Expression profiling using DNA microarrays is a powerful technique that is widely used in the life sciences. How reliable are microarray-derived measurements? The assessment of performance is challenging because of the complicated nature of microarray experiments and the many different technology platforms. There is a mounting call for standards to be introduced, and this review addresses some of the issues that are involved. Two important characteristics of performance are accuracy and precision. The assessment of these factors can be either for the purpose of technology optimization or for the evaluation of individual microarray hybridizations. Microarray performance has been evaluated by at least four approaches in the past. Here, we argue that external RNA controls offer the most versatile system for determining performance and describe how such standards could be implemented. Other uses of external controls are discussed, along with the importance of probe sequence availability and the quantification of labelled material.
Introduction
DNA microarrays are universal tools that can be applied throughout the life sciences (Brown & Botstein, 1999; Lockhart & Winzeler, 2000; Young, 2000). mRNA-expression profiling is the most frequent application. Such microarray hybridizations determine changes in mRNA levels between two samples or result in an absolute quantification that is correlated to mRNA levels. How reliable are these measurements? Given the widespread interest, it is surprising that there have been relatively few systematic analyses of microarray performance.
One reason for this lack of assessment is the complicated nature of microarray technology; there is no single 'microarray technology', but rather a collection of different technology platforms. Established platforms include Affymetrix GeneChips (Santa Clara, CA, USA), PCR-product-based cDNA arrays and long oligomer arrays that are manufactured in-house or by Agilent (Palo Alto, CA, USA). New platforms are still being introduced, such as the Illumina Beadarray (San Diego, CA, USA; Fan et al, 2004) or the Universal Hexamer Array from Agilix (New Haven, CT, USA; Roth et al, 2004). To complicate matters further, many technical alternatives are possible within each platform for each of the numerous steps between sample preparation and data analysis. These include diverse methods of generating labelled material, various hybridization conditions, different microarray scanners and settings, a range of image-quantification techniques, and several approaches for determining statistically and biologically significant differential gene expression. Microarray technology is therefore an amalgamation of many different techniques, even within individual technology platforms.
This complexity makes the need for comparing performance even stronger, whilst confounding such comparisons. Determining reliability is a complicated undertaking if all aspects are to be assessed in a non-arbitrary way across the different platforms and their variants. In addition, reliability is a sensitive issue for those groups that provide the technology. Finally, not every application requires reliable estimates of mRNA level changes. This should be interpreted as an indication of the power of microarray technology, as even lower quality data can yield important results.
Improved performance would nevertheless benefit all applications. A high degree of reliability is a requirement if certain fields, such as systems biology (Ideker et al, 2001) or diagnostic mRNA-expression profiling (van de Vijver et al, 2002) are to mature. A strong argument can be made for investigating how the technology can be systematically assessed, given its increased usage, the costs that are involved and the fact that the aim is to determine the mRNA levels of all genes, including those that are expressed at nearly zero levels. Here, we describe approaches for determining microarray performance and propose that the use of external control RNAs is a versatile and robust method for achieving this goal.
Accuracy and precision
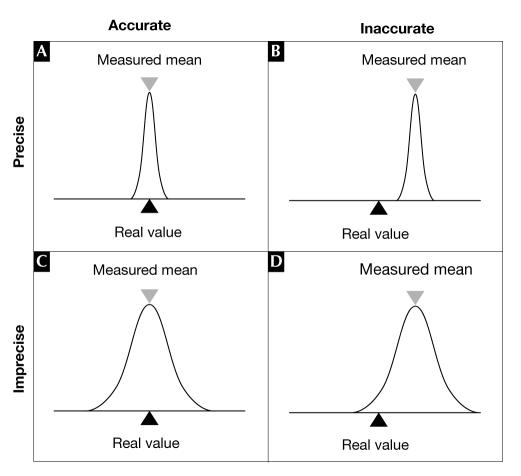
Which performance parameters should be assessed? The two main characteristics of data quality are accuracy and precision. Whereas accuracy refers to how close a measurement is to the real value, precision indicates how often a measurement yields the same result (Fig 1). When microarray data are discussed, the focus is often on precision; that is, reproducibility rather than accuracy. Reproducibility is easier to assess, by taking repeated measurements. Previous reviews have discussed the pitfalls that are involved in determining reproducibility, such as the confusion between biological and technical variation or the requirement for dyeswap hybridizations (Churchill, 2002; Quackenbush, 2002; Yang & Speed, 2002a). The emphasis of this paper is therefore on accuracy. When referring to both accuracy and precision together, the terms data quality, reliability or performance are used. The term 'microarray experiment' refers to a collection of individual microarray hybridizations that are generated for a single purpose.
