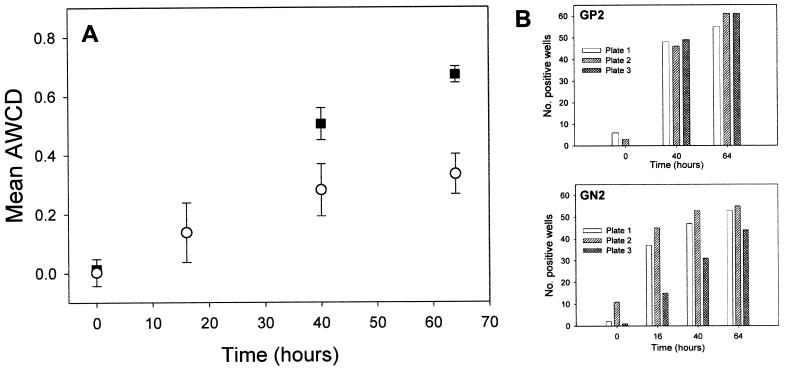
Biolog GP2 anaerobic and GN2 aerobic metabolic patterns were found to be reproducible and discriminatory. There was better reproducibility of AWCD (Fig. 2A) and the number of positive wells (Fig. 2B) in triplicate GP2 plates compared to GN2 plates. The between-sample variability observed might be due principally to minor differences in the inoculum density or sample dispersion (7). For these experiments, microcosm plaque suspension densities of 8 mg/ml were based on the initial wet weight of the sample. Variation in the biomass per wet weight (17) may also account for some of the variation observed in metabolic response. Once the AWCD exceeded the threshold level of 0.5, the reproducibility of response between the plates improved considerably.
FIG. 2.
Reproducibility of Biolog GP2 and GN2 plates. (A) Calculated mean AWCD from triplicate GP2 (▪) and GN2 (○) plates by using a single plaque suspension. Error bars indicate standard deviation. (B) Number of positive wells [OD595 > (OD595 of control + 25%)] from each of the triplicate GP2 and GN2 plates.
Overall, the reproducibility and rate of substrate utilization of the microcosm plaque in the Biolog GP2 plate incubated anaerobically (H2 free) were found to be the most suitable attributes for characterization of the metabolic diversity of microcosm dental plaque. This analysis can be supplemented by using GN2 plates incubated aerobically or ECO plates incubated under 10% CO2 in air. These findings highlight the necessity for empirically optimizing the gas incubation conditions in Biolog microplates for comparison of microbial communities from different environments, especially if anaerobic bacteria are likely to be important.
The utility of the Biolog GP2 plate response for discriminating between plaque microcosms was tested in an experiment in which the frequency of sucrose application was varied, which should influence metabolic properties and microbial composition (Fig. 3). Ten plaque biofilms were cultured in DMM containing 5 mM glucose, with triplicate plaques receiving 10% (wt/vol) sucrose applications at 4-, 6-, and 8-h intervals. No sucrose was supplied to one plaque. In addition to Biolog GP2 analyses, a partial cultural analysis of composition was carried out.
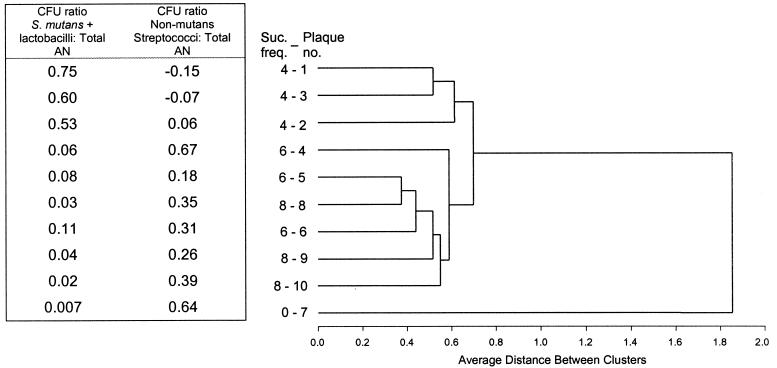
FIG. 3.
Cluster analysis of Biolog GP2 metabolic patterns from 10 microcosm plaques receiving 10% (wt/vol) sucrose treatments at 0-h (plaque 7), 8-h (plaques 8 to 10), 6-h (plaques 4 to 6), or 4-h (plaques 1 to 3) intervals (Suc. freq.). The ratio of aciduric bacteria (S. mutans and lactobacilli) and non-mutans streptococci to total anaerobic CFU (Total AN) is shown.
Cluster analysis of Biolog GP2 responses in which GP2 plates were incubated anaerobically showed a separation of the triplicate plaque microcosms into three distinct groups related to sucrose frequency (Fig. 3). Plaques supplied with sucrose at 4-h intervals formed a distinct cluster. Plaques receiving sucrose at 6- and 8-h intervals were grouped together. The plaque receiving no sucrose (plaque 7) was well separated from all plaques receiving periodic sucrose. These clusters correlated to changes in the abundance of the aciduric bacteria S. mutans and lactobacilli, species that predominated in the microcosm plaques receiving a high frequency of sucrose (plaques 1 to 3), but were at low frequency from 6- or 8-h sucrose plaques, and were extremely rare in the no-sucrose plaque (plaque 7) (Fig. 3). Non-mutans streptococci showed the reverse relationship to sucrose frequency. The relationships derived by Biolog GP2 analysis lend support to the observation that increasing carbohydrate frequency changes plaque microbial community structure and metabolic capabilities. The mixed clustering of the 6- and 8-h sucrose plaques suggests that these treatment regimes did not differentially affect plaque metabolism and species composition to the same extent.
One limitation of the Biolog procedure is the amount of sample required for analysis. This assay for plaque is confined to the scaled-up amounts available from laboratory microcosms, such as those cultured in the artificial mouth, but impossible to obtain from a human mouth.
The pattern of substrate utilization obtained with the Biolog microplate system was developed to assess the similarities between microbial ecosystems in soil and related environments at low to moderate levels of substrate. The present findings show that with careful attention to incubation conditions, this analysis can be applied to measure the metabolic similarities between cultured microcosm plaques, which are dominated by anaerobes and cultured in a periodically high-nutrient environment simulating that of the oral cavity. Due to the microbial complexity of plaque, a simple quantitative measure of the degree of functional relatedness between two plaque microcosms has been difficult to establish. CLPP analysis yields quantitative measures of similarity for microbiologically complex dental plaque biofilm microcosms. A similar approach may prove useful in other high-nutrient or anaerobic microbial ecosystems where sufficient sample is available for analysis.
Acknowledgments
This work was supported by NIH grant R01 DE12752-01A1, the Health Research Council of New Zealand, and the Wellington Medical Research Foundation.
REFERENCES
- 1.Becker, M. R., B. J. Paster, E. J. Leys, M. L. Moeschberger, S. G. Kenyon, J. L. Galvin, S. K. Boches, F. E. Dewhirst, and A. L. Griffen. 2002. Molecular analysis of bacterial species associated with childhood caries. J. Clin. Microbiol. 40:1001-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gamo, M., and T. Shoji. 1999. A method of profiling microbial communities based on a most-probable-number assay that uses BIOLOG plates and multiple sole carbon sources. Appl. Environ. Microbiol. 65:4419-4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garland, J. L. 1997. Analysis and interpretation of community-level physiological profiles in microbial ecology. FEMS Microbiol. Ecol. 24:289-300. [Google Scholar]
- 4.Garland, J. L. 1996. Analytical approaches to the characterization of samples of microbial communities using patterns of potential C source utilization. Soil Biol. Biochem. 28:213-221. [Google Scholar]
- 5.Garland, J. L., and R. M. Lehman. 1999. Dilution/extinction of community phenotypic characters to estimate relative structural diversity in mixed communities. FEMS Microbiol. Ecol. 30:333-343. [DOI] [PubMed] [Google Scholar]
- 6.Garland, J. L., and A. L. Mills. 1991. Classification and characterization of heterotrophic microbial communities on the basis of patterns of community-level sole-carbon-source utilization. Appl. Environ. Microbiol. 57:2351-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haack, S. K., H. Garchow, M. J. Klug, and L. J. Forney. 1995. Analysis of factors affecting the accuracy, reproducibility, and interpretation of microbial community carbon source utilization patterns. Appl. Environ. Microbiol. 61:1458-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Konopka, A., L. Oliver, and R. F. Turco. 1998. The use of carbon substrate utilization patterns in environmental and ecological microbiology. Microb. Ecol. 35:103-115. [DOI] [PubMed] [Google Scholar]
- 9.Marsh, P., and M. V. Martin. 1999. Oral microbiology, 4th ed. Wright, Oxford, United Kingdom.