Figure 1.
Accuracy and precision. (A) A method is accurate and precise when it repeatedly returns a measurement close to the real value. (B) If a method contains a systematic error, it might frequently return an identical measurement that is lower than the actual value; this allegation is frequently made against microarray data (Yuen et al, 2002). In such a case, the measurement is precise but inaccurate. (C) If measurements suffer from noise, the average of a series of measurements might still return the real value but with a large standard deviation; in this case, the measurement is accurate but not precise. (D) The worst case is when measurements report the incorrect value with a large standard deviation.
Performance-assessment models
There are at least two situations that require the systematic determination of performance: technology assessment and individual hybridization assessment. An example of the first case is determining which platform works best. This approach can be expanded to include technology optimization. For example, how is the accuracy of a microarray hybridization influenced by various labelling strategies? This question can be asked for every step of microarray production and experimentation, whether it is testing oligomer-probe design, assessing hybridization protocols, equipment, or even evaluating data-processing approaches.
The second issue that could benefit from performance assessment is at the level of individual hybridizations: do all of the microarray hybridizations in a large experiment or project yield results of similar quality? This comes after technology optimization and is important because of the complicated nature of microarray methodology. Proceeding from biological material to differential mRNA levels involves several steps, many of which have not yet been stably optimized. Confounding artefacts are still being uncovered (Diehl et al, 2001; Ramdas et al, 2001; Chuaqui et al, 2002; Fare et al, 2003; Martinez et al, 2003; Raghavachari et al, 2003; t Hoen et al, 2003; Lyng et al, 2004). Therefore, monitoring quality would benefit individual hybridizations and projects. This could also aid in analyses of the data that are now being collected in public databases (Edgar et al, 2002; Brazma et al, 2003). In these cases, internal quality control would allow the refinement of decisions about which data to use, depending on the requirement for different quality parameters.
Approaches to determining performance
One method that can be used to optimize protocols is to measure and increase the signal intensity (Rickman et al, 2003; Wrobel et al, 2003). The underlying assumption is that increased signal-to-noise ratios will yield better quality hybridizations. However, an increase in signal might be aspecific; for example, owing to increased cross-hybridization or the nonspecific binding of fluorophores to nucleic-acid probes (Chuaqui et al, 2002). It is therefore risky to optimize signal-to-noise ratios without knowing whether specificity is being maintained.
A second approach is to determine the correlation between new methods and an approach that is already in use. Different amplification and labelling techniques are usually assessed by comparison to a standard cDNA-synthesis protocol (Mahadevappa & Warrington, 1999; Manduchi et al, 2002; Gupta et al, 2003; t Hoen et al, 2003; Kenzelmann et al, 2004). A correlation coefficient only shows how similarly two protocols behave; it does not give information on their individual accuracy. A high correlation (Barczak et al, 2003) might therefore mean that the technologies that are being compared both suffer from the same error. Moreover, a low correlation (Tan et al, 2003) still begs the question of which technique is better. Another use of correlation is to monitor reproducibility; for example, between the two dye channels of cDNA arrays. The drawback is that the technology is being optimized for yielding identical intensities, rather than for accurately reporting what most users are interested in: differences in mRNA levels. Perfectly tight same-versussame scatter plots, which are often touted in publications or advertisements as proof of superior performance, should be treated with caution. Optimization that is based on achieving tight scatter plots can lead to a decreased ability to report changes in mRNA levels. Ideally, optimization should focus on reporting relative or absolute mRNA levels and should take into account the entire range of expression levels.
A third method for performance evaluation is to use an established cell-culture experiment in which changes in mRNA levels are verified by other means, such as northern blotting analysis or quantitative reverse transcription (RT)-PCR (Taniguchi et al, 2001; Yuen et al, 2002; Polacek et al, 2003; Loguinov et al, 2004; Roth et al, 2004). Using such established differentials is a good method because it optimizes the reporting of differences in expression, which is the goal of most microarray hybridizations. One disadvantage is that verification and optimization are driven by the differences that are reported by the microarrays, rather than by all of the mRNA-level differences that are present in the experimental system. There is no test for false-negative differentials unless RT-PCR, for example, is carried out on many hundreds of genes that are not reported as being differentially expressed in the microarray experiment. A further drawback is that this method, similar to those described above, does not lend itself to the routine assessment of each individual microarray hybridization before optimization.
External controls
A fourth approach for assessing microarray technology is based on the use of external RNA controls, which are also known as spikes, spike-in controls, exogenous controls or standards (Lockhart et al, 1996; Eickhoff et al, 1999; Girke et al, 2000; Hughes et al, 2001; Yue et al, 2001; Dorris et al, 2002; Relogio et al, 2002; Badiee et al, 2003; Benes & Muckthaler, 2003; Wang et al, 2003). An external control approach does not have to suffer from the disadvantages discussed above; it can also lend itself to reporting accuracy in each microarray experiment after optimization (see below).
External controls are RNA molecules that are synthetically produced by in vitro transcription. Control RNAs can be added in defined amounts to biological samples that are of interest. Microarray probes that correspond to the controls report the fidelity of the technology in determining the presence of the controls. The crucial feature of external RNAs is that the user has absolute control over how they are added. When they are spiked differentially between two identical RNA samples, external RNAs can mimic differentially expressed genes. The user knows that only the control RNAs are differentially present and at exactly what amounts. Owing to this versatility, external controls form an ideal benchmarking system for microarray technology.
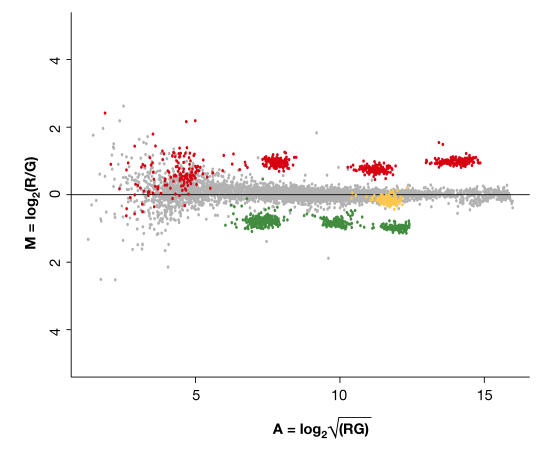
To illustrate their convenience, Figure 2 shows an example of an experiment with external controls. Here, two mixes with different combinations of external RNA controls are spiked into identical total RNA samples. This results in twofold change differentials that cover the entire range of mRNA levels. In this experiment, changes up to twofold are reported reasonably accurately, with decreased precision and accuracy in the region of the graph that is covered by the lowest spiked control. Such a design can be used for both technology and hybridization assessment.
Figure 2.
Assessing spiked ratios. A self-versus-self hybridization with nine external control RNAs that are spiked in at different ratios. The average expression ratio from a merged dye-swap experiment is plotted as a function of the average background-subtracted intensities of the two channels (R and G). The two channels were lowess-normalized on genes per print-tip (Yang et al, 2002b). Genes are indicated in grey. Ratio controls were spiked twofold up (red) or down (green). One control was added in equal amounts to both channels (yellow). Each control is represented at least 96 times on the arrays that are used, which results in the different clusters of control spots. The spread of each cluster is dependent on both hybridization and spotting-pin uniformity.