- 10.Marsh, P. D., and D. J. Bradshaw. 1999. Microbial community aspects of dental plaque, p. 237-253. In H. N. Newman and M. Wilson (ed.), Dental plaque revisited: oral biofilms in health and disease. BioLine, Cardiff, United Kingdom.
- 11.O'Connell, S. P., and J. L. Garland. 2002. Dissimilar response of microbial communities in Biolog GN and GN2 plates. Soil Biol. Biochem. 34:413-416. [Google Scholar]
- 12.Paster, B. J., S. K. Boches, J. L. Galvin, R. E. Ericson, C. N. Lau, V. A. Levanos, A. Sahasrabudhe, and F. E. Dewhirst. 2001. Bacterial diversity in human subgingival plaque. J. Bacteriol. 183:3770-3783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shu, M., L. Wong, J. H. Miller, and C. H. Sissons. 2000. Development of multi-species consortia biofilms of oral bacteria as an enamel and root caries model system. Arch. Oral Biol. 45:27-40. [DOI] [PubMed] [Google Scholar]
- 14.Sissons, C. H. 1997. Artificial dental plaque biofilm model systems. Adv. Dent. Res. 11:110-126. [DOI] [PubMed] [Google Scholar]
- 15.Sissons, C. H., T. W. Cutress, G. Faulds, and L. Wong. 1992. pH responses to sucrose and the formation of pH gradients in thick “artificial mouth” microcosm plaques. Arch. Oral Biol. 37:913-922. [DOI] [PubMed] [Google Scholar]
- 16.Sissons, C. H., T. W. Cutress, M. P. Hoffman, and J. St. J. Wakefield. 1991. A multi-station dental plaque microcosm (artificial mouth) for the study of plaque growth, metabolism, pH, mineralisation. J. Dent. Res. 70:1409-1416. [DOI] [PubMed] [Google Scholar]
- 17.Sissons, C. H., L. Wong, and T. W. Cutress. 1995. Patterns and rates of growth of microcosm dental plaque biofilms. Oral Microbiol. Immunol. 10:160-167. [DOI] [PubMed] [Google Scholar]
- 18.Sissons, C. H., L. Wong, and T. W. Cutress. 1996. Inhibition by ethanol of the growth of biofilm and dispersed microcosm dental plaques. Arch. Oral Biol. 41:27-34. [DOI] [PubMed] [Google Scholar]
- 19.Sissons, C. H., L. Wong, and M. Shu. 1998. Factors affecting the resting pH of in vitro human microcosm dental plaque and Streptococcus mutans biofilms. Arch. Oral Biol. 43:93-102. [DOI] [PubMed] [Google Scholar]
- 20.Tanner, A., and M. F. G. Maiden. 1996. Gingivitis and the initial periodontal lesion. Microb. Ecol. Health Dis. 9:359-365. [Google Scholar]
- 21.Wong, L., and C. H. Sissons. 2001. A comparison of human dental plaque microcosm biofilms grown in an undefined medium and chemically defined artificial saliva. Arch. Oral Biol. 46:477-486. [DOI] [PubMed] [Google Scholar]