Design and handling of external controls
The most important requirement for external controls is that they are representative of the endogenous mRNAs with regard to length and sequence characteristics. Similarly, the microarray probes that are used for monitoring the external control RNAs should be typical of all of the probes that are present on the microarray with regard to design, manufacture and cross-hybridization potential. Excessive cross-hybridization towards and from endogenous transcripts must be avoided. Detailed recommendations on how to achieve representative controls and probes are described in the supplementary information online. Requirements for the vectors that are used to generate external control RNA, details of how to store and handle controls, measures to avoid pipetting errors and information about how to incorporate the controls on microarrays are also described. An important consequence of some of these criteria is that several controls and probes must be used.
Spiking strategies
External controls can be spiked at different stages during sample processing. However, routine use dictates that external controls should be spiked early during sample processing so that as many steps as possible are monitored. The easiest way to achieve this is to spike external control mixes into total RNA samples, thereby controlling all of the downstream steps.
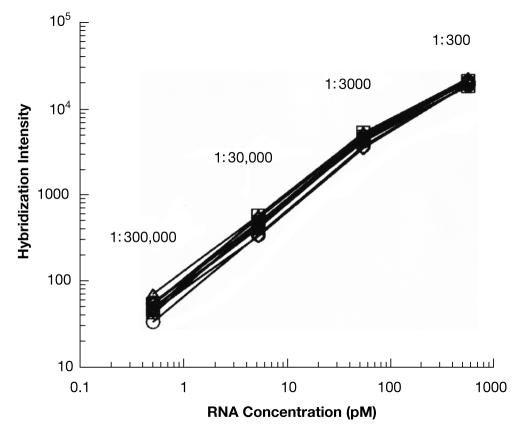
The example in Figure 2 illustrates how external controls can be used in both technology and hybridization-assessment techniques. Several controls are required to cover the entire range of expression levels. It is advantageous to include spikes at levels that are both higher and lower than the range that is strictly required. It is also better to include more than the single non-differential control that is shown in the example. An ideal design would include both low (twofold) and high (tenfold) differentials in a single experiment. However, the number of controls that are required can be significantly reduced by monitoring the absolute intensities of the probes in relation to the spiked amounts (Fig 3). The rationale behind this approach is that if the technology accurately and precisely measures absolute amounts of spiked controls, it will do the same for differential expression.
Figure 3.
Dose–response test. A log–log plot of the signal intensity versus concentration for 11 control RNAs. Reproduced with permission from Lockhart et al (1996) Nature Biotechnology © Nature Publishing Group.
RNA sample preparation
Stricter requirements for RNA sample preparation and the possibility of pipetting errors represent a potential 'Achilles' heel' for some applications. These are discussed below in increasing order of difficulty. Technology assessment and optimization present no challenge with regard to RNA preparation. Here, controls can be spiked differentially between aliquots of an identical total RNA sample. For hybridization assessment in a single project, heterogeneity in RNA quantification of different samples can be confounding. However, there is no reason why the quantification or quality of RNA should be heterogeneous, as long as the yields are high enough, and the preparation and handling methods are consistent. Recent improvements in low-yield RNA quantification include the introduction of microlitre-volume spectrophotometers. Provided that such samples can be used for repeat hybridizations, any inconsistencies will be obvious. The most difficult situation for hybridization assessment is when unique samples, such as biopsy material, are used that can only be obtained in low yields and are of heterogeneous quality. However, external controls could be used here as a control for differential sample quality; aberrant behaviour of the mRNA population relative to the controls indicates heterogeneity in the quality or quantification of the RNA sample.
External control availability
Some external control RNAs and their corresponding probes are already commercially obtainable. However, their use is restricted by cost and sequence availability. Plasmids for the external controls that are present on Affymetrix microarrays (Lockhart et al, 1996) can be obtained from the American Type Culture Collection (ATCC). Most of the controls used in the examples cited here were designed several years ago, when only limited genome sequence information was available with which to determine the cross-hybridization potential. At present, no sufficiently large set of external controls is available that fits all the criteria and can be used across several organisms and applications. This could soon change, as the industry-led External RNA Control Consortium (ERCC; http://www.affymetrix.com/community/standards/ercc.affx) is endeavouring to establish and develop a universal set of external RNA controls. Such an initiative should be wholeheartedly endorsed.
Alternative uses of external controls
Besides sensitivity testing (Fig 3; Lockhart et al, 1996; Girke et al, 2000; Hughes et al, 2001; Dorris et al, 2002; Ramakrishnan et al, 2002; Relogio et al, 2002) and comparison of labelling strategies (Badiee et al, 2003), external control probes can also be used to estimate background when the corresponding control RNA is not spiked (Dorris et al, 2002; Ramakrishnan et al, 2002). External controls could also be applied as standards to determine absolute mRNA levels, for example, as picomoles per sample or even as mRNA copies per cell. For such measurements to be accurate, further work is required to ensure that various probe properties—such as amount, melting temperature and cross-hybridization risk—also become standardized. Another use of controls is to normalize samples that are expected to show general and/or large unbalanced shifts in the mRNA population. As well as the inactivation of general transcription factors (Holstege et al, 1998), such external normalization approaches are required for studies of mRNA decay (Wang et al, 2002) and many other cellular processes that might show large changes in the mRNA population (Preiss et al, 2003; van de Peppel et al, 2003).
Insight into data quality is not always necessary
We believe that all microarray applications would benefit from the wider use of external controls, especially for technology assessment and optimization. However, not all applications require quality assessment. One such example is microarray screening with the sole purpose of identifying differentially expressed genes; as long as the differential expression of these genes is verified independently, there is no requirement for knowing the data quality of the microarrays. Many studies still fall under this category even though much more is feasible (Lockhart & Winzeler, 2000; Young, 2000). A common denominator of studies that go beyond screening is comparative analysis across many experiments for the purpose of examining entire metabolic, regulatory and pathological pathways. A wider implementation of performance assessment would increase the value of screening experiments for such comparative studies and would also benefit the success of the screens themselves.
Other issues that affect accuracy
The wider implementation of external controls will not address all of the data-reliability issues. An obvious example is the lack of microarray-probe sequence information. Owing to the necessary revisions of genome annotation, a lack of probe sequence information confounds the analysis of experiments at present, especially across different arrays. Without probe sequence information, there is no guarantee that an oligomer or cDNA probe actually represents the gene that it is supposed to. This problem could be addressed by making it compulsory to submit probe sequence information along with microarray data. The Microarray Gene Expression Data (MGED) Society (http://www.mged.org), which is responsible for coordinating the agreement on the annotation of microarray data, is now recommending that probe sequence information is also included as part of the Minimal Information About a Microarray Experiment (MIAME) criteria (Brazma et al, 2001).
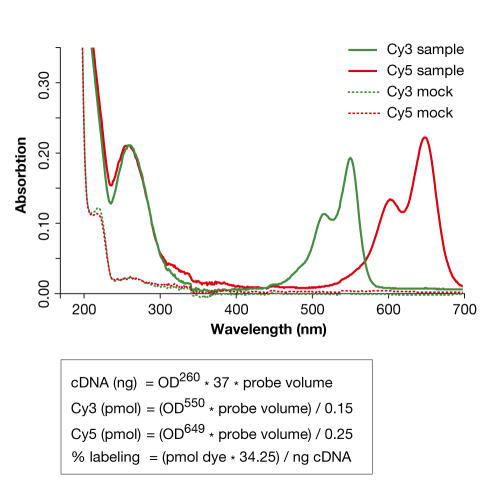
A different confounding issue for performance assessment is the poor description of labelled material. At present, the absolute amount and incorporation percentage of labelled material being applied to microarrays is usually not reported. Both are important determinants of hybridization success and can be measured relatively easily (Fig 4). This omission represents a step backwards from the practice that was once observed for the description of protocols using radioactive labelling. Information on the yield and activity of labelled material is important for assessing differences between labelling strategies, and has a bearing on background, quenching and dye-bias artefacts. It is therefore important to monitor yield and specific activity in relation to data quality.
Figure 4.
Quantification of labelled material. Absorption spectra for Cy3- and Cy5-labelled cDNA after the removal of unincorporated dye. Peaks for cDNA, and incorporated Cy3 and Cy5, are found at 260, 550 and 649 nm, respectively. Two mock-labelled samples are included as negative controls in which reverse transcriptase was left out of the cDNA-synthesis reaction. This is important for determining the success of purification and the removal of RNA template by hydrolysis. The cDNA yields and dye incorporation can be calculated using the indicated formulas.
The data-reliability issues discussed here should not discourage the use of microarrays; rather, this is a review of approaches that might further advance an important technology. Although they were first introduced at an early stage (Lockhart et al, 1996), the widespread use of external RNA controls has been held back by several factors. Incomplete genome sequences were one such barrier, but this is no longer an issue. Another factor is the additional work that is required, many aspects of which are discussed above. The most feasible application of external controls is in technology assessment and optimization, which on its own would contribute markedly to increased microarray accuracy.
Supplementary information is available at EMBO reports online (http://www.emboreports.org).
Supplementary Material
Design and use of external controls in DNA microarray experiments
Frank C. P. Holstege & Harm van Bakel. FCPH is the recipient of an EMBO Young Investigator Award
Acknowledgments
We thank E. Brendeford for providing Figure 2 and N. Kettelarij for providing the data for Figure 4. We acknowledge the technology-development work of D. van Leenen, T. Miles, M. Groot-Koerkamp and J. van Helvoort, as well as fruitful discussions with R. Kerkhoven, P. van Hummelen, M. Kuiper, T. van der Lende and T. Freeman. Work in the authors' laboratory is supported by grants from the Netherlands Organization for Scientific Research (NWO), the European Union Fifth Framework Project (TEMBLOR) and the EMBO Young Investigator Programme.
References
- Badiee A, Eiken HG, Steen VM, Lovlie R (2003) Evaluation of five different cDNA labeling methods for microarrays using spike controls. BMC Biotechnol 3: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barczak A, Rodriguez MW, Hanspers K, Koth LL, Tai YC, Bolstad BM, Speed TP, Erle DJ (2003) Spotted long oligonucleotide arrays for human gene expression analysis. Genome Res 13: 1775–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes V, Muckthaler M (2003) Standardization of protocols in cDNA microarray analysis. Trends Biochem Sci 28: 244–249 [DOI] [PubMed] [Google Scholar]
- Brazma A et al. (2001) Minimum information about a microarray experiment (MIAME)-toward standards for microarray data. Nat Genet 29: 365–371 [DOI] [PubMed] [Google Scholar]
- Brazma A et al. (2003) ArrayExpress: a public repository for microarray gene expression data at the EBI. Nucleic Acids Res 31: 68–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown PO, Botstein D (1999) Exploring the new world of the genome with DNA microarrays. Nat Genet 21: 33–37 [DOI] [PubMed] [Google Scholar]
- Chuaqui RF et al. (2002) Post-analysis follow-up and validation of microarray experiments. Nat Genet 32 (Suppl): 509–514 [DOI] [PubMed] [Google Scholar]
- Churchill GA (2002) Fundamentals of experimental design for cDNA microarrays. Nat Genet 32 (Suppl): 490–495 [DOI] [PubMed] [Google Scholar]
- Diehl F, Grahlmann S, Beier M, Hoheisel JD (2001) Manufacturing DNA microarrays of high spot homogeneity and reduced background signal. Nucleic Acids Res 29: E38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorris DR, Ramakrishnan R, Trakas D, Dudzik F, Belval R, Zhao C, Nguyen A, Domanus M, Mazumder A (2002) A highly reproducible, linear, and automated sample preparation method for DNA microarrays. Genome Res 12: 976–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickhoff B, Korn B, Schick M, Poustka A, van der Bosch J (1999) Normalization of array hybridization experiments in differential gene expression analysis. Nucleic Acids Res 27: E33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan JB et al. (2004) A versatile assay for high-throughput gene expression profiling on universal array matrices. Genome Res 14: 878–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fare TL et al. (2003) Effects of atmospheric ozone on microarray data quality. Anal Chem 75: 4672–4675 [DOI] [PubMed] [Google Scholar]
- Girke T, Todd J, Ruuska S, White J, Benning C, Ohlrogge J (2000) Microarray analysis of developing Arabidopsis seeds. Plant Physiol 124: 1570–1581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V et al. (2003) Directly labeled mRNA produces highly precise and unbiased differential gene expression data. Nucleic Acids Res 31: E13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstege FC, Jennings EG, Wyrick JJ, Lee TI, Hengartner CJ, Green MR, Golub TR, Lander ES, Young RA (1998) Dissecting the regulatory circuitry of a eukaryotic genome. Cell 95: 717–728 [DOI] [PubMed] [Google Scholar]
- Hughes TR et al. (2001) Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol 19: 342–347 [DOI] [PubMed] [Google Scholar]
- Ideker T, Galitski T, Hood L (2001) A new approach to decoding life: systems biology. Annu Rev Genomics Hum Genet 2: 343–372 [DOI] [PubMed] [Google Scholar]
- Kenzelmann M, Klaren R, Hergenhahn M, Bonrouhi M, Grone HJ, Schmid W, Schutz G (2004) High-accuracy amplification of nanogram total RNA amounts for gene profiling. Genomics 83: 550–558 [DOI] [PubMed] [Google Scholar]
- Lockhart DJ, Winzeler EA (2000) Genomics, gene expression and DNA arrays. Nature 405: 827–836 [DOI] [PubMed] [Google Scholar]
- Lockhart DJ et al. (1996) Expression monitoring by hybridization to high-density oligonucleotide arrays. Nat Biotechnol 14: 1675–1680 [DOI] [PubMed] [Google Scholar]
- Loguinov AV, Mian IS, Vulpe CD (2004) Exploratory differential gene expression analysis in microarray experiments with no or limited replication. Genome Biol 5: R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyng H, Badiee A, Svendsrud DH, Hovig E, Myklebost O, Stokke T (2004) Profound influence of microarray scanner characteristics on gene expression ratios: analysis and procedure for correction. BMC Genomics 5: 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahadevappa M, Warrington JA (1999) A high-density probe array sample preparation method using 10- to 100-fold fewer cells. Nat Biotechnol 17: 1134–1136 [DOI] [PubMed] [Google Scholar]
- Manduchi E, Scearce LM, Brestelli JE, Grant GR, Kaestner KH, Stoeckert CJ (2002) Comparison of different labeling methods for two-channel high-density microarray experiments. Physiol Genomics 10: 169–179 [DOI] [PubMed] [Google Scholar]
- Martinez MJ et al. (2003) Identification and removal of contaminating fluorescence from commercial and in-house printed DNA microarrays. Nucleic Acids Res 31: E18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacek DC et al. (2003) Fidelity and enhanced sensitivity of differential transcription profiles following linear amplification of nanogram amounts of endothelial mRNA. Physiol Genomics 13: 147–156 [DOI] [PubMed] [Google Scholar]
- Preiss T, Baron-Benhamou J, Ansorge W, Hentze MW (2003) Homodirectional changes in transcriptome composition and mRNA translation induced by rapamycin and heat shock. Nat Struct Biol 10: 1039–1047 [DOI] [PubMed] [Google Scholar]
- Quackenbush J (2002) Microarray data normalization and transformation. Nat Genet 32 (Suppl): 496–501 [DOI] [PubMed] [Google Scholar]
- Raghavachari N, Bao YP, Li G, Xie X, Muller UR (2003) Reduction of autofluorescence on DNA microarrays and slide surfaces by treatment with sodium borohydride. Anal Biochem 312: 101–105 [DOI] [PubMed] [Google Scholar]
- Ramakrishnan R et al. (2002) An assessment of Motorola CodeLink microarray performance for gene expression profiling applications. Nucleic Acids Res 30: E30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdas L, Coombes KR, Baggerly K, Abruzzo L, Highsmith WE, Krogmann T, Hamilton SR, Zhang W (2001) Sources of nonlinearity in cDNA microarray expression measurements. Genome Biol 2: R0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Relogio A, Schwager C, Richter A, Ansorge W, Valcarcel J (2002) Optimization of oligonucleotide-based DNA microarrays. Nucleic Acids Res 30: E51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman DS, Herbert CJ, Aggerbeck LP (2003) Optimizing spotting solutions for increased reproducibility of cDNA microarrays. Nucleic Acids Res 31: E109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth ME et al. (2004) Expression profiling using a hexamer-based universal microarray. Nat Biotechnol 22: 418–426 [DOI] [PubMed] [Google Scholar]
- t Hoen PA, de Kort F, van Ommen GJ, den Dunnen JT (2003) Fluorescent labelling of cRNA for microarray applications. Nucleic Acids Res 31: E20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan PK, Downey TJ, Spitznagel EL, Xu P, Fu D, Dimitrov DS, Lempicki RA, Raaka BM, Cam MC (2003) Evaluation of gene expression measurements from commercial microarray platforms. Nucleic Acids Res 31: 5676–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi M, Miura K, Iwao H, Yamanaka S (2001) Quantitative assessment of DNA microarrays: comparison with northern blot analyses. Genomics 71: 34–39 [DOI] [PubMed] [Google Scholar]
- van de Peppel J, Kemmeren P, van Bakel H, Radonjic M, van Leenen D, Holstege FC (2003) Monitoring global messenger RNA changes in externally controlled microarray experiments. EMBO Rep 4: 387–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van de Vijver MJ et al. (2002) A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med 347: 1999–2009 [DOI] [PubMed] [Google Scholar]
- Wang HY, Malek RL, Kwitek AE, Greene AS, Luu TV, Behbahani B, Frank B, Quackenbush J, Lee NH (2003) Assessing unmodified 70-mer oligonucleotide probe performance on glassslide microarrays. Genome Biol 4: R5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Liu CL, Storey JD, Tibshirani RJ, Herschlag D, Brown PO (2002) Precision and functional specificity in mRNA decay. Proc Natl Acad Sci USA 99: 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrobel G, Schlingemann J, Hummerich L, Kramer H, Lichter P, Hahn M (2003) Optimization of high-density cDNA-microarray protocols by 'design of experiments'. Nucleic Acids Res 31: E67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YH, Speed T (2002a) Design issues for cDNA microarray experiments. Nat Rev Genet 3: 579–588 [DOI] [PubMed] [Google Scholar]
- Yang YH, Dudoit S, Luu P, Lin DM, Peng V, Ngai J, Speed TP (2002b) Normalization for cDNA microarray data: a robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res 30: E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RA (2000) Biomedical discovery with DNA arrays. Cell 102: 9–15 [DOI] [PubMed] [Google Scholar]
- Yue H et al. (2001) An evaluation of the performance of cDNA microarrays for detecting changes in global mRNA expression. Nucleic Acids Res 29: E41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuen T, Wurmbach E, Pfeffer RL, Ebersole BJ, Sealfon SC (2002) Accuracy and calibration of commercial oligonucleotide and custom cDNA microarrays. Nucleic Acids Res 30: E48. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Design and use of external controls in DNA microarray experiments